Key Points
The NF-κB subunits p52 and RelB increase IRF4 promoter activity and expression in PTCL cells.
A positive feedback loop involving CD30, NF-κB, and IRF4 drives PTCL cell proliferation and can be blocked by NF-κB inhibitors.
Abstract
Peripheral T-cell lymphomas (PTCLs) are generally aggressive non-Hodgkin lymphomas with poor overall survival rates following standard therapy. One-third of PTCLs express interferon regulatory factor-4 (IRF4), a tightly regulated transcription factor involved in lymphocyte growth and differentiation. IRF4 drives tumor growth in several lymphoid malignancies and has been proposed as a candidate therapeutic target. Because direct IRF4 inhibitors are not clinically available, we sought to characterize the mechanism by which IRF4 expression is regulated in PTCLs. We demonstrated that IRF4 is constitutively expressed in PTCL cells and drives Myc expression and proliferation. Using an inhibitor screen, we identified nuclear factor κB (NF-κB) as a candidate regulator of IRF4 expression and cell proliferation. We then demonstrated that the NF-κB subunits p52 and RelB were transcriptional activators of IRF4. Further analysis showed that activation of CD30 promotes p52 and RelB activity and subsequent IRF4 expression. Finally, we showed that IRF4 transcriptionally regulates CD30 expression. Taken together, these data demonstrate a novel positive feedback loop involving CD30, NF-κB, and IRF4; further evidence for this mechanism was demonstrated in human PTCL tissue samples. Accordingly, NF-κB inhibitors may represent a clinical means to disrupt this feedback loop in IRF4-positive PTCLs.
Introduction
Peripheral T-cell lymphomas (PTCLs) are aggressive non-Hodgkin lymphomas (NHLs) characterized by poor overall survival rates.1-3 Most patients are treated with anthracycline-based chemotherapy regimens originally designed for B-cell lymphomas. Five-year overall survival rates vary somewhat by subtype but average ∼35%. The possibility of individualized targeted therapy holds promise, but advances have been hindered by the fact that few therapeutic targets have been identified and validated in PTCLs. Our group’s approach to this challenge has been to characterize the genetics of PTCLs to identify novel biomarkers and therapeutic targets. We have focused particularly on chromosomal translocations, which have played a major role in identifying diagnostic, prognostic, and theranostic biomarkers in hematologic neoplasms in general (eg, BCR-ABL) and PTCLs specifically (ALK rearrangements).4,5
We previously discovered a recurrent translocation in PTCL, t(6;14)(p25.3;q11.2), that juxtaposes the interferon regulatory factor-4 (IRF4) and T-cell receptor-α (TRA) genes and is associated with increased IRF4 expression.6 Because TRA translocation partners typically function as oncogenes in T-cell malignancies, we postulated that IRF4 plays an oncogenic role in PTCLs.7 IRF4 is a tightly regulated transcription factor involved in growth and differentiation of normal T and B lymphocytes.8,9 IRF4 is also expressed in B-cell NHLs and multiple myeloma, in which it drives tumor growth and has been proposed as a candidate therapeutic target.10-12 Although t(6;14)(p25.3;q11.2) is present in <1% of PTCLs, we and others have shown that IRF4 protein is expressed in approximately one-third of cases.6,13,14 These data suggest that alternative mechanisms drive IRF4 expression in most PTCLs. Direct inhibitors to IRF4 are not yet clinically available.15 Therefore, we undertook this study to characterize the functional role of IRF4 in PTCL cells, to identify the mechanisms driving IRF4 expression in PTCL, and to develop a clinically feasible strategy to target those mechanisms.
Materials and methods
Clinical samples and cell lines
Tissue-based studies were performed on paraffin-embedded and/or frozen PTCLs diagnosed by standard World Health Organization criteria.16 PTCLs from 277 patients (175 male, 102 female, median age 61 years; for subtypes, see supplemental Table 1 on the Blood Web site) were examined by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH). Peripheral blood samples from healthy donors were enriched for T cells by negative selection using the RosetteSep Human T Cell Enrichment Cocktail (Stem Cell, Vancouver, BC, Canada) following the manufacturer’s instructions. T cells were stimulated by incubation with 1 µg/mL phorbol myristate acetate (PMA) and 1 µM ionomycin for 24 hours at 37°C and 5% CO2. DNA for germ-line analysis was extracted from peripheral blood mononuclear cells from PTCL patients under protocols conducted under the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence Molecular Epidemiology Resource.17 The study was approved by the Mayo Clinic and University of Iowa Institutional Review Boards. Research was conducted in accordance with the Declaration of Helsinki.
Cell lines used in the study included Karpas 299 (ATCC, Gaithersburg, MD), SU-DHL-1 and SR-786 (DSMZ, Braunschweig, Germany), and FE-PD (generously provided by K. Pulford, Oxford, United Kingdom, with kind permission from A. Del Mistro, Padova, Italy). Cells were maintained in RPMI 1640 (Gibco, Grand Island, NY) containing 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco).
Genetic studies
FISH was performed on paraffin tissue sections using a recently developed break-apart probe to the IRF4 locus on 6p25.3 that lacks the previously described cross-hybridization to 16p11.6 Cases with 3 or more intact fusion signals were considered to have extra copies of IRF4. In a subset of cases, polymerase chain reaction and Sanger sequencing were performed on DNA extracted from frozen PTCL tumor tissue with primer sets that amplified exons 2 to 10 of the IRF4 gene (supplemental Table 2). IRF4 single nucleotide polymorphisms (SNPs) were genotyped in peripheral blood DNA from nonleukemic PTCL patients as part of a larger project17 conducted by the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence Molecular Epidemiology Resource using a custom Illumina (San Diego, CA) Infinium iSelect array.
Luciferase reporter assays
The 2.4-kb IRF4 promoter18 was polymerase chain reaction amplified using the primers forward: 5′-ACGGTACCTCGTGGAATATCACGGTCAGCCTT-3′ and reverse: 5′-ACGAGCTCTGCGAGGTGGGAAAGAGGAACTTT-3′ and cloned into the pGL3 luciferase reporter vector (Promega, Madison, WI) following KpnI and SacI digestion. The MYC luciferase reporter contained −2446/+334 of the MYC promoter/gene cloned into the pGL2 vector (Promega). A pGL3 construct containing the TNFRSF8 (CD30) promoter was generously provided by R. Horie (Kanagawa, Japan).19 For reporter assays, cells were transfected with the relevant pGL3 construct by electroporation and plated in 6-well plates containing RPMI 1640 with 10% serum. For concurrent gene knockdown or overexpression, small interfering RNA (siRNA) or pcDNA3-HA-IRF4 (generously provided by H. Hayashi and T. Matsuyama, Nagasaki, Japan20 ) was cotransfected with the reporter construct, and promoter binding was determined 24 hours postelectroporation. For drug treatment, inhibitors were administered 4 hours postelectroporation, and reporter activity was determined 3 hours after drug administration. Cell lysis and quantification of promoter binding was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Total protein was used for normalization of cell number. At least 3 separate transfections were performed for each treatment.
For additional methods, see supplemental Materials.
Results
IRF4 is constitutively expressed in PTCL and drives proliferation
In normal T cells, IRF4 protein expression was absent under basal conditions and induced upon external activation, as anticipated from previous studies.21 In contrast, PTCL cell lines constitutively expressed IRF4 in the absence of external stimulation (Figure 1A; supplemental Figure 1A). Because PMA stimulates T cells through activation of protein kinase C (PKC), we also treated stimulated T cells with the PKCβ inhibitor enzastaurin (LY317615) and demonstrated inhibition of the induction of IRF4 expression (Figure 1A). Enzastaurin also has been reported to downregulate IRF4 expression in multiple myeloma cells.22 Unlike both normal T cells and myeloma cells, however, PTCL cells were resistant to the effects of enzastaurin in inhibiting IRF4 expression.
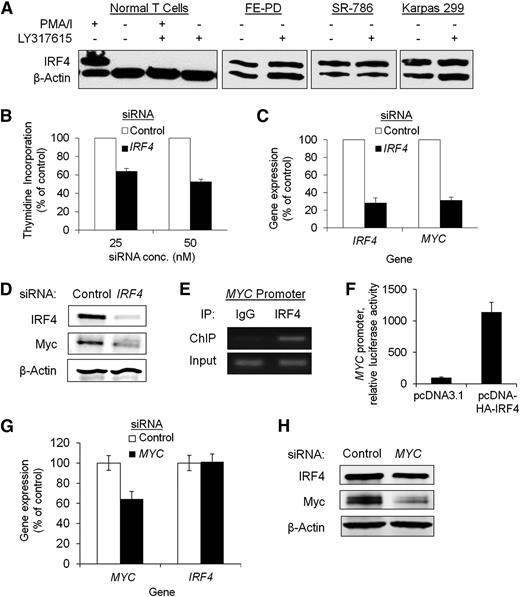
IRF4 is highly expressed and drives Myc expression and proliferation in PTCL cell lines. (A) IRF4 expression was absent in normal resting T cells and was induced upon stimulation with PMA/ionomycin (PMA/I). This induction was blocked by the PKCβ inhibitor LY317615 (enzastaurin). PTCL cells constitutively expressed IRF4, and this expression was resistant to inhibition by LY317615. (B) siRNA-based knockdown of IRF4 inhibited cell proliferation (25 nM siRNA, P < .0001; 50 nM siRNA, P < .0001) in Karpas 299 (B); IRF4 (P = .0009) and MYC (P = .0048) gene expression in Karpas 299 (C); and IRF4 and MYC protein expression in Karpas 299 (D). (E) ChIP for IRF4 demonstrated binding to the MYC promoter in Karpas 299 cells. (F) Overexpression of IRF4 increased MYC promoter activity in Karpas 299 (luciferase assay, P = .0032). (G) MYC knockdown did not inhibit IRF4 gene expression (P = .91) (G), and had only minimal effect on IRF4 protein expression in Karpas 299 (H).
IRF4 is highly expressed and drives Myc expression and proliferation in PTCL cell lines. (A) IRF4 expression was absent in normal resting T cells and was induced upon stimulation with PMA/ionomycin (PMA/I). This induction was blocked by the PKCβ inhibitor LY317615 (enzastaurin). PTCL cells constitutively expressed IRF4, and this expression was resistant to inhibition by LY317615. (B) siRNA-based knockdown of IRF4 inhibited cell proliferation (25 nM siRNA, P < .0001; 50 nM siRNA, P < .0001) in Karpas 299 (B); IRF4 (P = .0009) and MYC (P = .0048) gene expression in Karpas 299 (C); and IRF4 and MYC protein expression in Karpas 299 (D). (E) ChIP for IRF4 demonstrated binding to the MYC promoter in Karpas 299 cells. (F) Overexpression of IRF4 increased MYC promoter activity in Karpas 299 (luciferase assay, P = .0032). (G) MYC knockdown did not inhibit IRF4 gene expression (P = .91) (G), and had only minimal effect on IRF4 protein expression in Karpas 299 (H).
In multiple myeloma cells, IRF4 and Myc regulate each other’s expression to drive tumor cell proliferation.10 We therefore examined whether this autoregulatory circuit was present in PTCL cells. As in multiple myeloma, silencing of IRF4 by siRNA in Karpas 299 cells inhibited proliferation (Figure 1B, P < .0001), downregulated MYC gene expression (Figure 1C, P = .0048), and attenuated Myc protein expression (Figure 1D). Further, direct interaction between IRF4 and the MYC promoter was demonstrated by chromatin immunoprecipitation (ChIP) (Figure 1E), and overexpression of IRF4 resulted in increased MYC promoter activity (Figure 1F, P = .0032). However, in contrast to multiple myeloma, IRF4 gene expression was unaffected (P = .91), and IRF4 protein expression was only mildly diminished by knockdown of MYC in PTCL cells (Figure 1G-H). Therefore, although the functional role of IRF4 in PTCL cells shares features with its role in myeloma cells, the lack of response to PKC inhibition and minimal impact of MYC knockdown suggest differences in the mechanism of IRF4 regulation.
IRF4 expression in PTCLs is associated with IRF4 CNAs and a germ-line IRF4 polymorphism
We previously identified a translocation, t(6;14)(p25.3;q11.2), leading to IRF4/TRA gene fusion and IRF4 overexpression in PTCLs.6 However, this translocation was rare (<1%), prompting us to explore other potential genetic mechanisms for IRF4 expression in PTCL. IRF4 mutations have been reported in both multiple myeloma and B-cell NHLs23,24 ; therefore, we selected 30 IRF4-positive PTCLs for Sanger sequencing. We identified only 1 nonsynonymous variant in exon 5 (S309R) in a single PTCL (3%). In addition, IRF4 coding mutations were not observed in a next-generation sequencing–based study of 68 PTCLs (A.L.F., unpublished data), suggesting that IRF4 coding mutations rarely, if ever, contribute to IRF4 protein expression in PTCL.
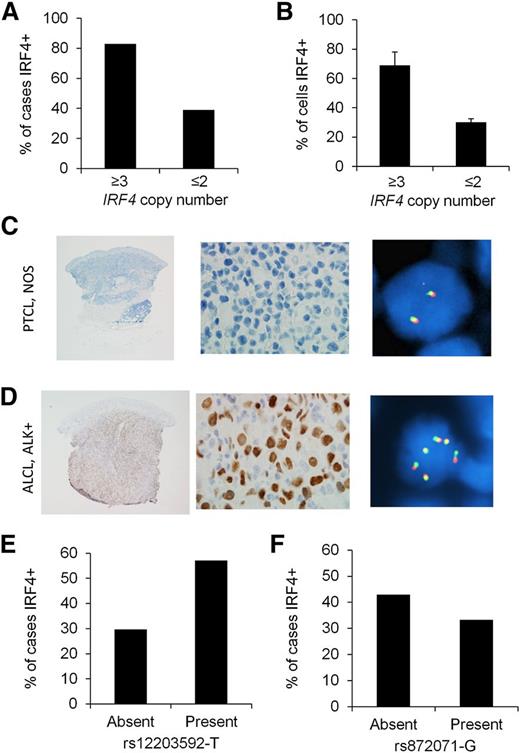
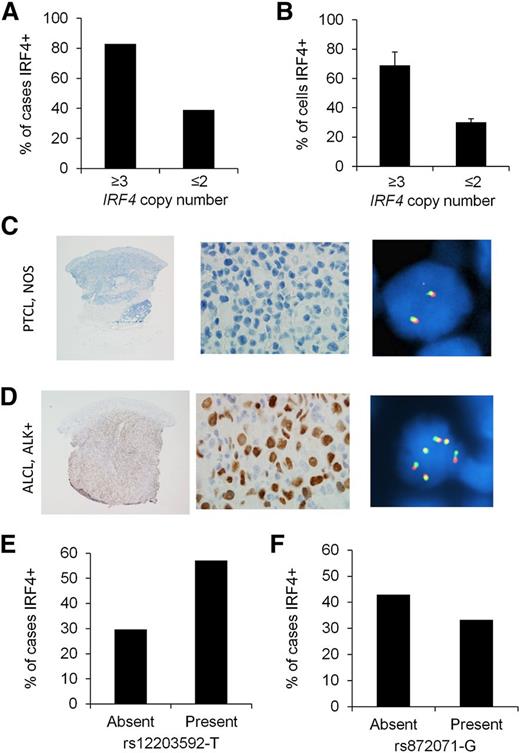
Copy number abnormalities (CNAs) of oncogenes can lead to oncoprotein overexpression in cancer. Although recurrent IRF4 CNAs have not been reported to our knowledge, we have observed extra (≥3) copies of the intact IRF4 locus in occasional PTCLs evaluated for IRF4 translocations by break-apart FISH (A.L.F., unpublished data). Therefore, we determined whether extra copies of the IRF4 gene were associated with IRF4 protein expression. Extending our previous results,6 we examined tissue samples from 277 PTCL patients for IRF4 protein expression by IHC and for IRF4 copy number by FISH (supplemental Table 1). IRF4 was positive by IHC in 116/277 cases (42%). Extra copies of IRF4 were seen in 18/277 cases (6%). Among PTCLs with extra copies of IRF4, 83% were IRF4 protein positive, compared with 39% of cases without extra copies of IRF4 (Figure 2A, P = .0002). The mean percentage of tumor cells with IRF4 staining was 69 ± 9% in cases with extra copies of IRF4, compared with 30 ± 2% in cases without extra copies (Figure 2B, P = .0001). In addition, the distribution among PTCL subtypes of IRF4 protein expression and extra copies of IRF4 were proportionally similar (highest in anaplastic large cell lymphoma [ALCL] and absent in angioimmunoblastic T-cell lymphoma; supplemental Table 1). Representative PTCLs with and without extra copies of IRF4 are shown in Figure 2C-D.
IRF4 protein expression is associated with IRF4 CNAs in PTCLs. (A) Eighty-three percent of PTCLs with extra copies of IRF4 were IRF4 protein positive, compared with 39% of cases without extra copies (P = .0002). (B) PTCLs with extra copies of IRF4 had a higher mean percentage of tumor cells with IRF4 staining than those without extra copies (P = .0001). (C) PTCL, not otherwise specified (NOS), involving the skin. IHC shows no evidence of IRF4 expression (left panel, ×20; middle panel, ×400). IRF4 FISH using a break-apart probe shows a normal signal pattern (2 intact red/green fusion signals; right panel, ×600). (D) ALCL, anaplastic lymphoma kinase (ALK) positive, involving the skin. IHC shows nuclear IRF4 expression (left panel, ×20; middle panel, ×400). IRF4 FISH shows multiple intact copies of the IRF4 gene (6 fusion signals in cell shown; right panel, ×600). (E) The germ-line SNP rs12203592-T was significantly associated with IRF4 positivity in PTCLs (P = .03). (F) The germ-line SNP rs872071 was not significantly associated with IRF4 positivity (P = .39).
IRF4 protein expression is associated with IRF4 CNAs in PTCLs. (A) Eighty-three percent of PTCLs with extra copies of IRF4 were IRF4 protein positive, compared with 39% of cases without extra copies (P = .0002). (B) PTCLs with extra copies of IRF4 had a higher mean percentage of tumor cells with IRF4 staining than those without extra copies (P = .0001). (C) PTCL, not otherwise specified (NOS), involving the skin. IHC shows no evidence of IRF4 expression (left panel, ×20; middle panel, ×400). IRF4 FISH using a break-apart probe shows a normal signal pattern (2 intact red/green fusion signals; right panel, ×600). (D) ALCL, anaplastic lymphoma kinase (ALK) positive, involving the skin. IHC shows nuclear IRF4 expression (left panel, ×20; middle panel, ×400). IRF4 FISH shows multiple intact copies of the IRF4 gene (6 fusion signals in cell shown; right panel, ×600). (E) The germ-line SNP rs12203592-T was significantly associated with IRF4 positivity in PTCLs (P = .03). (F) The germ-line SNP rs872071 was not significantly associated with IRF4 positivity (P = .39).
Next, we examined the germ-line genotypes of 2 IRF4 SNPs, rs12203592 (C/T) and rs872071 (A/G). rs12203592-T has been associated with increased risk of childhood acute lymphoblastic leukemia and increased IRF4 promoter activity in vitro.18 rs872071-G has been associated with increased risk of chronic lymphocytic leukemia and mildly decreased IRF4 gene expression in lymphoblastoid cell lines.25 Based on the dominant model, 57% of PTCL patients with rs12203592-T had IRF4-positive tumors, compared with 30% of cases without rs12203592-T (odds ratio, 3.16; 95% confidence interval, 1.14-8.73; P = .03; Figure 2E). We did not identify a significant association between rs872071-G and IRF4 expression (odds ratio, 0.67; 95% confidence interval, 0.26-1.69; P = .39; Figure 2F). The associations between IRF4 protein expression and both IRF4 copy number and the germ-line SNP rs12203592 suggest that IRF4 promoter activity is critical in the constitutive expression of IRF4 in PTCLs. Our genetic findings also suggest, however, that genetic abnormalities of the IRF4 gene are not the predominant cause of IRF4 expression. Therefore, we next aimed to identify upstream pathways leading to transcriptional activation of IRF4.
IRF4 is a direct transcriptional target of NF-κB in PTCL
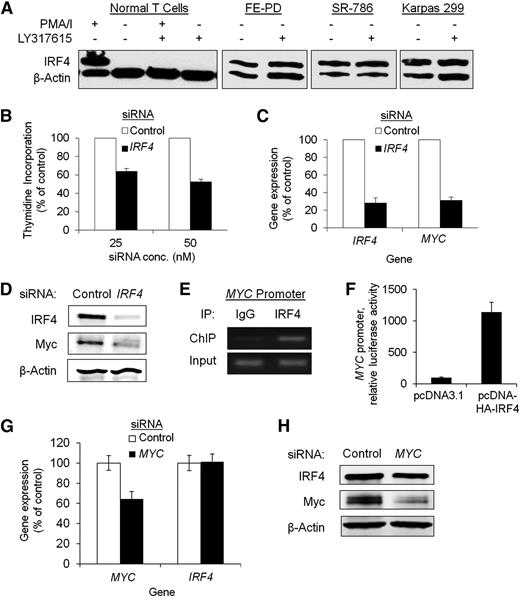
Because IRF4 expression in PTCL cells was constitutive and insensitive to PKC inhibition, we hypothesized that dysregulation of a pathway downstream of PKC might drive IRF4 expression in these cells. Therefore, we performed a pharmacologic screen using inhibitors that targeted 3 pathways downstream of PKC: mitogen-activated protein kinases, phosphatidylinositol 3-kinase/protein kinase B/mechanistic target of rapamycin, and nuclear factor κB (NF-κB) (supplemental Figure 1B). Although several of the inhibitors tested diminished PTCL cell proliferation, only bortezomib blocked IRF4 expression. Bortezomib inhibited both IRF4 expression and proliferation in a time- and dose-dependent fashion (Figure 3A-B). As a proteasome inhibitor, bortezomib prevents NF-κB activation but also has substantial other biological effects. Therefore, we used the inhibitory NF-κB (IκB) kinase inhibitor BMS-345541 to investigate the role of NF-κB in IRF4 expression more specifically. BMS-345541 diminished IκBα phosphorylation as expected (Figure 3C) and also reduced IRF4 gene expression by 96% at 4 hours posttreatment (Figure 3D, P < .0001). Furthermore, BMS-345541 inhibited IRF4 protein expression in all 4 IRF4-positive PTCL cell lines tested (Figure 3E). NF-κB inhibition was confirmed by observing reduced expression of X-linked inhibitor of apoptosis, a known NF-κB target.26
NF-κB inhibition attenuates IRF4 expression and proliferation in PTCL cells. (A) The proteasome and NF-κB inhibitor bortezomib inhibited IRF4 protein expression in Karpas 299 cells in a time-dependent manner. (B) Bortezomib inhibited cell proliferation in a dose-dependent manner in Karpas 299 cells. (C) The IκB kinase inhibitor BMS-34554 (dissolved in dimethylsulfoxide [DMSO]) inhibited IκBα Ser32 phosphorylation 10 minutes postadministration compared with vehicle alone (0.5% DMSO) or no treatment in Karpas 299 cells. (D) BMS-345541 decreased IRF4 mRNA expression 4 hours postadministration (P < .0001, Karpas 299). (E) BMS-345541 inhibited protein expression of IRF4 and the NF-κB target, X-linked inhibitor of apoptosis (XIAP), 24 hours postadministration in 4 IRF4-positive PTCL cell lines.
NF-κB inhibition attenuates IRF4 expression and proliferation in PTCL cells. (A) The proteasome and NF-κB inhibitor bortezomib inhibited IRF4 protein expression in Karpas 299 cells in a time-dependent manner. (B) Bortezomib inhibited cell proliferation in a dose-dependent manner in Karpas 299 cells. (C) The IκB kinase inhibitor BMS-34554 (dissolved in dimethylsulfoxide [DMSO]) inhibited IκBα Ser32 phosphorylation 10 minutes postadministration compared with vehicle alone (0.5% DMSO) or no treatment in Karpas 299 cells. (D) BMS-345541 decreased IRF4 mRNA expression 4 hours postadministration (P < .0001, Karpas 299). (E) BMS-345541 inhibited protein expression of IRF4 and the NF-κB target, X-linked inhibitor of apoptosis (XIAP), 24 hours postadministration in 4 IRF4-positive PTCL cell lines.
Because bortezomib has been shown to induce apoptosis in ALCL cells,27 we examined whether BMS-345541 and bortezomib also induced apoptosis in our model. Indeed, at higher concentrations (5-10 μM BMS-345541 and 5-10 nM bortezomib), both drugs led to apoptosis as assessed by poly (ADP-ribose) polymerase cleavage (supplemental Figure 2A). This induction was at least partially IRF4 independent because IRF4 knockdown alone did not induce apoptosis (supplemental Figure 2B-C). However, a construct in which IRF4 expression was driven by a cytomegalovirus promoter partially rescued the effect of BMS-345541 at a dose that inhibited native IRF4 expression and proliferation but did not induce apoptosis (supplemental Figure 3). These results suggest a contribution of IRF4 to NF-κB function in PTCL cells and, together with the antiproliferative effect of IRF4 knockdown (Figure 1B), indicate that IRF4 preferentially promotes proliferation more than inhibiting apoptosis in these cells.
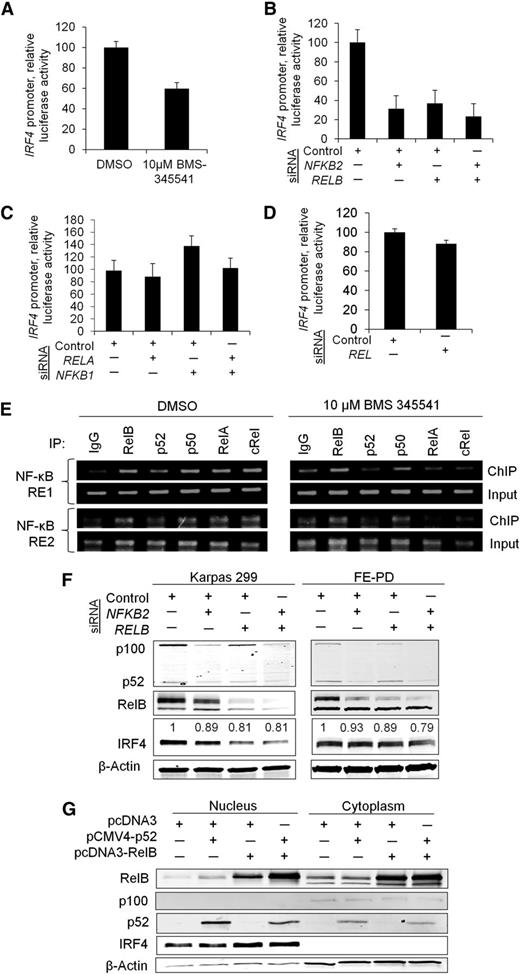
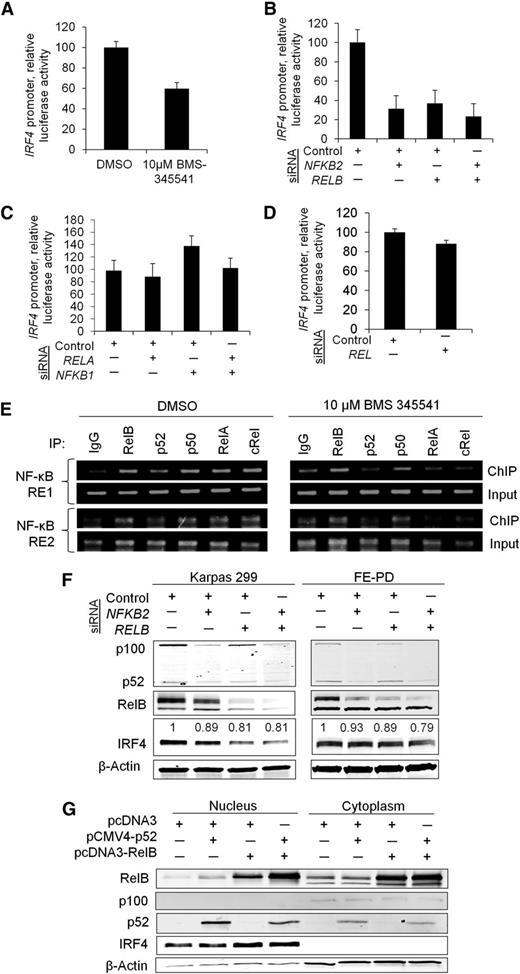
To investigate the transcriptional regulation of IRF4 by NF-κB further, we first confirmed that the decrease in IRF4 expression induced by BMS-345541 was associated with decreased IRF4 promoter activity. A luciferase reporter assay demonstrated reduced IRF4 promoter activity in the presence of BMS-345541 for 3 hours (Figure 4A, P = .0086). We then investigated which NF-κB subunits were functionally responsible for driving IRF4 expression. Promoter activity was assessed following knockdown of the genes encoding the individual NF-κB subunits: RELA (p65), RELB, NFKB1 (p105/p50), NFKB2 (p100/p52), and REL (c-Rel). siRNA knockdown of RELB and NFKB2 resulted in marked decreases in IRF4 promoter activity to 31% and 37% of control, respectively (P = .014), and the combined knockdown of RELB and NFKB2 inhibited IRF4 promoter luciferase activity to 23% of control (Figure 4B). However, knockdown of RELA, NFKB1, and REL had no significant effect on IRF4 promoter activity (Figure 4C-D). ChIP identified interaction of the RelB and p52 as well as p65, p50, and c-Rel NF-κB subunits with the 2 NF-κB response elements (REs) located 1166 bp (NF-κB RE1) and 103 bp (NF-κB RE2) upstream of the IRF4 transcription start site. Of note, this binding diminished in the presence of BMS-345541 (Figure 4E).
RelB and p52 transcriptionally regulate IRF4. (A) The IκB kinase inhibitor BMS-34554 (“BMS”) decreased IRF4 promoter activity 3 hours postadministration (P = .0086; Karpas 299). (B) Individual and concurrent knockdown of NFKB2 (p52) and RELB attenuated IRF4 promoter activity (P = .014; Karpas 299). (C) Knockdown of RELA (p65) alone or in combination with NFKB1 (p50) had no effect on IRF4 promoter activity (P = .25; Karpas 299). (D) Knockdown of REL (c-Rel) did not significantly inhibit IRF4 promoter activity (P = .063; Karpas 299). (E) ChIP demonstrated that all NF-κB subunits interacted with NF-κB REs 1 and 2 in the IRF4 promoter, located 1166 bp and 103 bp upstream of the IRF4 transcription start site, respectively (Karpas 299). These interactions were diminished to varying degrees by BMS-345541 3 hours postadministration. (F) Concurrent knockdown of NFKB2 and RELB resulted in decreased IRF4 protein expression in Karpas 299 and FE-PD cell lines. Densitometry values for IRF4 bands normalized to β-actin are shown. (G) Simultaneous overexpression of p52 and RelB increased nuclear IRF4 protein abundance in Karpas 299.
RelB and p52 transcriptionally regulate IRF4. (A) The IκB kinase inhibitor BMS-34554 (“BMS”) decreased IRF4 promoter activity 3 hours postadministration (P = .0086; Karpas 299). (B) Individual and concurrent knockdown of NFKB2 (p52) and RELB attenuated IRF4 promoter activity (P = .014; Karpas 299). (C) Knockdown of RELA (p65) alone or in combination with NFKB1 (p50) had no effect on IRF4 promoter activity (P = .25; Karpas 299). (D) Knockdown of REL (c-Rel) did not significantly inhibit IRF4 promoter activity (P = .063; Karpas 299). (E) ChIP demonstrated that all NF-κB subunits interacted with NF-κB REs 1 and 2 in the IRF4 promoter, located 1166 bp and 103 bp upstream of the IRF4 transcription start site, respectively (Karpas 299). These interactions were diminished to varying degrees by BMS-345541 3 hours postadministration. (F) Concurrent knockdown of NFKB2 and RELB resulted in decreased IRF4 protein expression in Karpas 299 and FE-PD cell lines. Densitometry values for IRF4 bands normalized to β-actin are shown. (G) Simultaneous overexpression of p52 and RelB increased nuclear IRF4 protein abundance in Karpas 299.
To confirm the role of the RelB and p52 subunits in regulating IRF4, we determined the effect of modulating RelB and p52 protein levels on IRF4 protein expression. Knockdown of NFKB2 or RELB individually or in combination reduced IRF4 protein expression in 2 IRF4-positive cell lines, Karpas 299 and FE-PD (Figure 4F). The contribution of NFKB2 was less than that seen in the luciferase assay (Figure 4B), possibly because of more limited effects of p52 on the native chromatin configuration than on the promoter-luciferase construct. We also found that the combined overexpression of RelB and p52 in K299 and SU-DHL-1 resulted in increased nuclear IRF4 protein expression (Figure 4G and supplemental Figure 4A). Combined overexpression of p50/p65 and p50/c-Rel resulted only in a slight induction of IRF4 expression (supplemental Figure 4B). Taken together, these data indicate that IRF4 is a direct target of NF-κB transcription factors and define the p52 and RelB subunits as major regulators of IRF4 in PTCL cells.
Identification of a novel positive feedback loop involving CD30, NF-κB, and IRF4 in PTCL
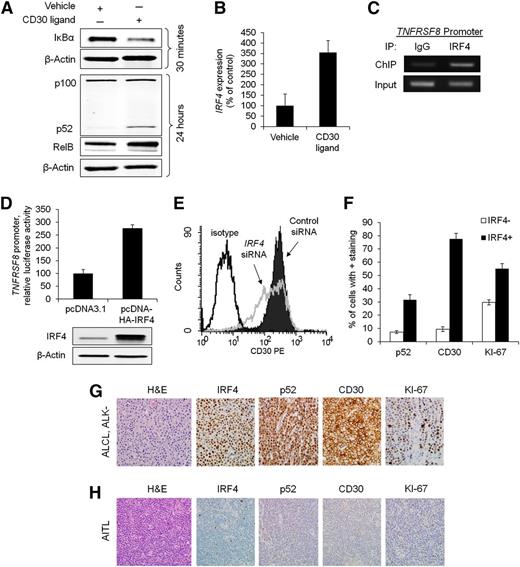
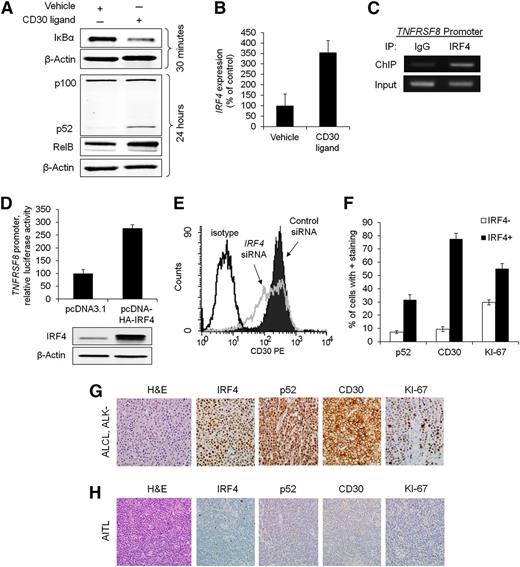
We and others have previously shown that IRF4 expression in PTCLs generally parallels expression of CD30, a transmembrane cytokine receptor encoded by the TNFRSF8 gene.6,13 Because CD30 is a known activator of NF-κB,28-31 we examined the role of CD30 in NF-κB activation and subsequent IRF4 expression in PTCL. Activation of CD30 receptor by its ligand or an anti-CD30 cross-linking antibody resulted in rapid degradation of IκBα, followed by cleavage of p100 to p52 and increased RelB expression (Figure 5A; supplemental Figure 5A). CD30 activation also resulted in a marked increase in IRF4 gene expression (355% of control, P = .033) at 24 hours poststimulation (Figure 5B; supplemental Figure 5B).
Reciprocal regulation of IRF4 and CD30 in PTCL cells. (A) CD30 ligand (100 ng/mL) activated the NF-κB pathway in Karpas 299 cells, as evidence by depletion of IκBα, increased RelB expression (156% of control), and cleavage of p100 into p52. Karpas 299 and the other cell lines studied do not express endogenous CD30 ligand (supplemental Figure 6B). (B) Stimulation with CD30 ligand for 24 hours increased IRF4 gene expression (P = .033). (C) IRF4 ChIP demonstrated interaction of IRF4 with the TNFRSF8 promoter in Karpas 299 cells (see also supplemental Figure 6A). (D) Overexpression of IRF4 increased TNFRSF8 promoter activity (luciferase assay, P = .0029). (E) Knockdown of IRF4 decreased surface CD30 protein expression, as detected by flow cytometry. (F) IRF4-positive PTCLs had significantly higher percentages of cells staining for p52 (nuclear; P < .0001), CD30 (P < .0001), and KI-67 (P < .0001). (G) ALCL, ALK negative. IHC showed strong nuclear expression of IRF4 and p52, uniform staining for CD30, and a high proliferative rate by KI-67 (all images, ×400). (H) Angioimmunoblastic T-cell lymphoma (AITL). The tumor cells were negative for IRF4, p52, and CD30 and showed a low proliferative rate by KI-67 (×400).
Reciprocal regulation of IRF4 and CD30 in PTCL cells. (A) CD30 ligand (100 ng/mL) activated the NF-κB pathway in Karpas 299 cells, as evidence by depletion of IκBα, increased RelB expression (156% of control), and cleavage of p100 into p52. Karpas 299 and the other cell lines studied do not express endogenous CD30 ligand (supplemental Figure 6B). (B) Stimulation with CD30 ligand for 24 hours increased IRF4 gene expression (P = .033). (C) IRF4 ChIP demonstrated interaction of IRF4 with the TNFRSF8 promoter in Karpas 299 cells (see also supplemental Figure 6A). (D) Overexpression of IRF4 increased TNFRSF8 promoter activity (luciferase assay, P = .0029). (E) Knockdown of IRF4 decreased surface CD30 protein expression, as detected by flow cytometry. (F) IRF4-positive PTCLs had significantly higher percentages of cells staining for p52 (nuclear; P < .0001), CD30 (P < .0001), and KI-67 (P < .0001). (G) ALCL, ALK negative. IHC showed strong nuclear expression of IRF4 and p52, uniform staining for CD30, and a high proliferative rate by KI-67 (all images, ×400). (H) Angioimmunoblastic T-cell lymphoma (AITL). The tumor cells were negative for IRF4, p52, and CD30 and showed a low proliferative rate by KI-67 (×400).
We then investigated a potential feedback mechanism between IRF4 and CD30. Previous IRF4 ChIP-Seq studies have demonstrated cooperative binding between IRF4 and activator protein 1 (AP-1) to recognize TGAnTCA/GAAA motifs in normal activated T cells.32,33 Because this AP-1/IRF4 composite element is present in the TNFRSF8 promoter,34 we sought to determine whether IRF4 is bound to this element in PTCL cells. Indeed, IRF4 ChIP identified binding to the TNFRSF8 promoter in both ALK-positive and ALK-negative ALCL cell lines (Figure 5C and supplemental Figure 6A). Moreover, IRF4 overexpression resulted in increased TNFRSF8 promoter activity (Figure 5D, P = .0029). Finally, siRNA knockdown of IRF4 reduced cell surface expression of CD30 protein (Figure 5E). Taken together, these data support a novel mechanism for IRF4 regulation in PTCL that is different from that previously reported in multiple myeloma.10 Although IRF4 ChIP has identified some interaction of IRF4 with the TNFRSF8 promoter in myeloma cells, CD30 protein is expressed infrequently in multiple myeloma clinical samples and cell lines (supplemental Figure 7).10,35
Having demonstrated regulation of IRF4 by a CD30/NF-κB positive feedback loop in PTCL cell lines, we sought to determine whether IRF4 expression was associated with expression of p52 and CD30 in PTCL tissue samples. Immunohistochemical staining for both p52 and CD30 was significantly higher in IRF4-positive PTCLs compared with IRF4-negative PTCLs (Figures 5F-H). In keeping with the in vitro effects of IRF4 on PTCL cell proliferation, the proliferation marker, KI-67, also was significantly higher in IRF4-positive PTCLs. When stratified by histologic classification, the pathway is enriched in ALCLs, a PTCL subtype that consistently expresses CD30 by definition (supplemental Tables 5-7).
Discussion
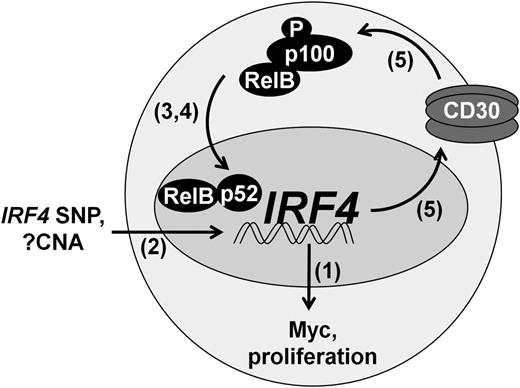
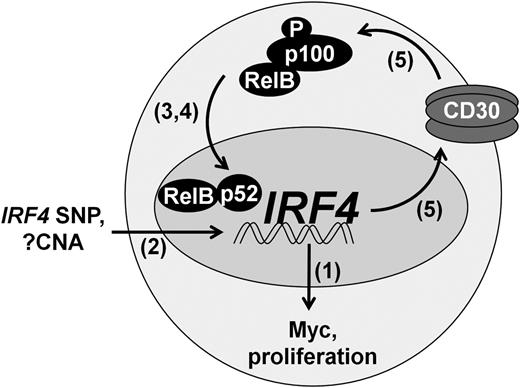
Here, we demonstrate a growth promoting role for IRF4 in PTCL and define a novel mechanism regulating its expression, a model of which is depicted in Figure 6. IRF4 expression is regulated by binding of NF-κB, particularly the p52 and RelB subunits, to NF-κB REs in the IRF4 promoter. Furthermore, IRF4 expression is associated with the presence of extra copies of the IRF4 gene, as well an intronic SNP known to increase IRF4 promoter activity. The resulting IRF4 protein binds the MYC promoter, thereby driving Myc expression and tumor cell proliferation. In addition, IRF4 binds the TNFRSF8 promoter, resulting in increased expression of CD30. CD30 subsequently activates NF-κB, further increasing IRF4 expression in a positive feedback loop. This feedback loop is interrupted by clinically available NF-κB inhibitors. These findings advance the understanding of IRF4 in cancer and provide a potential strategy for targeted therapy of IRF4-positive PTCLs.
Positive feedback loop regulating IRF4 expression in PTCL.IRF4 is transcriptionally regulated by the alternative NF-κB pathway (RelB/p52), augmented in some cases by the intronic IRF4 SNP rs12203592-T. IRF4 CNAs are associated with IRF4 protein expression, but their contribution to IRF4 transcription is not known. IRF4 drives Myc expression and tumor proliferation. In addition, IRF4 increases CD30 expression, leading to activation of NF-κB and perpetuation of IRF4 expression. Numbers in parentheses indicate figure numbers corresponding to relevant experiments.
Positive feedback loop regulating IRF4 expression in PTCL.IRF4 is transcriptionally regulated by the alternative NF-κB pathway (RelB/p52), augmented in some cases by the intronic IRF4 SNP rs12203592-T. IRF4 CNAs are associated with IRF4 protein expression, but their contribution to IRF4 transcription is not known. IRF4 drives Myc expression and tumor proliferation. In addition, IRF4 increases CD30 expression, leading to activation of NF-κB and perpetuation of IRF4 expression. Numbers in parentheses indicate figure numbers corresponding to relevant experiments.
The regulation and role of IRF4 in PTCLs demonstrated here shows both similarities and important differences to findings previously reported in other lymphoid neoplasms. In multiple myeloma,10 and as recently demonstrated in PTCL,36 IRF4 transcriptionally regulates MYC and drives tumor cell proliferation. The site at which IRF4 binds to the MYC promoter in PTCL (1486 bp upstream MYC) corresponds to the IRF4 binding site previously reported in myeloma.10 Although relatively little is known about the IRF4 transcriptional program in PTCLs, the mechanisms and domains for promoter binding by IRF4 have been studied more extensively in normal B and T cells, as well as B-cell neoplasms, and have been reviewed previously.11 Interestingly, analogous to rare IRF4/TRA translocations in PTCL, occasional myelomas and B-cell NHLs carry translocations fusing the IRF4 gene to one of the immunoglobulin gene loci.37-39 Unlike multiple myeloma, however, IRF4 expression is not strongly regulated by Myc in PTCL; rather, IRF4 expression in PTCL induces expression of CD30, absent in most cases of multiple myeloma.10,35 CD30 expression also is expressed only infrequently in diffuse large B-cell lymphomas (DLBCLs), despite common expression of IRF4 in the activated B-cell-like (ABC) subtype.40-42 Although these entities differ in the frequency of CD30 expression, activation of NF-κB is a feature common to PTCLs, multiple myeloma, and ABC-type DLBCLs.43-45 Among the NF-κB subunits, RelA (p65) has been implicated in DLBCL and multiple myeloma,10,46,47 whereas c-Rel has been implicated as a driver of IRF4 in activated lymphocytes, in human T-lymphotropic virus-I–transformed cells, and in the human T-lymphotropic virus-I–associated neoplasm adult T-cell leukemia/lymphoma.48-50 In PTCL, however, our data suggest that the alternative NF-κB pathway (p52 and RelB) plays a dominant role in IRF4 regulation, with an additional but lesser contribution of the classical pathway. This finding also is supported by a recent study on the role of NF-κB-inducing kinase in PTCL.45
NF-κB is a well-established therapeutic target in lymphoid malignancies.43 Proteasome inhibitors like bortezomib, which function in part by preventing NF-κB activation, are widely used in multiple myeloma.51 Among DLBCLs, responses to proteasome inhibitors have been heterogeneous; the best clinical response rates have been observed in the ABC subtype, which also shows greater IRF4 expression and NF-κB activation than the germinal center B-cell-like subtype.40,41,52,53 Similarly, we would hypothesize that responses of PTCLs to agents targeting NF-κB might vary based on the activity of the IRF4/CD30/NF-κB feedback loop we describe here. Indeed, PTCLs have demonstrated mixed responses in clinical trials of combined regimens containing bortezomib.54-56 It would be of interest to examine retrospectively whether these responses were associated with IRF4 expression because biomarker-driven patient selection might increase response rates to NF-κB-targeted therapies. Nevertheless, our data should not be interpreted to suggest that the functional effects of NF-κB inhibition in PTCL cells are entirely IRF4 dependent, as the NF-kB complex is a master transcriptional regulator with a myriad of targets.57 It also is important to note that the relevance of the IRF4/CD30/NF-κB feedback loop is supported most strongly in ALCL; the low expression of these proteins in PTCLs other than ALCL (supplemental Table 5) and the paucity of non-ALCL cell lines suggest the need for further study of rare other PTCLs that strongly express proteins involved in this feedback mechanism.
Binding of IRF4 to the TNFRSF8 promoter and subsequent regulation of CD30 expression has not been reported previously in PTCL. This finding is supported, however, by recent data demonstrating cotargeting of TNFRSF8 by IRF4 and AP-1; positive regulation of TNFRSF8 by IRF4 in T helper 17 cells; and IRF4 binding upstream of and within the TNFRSF8 gene in activated CD4+ T cells.33,58 CD30-induced activation of NF-κB is well established and involves activation of the tumor necrosis factor receptor-associated factor (TRAF) family of signaling intermediates. TRAF1, TRAF2, TRAF3, and TRAF5 interact with the intracellular domain of CD30 and then interact directly with either the NF-κB pathway to promote survival or with caspases to promote cell death.59-62 In PTCL cells in particular, this study and others have demonstrated that CD30 stimulation drives processing of p100 to p52 and increases RelB expression.28-31 Importantly, CD30 expression has become increasingly relevant based on promising results in PTCL with brentuximab vedotin, a CD30-directed antibody-drug conjugate containing the antimicrotubule agent, monomethylauristatin E.63,64
In addition to the novel IRF4 positive feedback loop involving NF-κB and CD30, we also found that IRF4 CNAs and the germ-line SNP rs12203592-T were associated with IRF4 expression in PTCLs. Although IRF4 translocations and mutations were rare in PTCLs, extra copies of IRF4 and rs12203592-T were present in appreciable numbers of patients (7% and 24%, respectively). rs12203592 is located in intron 4 of the IRF4 gene. Do et al demonstrated that the wild-type C allele has a repressive effect on IRF4 promoter activity compared with the variant T allele.18 rs12203592-T has been associated with risk of childhood acute lymphoblastic leukemia and pigmentation in melanocytes but has not been found to be associated with NHL risk.18,65,66 Our data do not establish whether these genetic findings directly contribute to IRF4 expression in PTCLs. For example, IRF4 CNAs in IRF4-positive PTCLs might relate to increased polysomy rather than focal IRF4 gains. In either case, however, our overall findings suggest that genetic abnormalities of IRF4 are not the major driver of IRF4 expression and implicate transcriptional regulation by NF-κB in at least most cases. Other genetic abnormalities that activate NF-κB might increase IRF4 expression in PTCLs, and/or the coordinated expression of NF-κB and IRF4 might be characteristics of the cell of origin (eg, T helper 17); both these phenomena have been described in B cells and B-cell neoplasms.11
In addition to the CD30/NF-κB/IRF4 feedback circuit identified in this study, other mechanisms may contribute to IRF4 expression in PTCL and merit further investigation. IRF4 regulation may involve cross talk between alternative and classical NF-κB pathways, as supported by our data and by previous studies demonstrating that CD30 activates both classical and alternative NF-κB pathways, and that NF-κB-inducing kinase regulates DNA-binding activity of classical and alternative NF-κB subunits in PTCL cells.31,45 Additionally, p52 and RelB form complexes such as RelB/p50 and a BCL3/p52 homodimer ternary complex that modulate CD30-mediated signaling.30,67 Also, IRF4 promoter elements such as those that interact with AP-1, NF-1/CD28, Stat6/kB, Sp1, and/or PU.1 may be involved in transcriptional regulation of IRF4 independent of or in conjunction with NF-κB.48,68 Finally, some clinically available drugs have been associated with decreased IRF4 expression, although the mechanisms involved are incompletely understood. For example, the hypolipidemic drug simvastatin has been shown to decrease IRF4 expression in T cells.69 We found that simvastatin also can decrease IRF4 expression in PTCL cells (supplemental Figure 8). The mechanism and potential clinical implications of this finding merit further study.
In summary, our data provide evidence to support a novel model of IRF4 regulation involving a CD30/NF-κB positive feedback loop in PTCL. This relationship was recapitulated in PTCL tumor specimens. We demonstrated that this feedback loop can be disrupted by classes of agents that are clinically available or in clinical trials, including proteasome inhibitors and IκB kinase inhibitors. Further investigation to determine whether presence of this feedback loop will predict patient response to NF-κB inhibitors is warranted.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Scott H. Kaufmann for helpful comments on the manuscript.
This work was supported by the American Cancer Society (Research Scholar grant RSG-12-193-01-TBE) (A.L.F.), the Karl-Erivan Haub Family Career Development Award in Cancer Research at the Mayo Clinic in Rochester (A.L.F.), the National Institutes of Health National Cancer Institute (R01 CA177734 [A.L.F.], R01 CA92153 [J.R.C.], R01 CA1144 [S.L.S.], P30 CA15083 [Mayo Clinic Cancer Center], and P50 CA97274 [University of Iowa/Mayo Clinic Lymphoma SPORE]), and the National Center for Advancing Translational Science (CTSA grant UL1 TR000135). X.X. and Y.Z. were supported by scholarship awards from the China Scholarship Council. A.L.F. is a Damon Runyon Clinical Investigator supported by the Damon Runyon Cancer Research Foundation (CI-48-09).
Authorship
Contribution: R.L.B., M.E.F.-Z., and A.L.F. designed the study and wrote the manuscript; R.L.B., N.S.K., X.X., Y.Z., Z.-Z.Y., J.C.P., D.M.G., J.-H.L., S.F.E., and A.L.F. conducted experiments; R.A.K., M.E.L., and Y.W. conducted genetic studies; R.L.B., J.-H.L., L.L.A., R.P.K., J.M.C., J.R.C., S.L.S., D.F.J., A.D., M.E.F.-Z., and A.L.F. analyzed data; M.M.O. and M.J.M. conducted statistical analyses; and B.K.L. and S.M.A. contributed clinical specimens.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for N.S.K. is Geisinger Medical Center, Danville, PA; the current affiliation for S.F.E. is Northern Illinois University, De Kalb, IL; and the current affiliation for A.D. is Memorial Sloan-Kettering Cancer Center, New York, NY.
Correspondence: Andrew L. Feldman, Department of Laboratory Medicine and Pathology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: feldman.andrew@mayo.edu.

![Figure 3. NF-κB inhibition attenuates IRF4 expression and proliferation in PTCL cells. (A) The proteasome and NF-κB inhibitor bortezomib inhibited IRF4 protein expression in Karpas 299 cells in a time-dependent manner. (B) Bortezomib inhibited cell proliferation in a dose-dependent manner in Karpas 299 cells. (C) The IκB kinase inhibitor BMS-34554 (dissolved in dimethylsulfoxide [DMSO]) inhibited IκBα Ser32 phosphorylation 10 minutes postadministration compared with vehicle alone (0.5% DMSO) or no treatment in Karpas 299 cells. (D) BMS-345541 decreased IRF4 mRNA expression 4 hours postadministration (P < .0001, Karpas 299). (E) BMS-345541 inhibited protein expression of IRF4 and the NF-κB target, X-linked inhibitor of apoptosis (XIAP), 24 hours postadministration in 4 IRF4-positive PTCL cell lines.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/20/10.1182_blood-2014-05-578575/4/m_3118f3.jpeg?Expires=1769157906&Signature=w2hk1j9px5z5jh9vvjHk9HJw9B-i7rDqDnk--oY630GnQCtUNdGK89NOyaVGF13jS91RrJ4kZTYicC7u-YHm8fAB4rG2SzsFHL8D4ffaxMy-st2Qx2Q5Rtkc5pPmf1h6kF3Gqqn3IzlUMU5r4FczaKROvu2Re4mLctoX9QndiMi1pklbd5FX1zDwLkpDQsK3h-p3og8lagUcurJ~PGHxXOFyocfyNIG-fuTU9rZruC1kBN86XfmvdN9bOdzuXU4~ndpgAuy5wS2ZZD-D4UrHeeDZbAbhd5ya3Uz2bzIF6Jk8HVHEvw8QvceAkM3qZs8AxY6GKn96XPf-ulSzKdCHrQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. NF-κB inhibition attenuates IRF4 expression and proliferation in PTCL cells. (A) The proteasome and NF-κB inhibitor bortezomib inhibited IRF4 protein expression in Karpas 299 cells in a time-dependent manner. (B) Bortezomib inhibited cell proliferation in a dose-dependent manner in Karpas 299 cells. (C) The IκB kinase inhibitor BMS-34554 (dissolved in dimethylsulfoxide [DMSO]) inhibited IκBα Ser32 phosphorylation 10 minutes postadministration compared with vehicle alone (0.5% DMSO) or no treatment in Karpas 299 cells. (D) BMS-345541 decreased IRF4 mRNA expression 4 hours postadministration (P < .0001, Karpas 299). (E) BMS-345541 inhibited protein expression of IRF4 and the NF-κB target, X-linked inhibitor of apoptosis (XIAP), 24 hours postadministration in 4 IRF4-positive PTCL cell lines.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/20/10.1182_blood-2014-05-578575/4/m_3118f3.jpeg?Expires=1769157907&Signature=qYX87pZot-JgD8E6drrLUQim3W0dy2gSGqziYsAy8Sn2vF3GMmHYJxmzZ1LK13zjMyAZgR-fL2DuFGE~ZyLpJ5OBLTSGeiCiEWG2uqev1kG~cDZyK3G4Sm~wClAyYD4tO4j4XG0xdsQdPufYl5xR5lBzesNa3cQYXr8B0h5yhp9gfjz3zx~OaKap7tU0sc4-u3DEG05tT64Bb8fmfaE14E0~rGJkbrlzszLCzVm7dfVJAPUCOuRE4syVh22wm5x7G6hHMek9Oj8nNfOKZ-Vwl7RUrGVA8EFn394qvG61jk2UPfDdeQzHC601-MnXTwf2zhaH34GgAt90QahnWwvaWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)