Key Points
Akt/FLNA/TIF-90 signaling regulates rRNA synthesis in acute myelogenous leukemia cells.
Direct targeting of Akt has potential therapeutic applications in acute myelogenous leukemia treatment.
Abstract
The transcription initiation factor I (TIF-IA) is an important regulator of the synthesis of ribosomal RNA (rRNA) through its facilitation of the recruitment of RNA polymerase I (Pol I) to the ribosomal DNA promoter. Activation of the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) pathway, which occurs commonly in acute myelogenous leukemia, enhances rRNA synthesis through TIF-IA stabilization and phosphorylation. We have discovered that TIF-IA coexists with a splicing isoform, TIF-90, which is expressed preferentially in the nucleolus and at higher levels in proliferating and transformed hematopoietic cells. TIF-90 interacts directly with Pol I to increase rRNA synthesis as a consequence of Akt activation. Furthermore, TIF-90 binds preferentially to a 90-kDa cleavage product of the actin binding protein filamin A (FLNA) that inhibits rRNA synthesis. Increased expression of TIF-90 overcomes the inhibitory effect of this cleavage product and stimulates rRNA synthesis. Because activated Akt also reduces FLNA cleavage, these results indicate that activated Akt and TIF-90 function in parallel to increase rRNA synthesis and, as a consequence, cell proliferation in leukemic cells. These results provide evidence that the direct targeting of Akt would be an effective therapy in acute leukemias in which Akt is activated.
Introduction
Alterations in the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) axis are both associated with and causal of malignant transformation in many cancer types including acute myeloid leukemias (AMLs).1-3 The PI3K/Akt pathway is frequently constitutively activated in leukemic blasts from AML patients and contributes to both cell survival and proliferation.4-6 Activation of the target of rapamycin (TOR) has emerged as a major effector of the PI3K/Akt pathway’s ability to regulate protein synthesis and as a contributor to oncogenesis.7-9 In addition to its effects on protein translation, the mammalian target of rapamycin (mTOR) coordinates the synthesis of ribosomal proteins and ribosomal RNA (rRNA) to modulate ribosome biogenesis.10-12 The importance of mTOR activation in cancer progression has been highlighted by the ability of rapamycin, a potent and selective inhibitor of mTORC1, to downregulate the synthesis of rRNA and to inhibit the growth of many tumors.10,13-16 Although there is evidence that activated Akt enhances rRNA synthesis independent of mTOR,17 the specific mechanisms through which Akt directly promotes ribosome biogenesis have not yet been fully elucidated.
The PI3K/Akt/mTOR pathway controls the transcription of rRNA by polymerase I (Pol I) through transcription initiation factor I (TIF-IA), the mammalian homolog of yeast Rrn3p.18-20 TIF-IA interacts with the TATA-binding protein (TBP)-containing factor TIF-IB/SL1, and both are required to recruit RNA polymerase I (Pol I) to the ribosomal DNA (rDNA) promoter and generate a productive transcription initiation complex.21,22 TIF-IA is itself phosphorylated at multiple sites by a variety of protein kinases with both positive and negative effects on its ability to initiate transcription at this locus.23-25 It has also been shown that the phosphorylation of TIF-IA by mTOR enhances rRNA transcription20 and that TIF-IA expression is essential to maintaining nucleolar architecture and cell viability.26 We have recently shown that Akt regulates TIF-IA activity in part through the phosphorylation of casein kinase 2 (CK2), enhancing rRNA synthesis through this mechanism.27 Hence, TIF-IA is a key intermediate in the regulation of cellular proliferation through the PI3K/Akt/mTOR signaling network.
We have discovered that TIF-IA coexists with a splicing isoform, TIF-90, which results from the loss of 30 amino acids encoded by exon 6 of the gene. TIF-90 is expressed preferentially in the nucleolus and at higher levels in proliferating and transformed hematopoietic cells. We therefore interrogated its role as a regulator of rRNA synthesis. TIF-90 selectively interacts with a 90-kDa cleavage fragment of the actin binding protein filamin A (FLNA), which has been shown to have a direct inhibitory effect on rRNA synthesis.28 Activated Akt inhibits FLNA cleavage, thereby enhancing rRNA synthesis.29 Based on our studies, we propose a new model in which TIF-90 functions in concert with Akt to enhance rRNA synthesis by directly binding with the FLNA cleavage product.
Methods
Human patient samples
Mononuclear cells were isolated from bone marrow or the peripheral blood of patients using Stanford Institutional Review Board–approved protocols by density-gradient centrifugation using Ficoll-Paque Plus (GE Healthcare, Waukesha, MI). Cells at the interface were removed and washed with phosphate-buffered saline. Cell pellets were viably frozen at −80°C until use. All cultures were carried out in defined medium as previously referenced.30 The study was conducted in accordance with the Declaration of Helsinki.
Synthetic siRNA oligonucleotides
The siGENOME SMARTpool for small interfering RNA (siRNA) was purchased from Thermo Scientific (Lafayette, CO). Scrambled control RNA (SCR) was used as a control. The target sequences for siRNA are shown in the supplemental Table 1 (available on the Blood Web site).
RNA isolation and qRT-PCR
Total cellular RNA was isolated using the RNAeasy Plus mini kit (Qiagen, Hilden, Germany). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) reactions were performed in triplicate with specific primers (specific primer sequences in supplemental Table 2). qRT-PCR was carried out on a 7900T Fast real-time PCR system (Applied Biosystems, Foster, CA). Glyceraldehyde-3-phosphate dehydrogenase RNA was used as the internal control, and relative gene expression levels were calculated as ΔΔCt. The results are presented as the fold increase over control.
ChIP assay
Chromatin immunoprecipitation (ChIP) was performed as described by the manufacturer (Pierce, Rockford, IL). Precleared chromatin was incubated overnight by rotation with 4 μg of Pol I antibody or IgG antibody as a negative control. Inmunoprecipitates were resuspended in 50 μL Tris-EDTA buffer. Inputs and immunoprecipitated DNA samples were quantified by quantitative polymerase chain reaction (qPCR) on a 7900T Fast real-time PCR system (Applied Biosystems). Primers are listed in supplemental Table 2.
RNA labeling and analysis
The cells were washed and incubated in phosphate-free Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum for 2 hours followed by 1-hour labeling with 0.5 mCi [32P] orthophosphate (Perkin Elmer). Total RNA was extracted with Trizol (Life Technology) following the manufacturer’s protocol. Equal amounts of RNA (10 μg) were separated on a 1.2% 4-morpholinepropanesulfonic acid formaldehyde gel. The gel was dried and visualized by autoradiography.
Statistical analysis
Where indicated, results were compared using the unpaired Student t test with values obtained from at least 3 independent experiments. P < .05 was considered significant.
Results
Expression of the TIF-90 splice variant in normal and leukemic cells
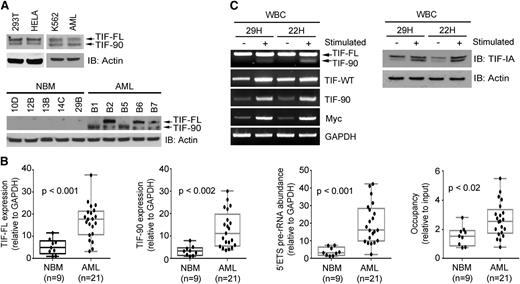
During an investigation of the expression of TIF-IA messenger RNA (mRNA) in cancer cell lines, we noted the presence of 2 distinct TIF-IA RNA transcripts (supplemental Figure 1A), suggesting the presence of a TIF-IA variant. This transcript lacks bp 253-342 of the TIF-IA mRNA, a region corresponding to exon 6 of the TIF-IA gene (supplemental Figure 1B). Primers flanking the entire open reading frame and the region corresponding to the splice variant (supplemental Figure 1B and supplemental Table 2) confirmed the expression of both variants in HeLa and 293T cells (supplemental Figure 2A). Loss of bp 253-342 results in an in-frame deletion of 30 amino acids from the mature protein. Western blot analysis using HeLa and 293T lysate demonstrates 2 bands, the smaller of which corresponds in size to the variant transcript (Figure 1A, top left). We conclude that the smaller protein represents a splice variant of TIF-IA that we term TIF-90.
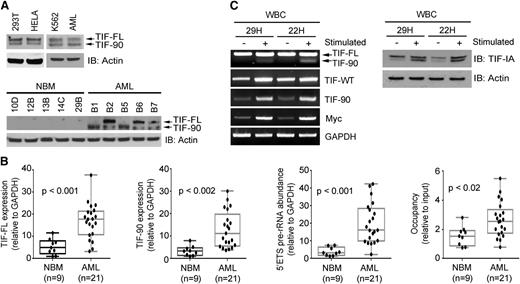
Expression of full-length TIF-IA protein (TIF-FL) and TIF-90 in normal, leukemic, and proliferating hematopoietic cells. (A) Expression of TIF-FL and the TIF-90 variant in cancer cell lines and primary leukemic cells (AML, n = 10) (top) and in representative individual normal bone marrow and leukemic cells (bottom) by western blot. (B) Quantitative expression of (1) TIF-FL mRNA, (2) TIF-90 mRNA, (3) pre-rRNA synthesis, and (4) Pol I occupancy of the rDNA promoter by qPCR in bone marrow mononuclear cells from normal individuals and from patients with AML. The bar indicates the average value, and the ends of the whiskers represent minimum and maximum values. Significance was determined using the Student t test. (C) Expression of TIF-FL and TIF-90 transcripts and protein in normal peripheral blood mononuclear cells from 2 individuals before and 22 hours after stimulation with phorbol ester and ionomycin: RT-PCR (left); western blot (right).
Expression of full-length TIF-IA protein (TIF-FL) and TIF-90 in normal, leukemic, and proliferating hematopoietic cells. (A) Expression of TIF-FL and the TIF-90 variant in cancer cell lines and primary leukemic cells (AML, n = 10) (top) and in representative individual normal bone marrow and leukemic cells (bottom) by western blot. (B) Quantitative expression of (1) TIF-FL mRNA, (2) TIF-90 mRNA, (3) pre-rRNA synthesis, and (4) Pol I occupancy of the rDNA promoter by qPCR in bone marrow mononuclear cells from normal individuals and from patients with AML. The bar indicates the average value, and the ends of the whiskers represent minimum and maximum values. Significance was determined using the Student t test. (C) Expression of TIF-FL and TIF-90 transcripts and protein in normal peripheral blood mononuclear cells from 2 individuals before and 22 hours after stimulation with phorbol ester and ionomycin: RT-PCR (left); western blot (right).
TIF-90 protein is expressed in the K562 leukemic cell line and in primary AML cells (Figure 1A, top right). To determine whether the expression of TIF-90 is a function of proliferation and/or transformation, we compared the expression of TIF-IA and TIF-90 in sorted CD34+ cells obtained from normal bone marrow with that in leukemic mononuclear cells obtained from patients with AML or a myeloproliferative disease using RT-PCR and qPCR. Although both isoforms were present in normal hematopoietic precursors, the expression of both TIF-FL and TIF-90 mRNA was significantly higher in the leukemic cells (supplemental Figure 2B-D and Figure 1B, left 2 panels). Western blot analysis of the corresponding cell lysates demonstrated that TIF-FL and TIF-90 protein expression levels are also elevated (Figure 1A, bottom). We then compared the relative levels of TIF-90 expression before and after stimulation of normal peripheral blood mononuclear cells with phorbol ester and ionomycin and found a pronounced increase in the expression of both TIF-90 and TIF-FL as a result of stimulation, as shown in Figure 1C. Because TIF-IA functions to recruit Pol I to the preinitiation complex at the rDNA promoter,31-33 we also determined the extent of Pol I recruitment to rDNA and the levels of pre-rRNA synthesis in normal and leukemic cells. Leukemic cells demonstrated significantly higher levels of Pol I binding to rDNA binding and of 5′external transcribed spacer (ETS) pre-rRNA synthesis compared with normal cells (Figure 1B, right 2 panels; supplemental Figure 2E-F). We therefore hypothesized that the TIF-90 splice variant might play an important role in the regulation of rRNA synthesis in AML cells.
TIF-90 regulates rRNA transcription
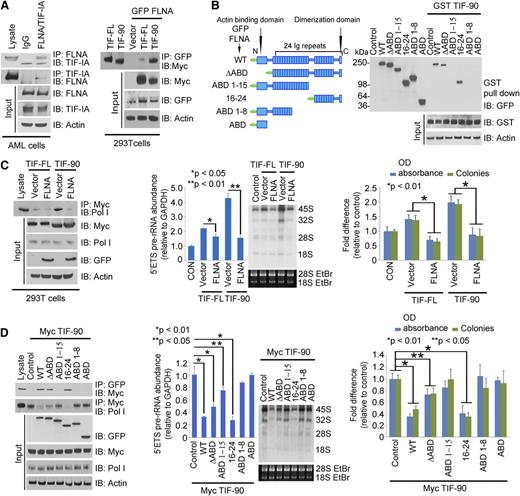
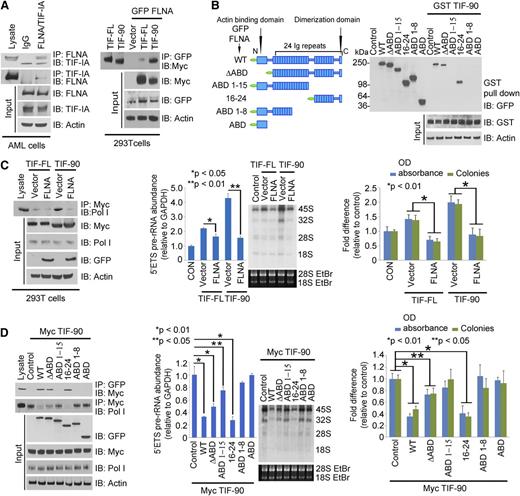
TIF-90 colocalizes with Pol I protein at the site of active rRNA synthesis (Figure 2A and supplemental Figure 3A). Line scans through 3-dimensional images show that the intensities of TIF-90 and Pol I immunostaining over the rDNA regions occur at similar locations throughout the nucleus, whereas TIF-FL does not consistently coincide with the location of Pol I (Figure 2A, right). TIF-90 also colocalizes with upstream binding factor (UBF) protein (supplemental Figure 3B). These data strongly suggest that TIF-90 preferentially binds to Pol I. A structural model of Saccharomycescerevisiae Rrn3 indicates that S145 and S185, corresponding to S138 and S199 of human TIF-IA, are essential for interaction with Pol I.34 These amino acids are conserved in TIF-90 (supplemental Figure 3C). An in vitro GST pull-down assay and coimmunoprecipitation experiments demonstrate that TIF-90 strongly interacts with Pol I, whereas TIF-FL does so to a lesser extent (Figure 2B).
TIF-90 regulates pre-rRNA synthesis. (A) Colocalization of TIF-FL and TIF-90 with Pol I and rDNA using immunostaining and fluorescence in situ hybridization assays. The 293T cells were transfected with Myc-TIF-FL or Myc-TIF-90 and costained with anti-Myc and anti-Pol I antibodies (left). rDNA was labeled with an rDNA probe as described in “Materials and methods” (right). Fluorescence intensity was measured along the line through three-dimensional pictures on the left. (B) Interaction of TIF-FL and TIF-90 with Pol I. Interaction of glutathione S-transferase (GST)-tagged TIF-FL and TIF-90 with Pol I in cell lysate (left). Coimmunoprecipitation of Pol I and Myc-TIF-FL or Myc-TIF-90 in transfected 293T cells (right). (C and D) Effects of TIF-FL and TIF-90 expression on rRNA synthesis in primary AML cells. Primary AML cell samples (n = 10) were combined and transfected with siSCR or siRNAs specific for TIF-FL or TIF-90 for 24 hours. (C) Levels of pre-rRNA expression relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample as determined by qPCR and RNA synthesis using [32P] labeling. (D) Effect of TIF depletion on rDNA promoter occupancy by Pol I. ChIP assays were performed as described in “Materials and methods” using anti-Pol I antibody. Values represent the mean ± standard deviation (SD) of triplicate determinations (n = 3) (left). The levels of expression of TIF-IA in corresponding samples in panel C are shown by western blot (right).
TIF-90 regulates pre-rRNA synthesis. (A) Colocalization of TIF-FL and TIF-90 with Pol I and rDNA using immunostaining and fluorescence in situ hybridization assays. The 293T cells were transfected with Myc-TIF-FL or Myc-TIF-90 and costained with anti-Myc and anti-Pol I antibodies (left). rDNA was labeled with an rDNA probe as described in “Materials and methods” (right). Fluorescence intensity was measured along the line through three-dimensional pictures on the left. (B) Interaction of TIF-FL and TIF-90 with Pol I. Interaction of glutathione S-transferase (GST)-tagged TIF-FL and TIF-90 with Pol I in cell lysate (left). Coimmunoprecipitation of Pol I and Myc-TIF-FL or Myc-TIF-90 in transfected 293T cells (right). (C and D) Effects of TIF-FL and TIF-90 expression on rRNA synthesis in primary AML cells. Primary AML cell samples (n = 10) were combined and transfected with siSCR or siRNAs specific for TIF-FL or TIF-90 for 24 hours. (C) Levels of pre-rRNA expression relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample as determined by qPCR and RNA synthesis using [32P] labeling. (D) Effect of TIF depletion on rDNA promoter occupancy by Pol I. ChIP assays were performed as described in “Materials and methods” using anti-Pol I antibody. Values represent the mean ± standard deviation (SD) of triplicate determinations (n = 3) (left). The levels of expression of TIF-IA in corresponding samples in panel C are shown by western blot (right).
To determine the relationship between localization of the TIF proteins and cell growth, cells were grown in medium with 10% fetal calf serum, serum starved for 12 hours, and then regrown in the presence of serum. As shown in supplemental Figure 4A, serum starvation results in the localization of total TIF-IA protein to the nucleus, whereas the subsequent addition of serum relocalizes TIF-IA to the nucleolus, as has been previously reported.27,35 These results suggest that the localization of TIF to the nucleolus is growth dependent.
We then examined the relative effects of TIF-FL and TIF-90 overexpression on rRNA synthesis in the presence or absence of endogenous TIF-IA expression. Given the high rate of rRNA turnover in cells, the levels of pre-rRNA transcript abundance are a valid approximation of the overall rate of rRNA transcription.16,36,37 We also used the incorporation of [32P] into newly synthesized RNA as an adjunctive assay. Analysis of pre-rRNA abundance using both 5′ETS qPCR and RNA labeling demonstrated that overexpression of TIF-90 enhances rRNA synthesis to a greater extent than does expression of TIF-FL (supplemental Figure 4B), as it also does in the context of siRNA-induced depletion of endogenous TIF-IA (supplemental Figure 4B). ChIP analysis revealed that overexpression of TIF-90 also enhances Pol I binding to rDNA (supplemental Figure 4C), consistent with the increase of pre-rRNA synthesis. To further determine the relative contributions of TIF-FL and TIF-90 to pre-rRNA synthesis in primary AML cells, TIF-FL and TIF-90 were selectively depleted. As shown in Figure 2C-D, a greater reduction of pre-rRNA synthesis and of Pol I binding to rDNA results from the selective knockdown of TIF-90 as opposed to a similar knockdown of TIF-FL. Similarly, depletion of TIF-90 but not of TIF-FL strongly reduces pre-rRNA synthesis and Pol I binding to rDNA in both 293T cells (supplemental Figure 4D-E) and K562 cells (supplemental Figure 4F-G).
FLNA interacts with TIF-90 to inhibit rRNA synthesis
The actin binding protein FLNA is a negative regulator of rRNA synthesis.28 We confirmed that depletion of FLNA enhances rRNA synthesis and cell proliferation (supplemental Figure 5A-B), suggesting that FLNA negatively regulates cell proliferation through inhibition of rRNA synthesis. The induction of the 90-kDa FLNA cleavage product reduced Pol I binding to rDNA and decreased pre-rRNA synthesis, as has been shown by Deng et al.28 Activated Akt, on the other hand, prevents FLNA cleavage by phosphorylating it at Ser 2152, thereby retaining it in the cytoplasm.29 Supplemental Figure 5C showed that overexpression of FLNA inhibits TIF-IA recruitment to rDNA, raising the possibility that FLNA might be an important intermediate.
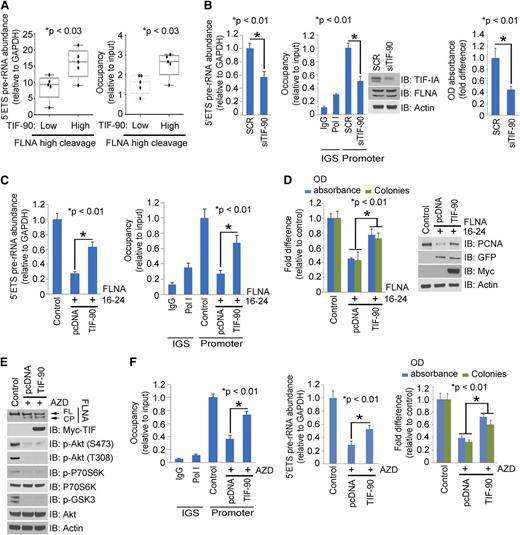
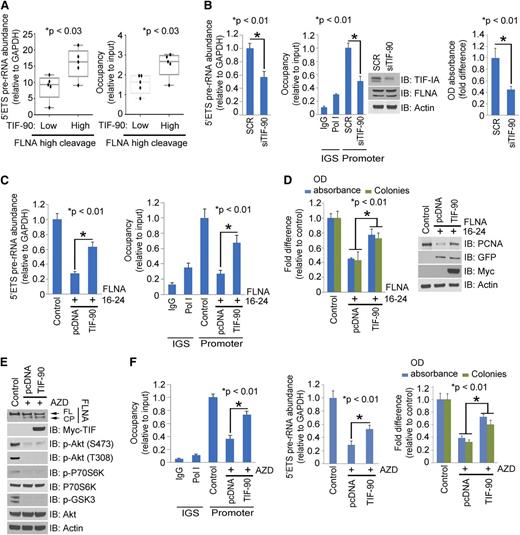
To investigate this hypothesis, we first examined the correlation between TIF-90 and FLNA cleavage with pre-rRNA synthesis and cell survival in primary AML cells. Indeed, the presence of the FLNA cleavage product correlates inversely with Pol I binding to rDNA, pre-rRNA synthesis level, and cell survival in 21 primary AML cells (Figure 3A-C). Moreover, the endogenous interaction of TIF-IA and Pol I is reduced in samples with FLNA cleavage (Figure 3D), whereas knockdown of FLNA in the high FLNA expression group increased this interaction (Figure 3D). The reduced interaction between TIF-IA and Pol I in AML cells with high levels of FLNA cleavage correlates with low levels of rRNA synthesis and shortened cell survival (Figure 3E). Increasing TIF-IA–Pol I binding by knocking down FLNA in cells with high FLNA cleavage restored rRNA synthesis and cell survival (Figure 3E). These data suggest that the expression of FLNA cleavage product inhibits TIF-90 functions on pre-rRNA synthesis in AML cells.
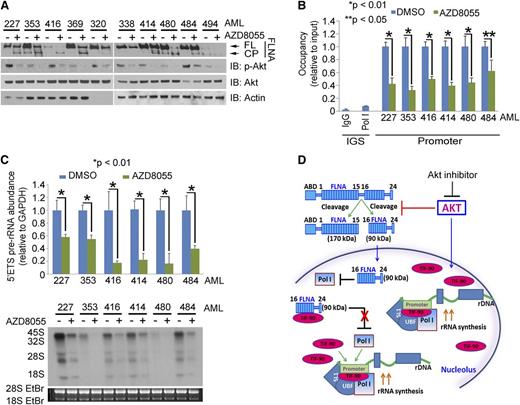
Relationship of FLNA cleavage and TIF-90 expression to pre-rRNA synthesis in AML cells. (A) Relative expression of phosphorylated Akt (p-Akt) and FLNA in primary AML cells. Cell lysate (30 μg) from 4 normal bone marrows and 21 AML samples was separated on sodium dodecyl sulfate gels and immunoblotted with the indicated antibodies (FL, full length; CP, cleavage product). (B and C) Correlation of FLNA cleavage and TIF-90 expression with rRNA synthesis and cell survival. Twenty-one AML patient samples were divided into 4 groups based on the expression of TIF-90 and presence of the FLNA cleavage product. Measurement of TIF-90 was performed by qPCR (supplemental Figure 2D), and measurements of FLNA cleavage were carried out using densitometry on western blots (Figure 3A). Samples are divided into low and high expression relative to the average value for all samples. The value of 5′ETS pre-rRNA in supplemental Figure 2E and Pol I recruited to rDNA promoter in supplemental Figure 2F for each sample was used for graphic representation. (B) Level of pre-rRNA synthesis and RNA labeling with [32P] was performed with the mixture of AML cells from each group; level of Pol I recruited to rDNA (C, left). The bar indicates the average value and the ends of the whiskers represent minimum and maximum values. Significance was determined using the Student t test; 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay (C, right). (D and E) Effects of FLNA cleavage on TIF-IA–Pol I binding, pre-rRNA synthesis, and cell survival in primary AML cells. Fourteen samples from AML patients were divided into low and high FNLA cleavage expression groups (each group, n = 7), and the level of FLNA cleavage was measured based on the densitometry results from panel A. (D) TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody (left); effects of knockdown of FLNA on TIF-IA and Pol I binding in primary AML cells (right). A mixture of AML cells (n = 7) that have FLNA cleavage levels above the average value was transfected with siSCR or siFLNA for 36 hours. TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody. (E) The 5′ETS pre-rRNA was measured by qPCR (left), ChIP assay (middle), and MTT assay (right).
Relationship of FLNA cleavage and TIF-90 expression to pre-rRNA synthesis in AML cells. (A) Relative expression of phosphorylated Akt (p-Akt) and FLNA in primary AML cells. Cell lysate (30 μg) from 4 normal bone marrows and 21 AML samples was separated on sodium dodecyl sulfate gels and immunoblotted with the indicated antibodies (FL, full length; CP, cleavage product). (B and C) Correlation of FLNA cleavage and TIF-90 expression with rRNA synthesis and cell survival. Twenty-one AML patient samples were divided into 4 groups based on the expression of TIF-90 and presence of the FLNA cleavage product. Measurement of TIF-90 was performed by qPCR (supplemental Figure 2D), and measurements of FLNA cleavage were carried out using densitometry on western blots (Figure 3A). Samples are divided into low and high expression relative to the average value for all samples. The value of 5′ETS pre-rRNA in supplemental Figure 2E and Pol I recruited to rDNA promoter in supplemental Figure 2F for each sample was used for graphic representation. (B) Level of pre-rRNA synthesis and RNA labeling with [32P] was performed with the mixture of AML cells from each group; level of Pol I recruited to rDNA (C, left). The bar indicates the average value and the ends of the whiskers represent minimum and maximum values. Significance was determined using the Student t test; 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay (C, right). (D and E) Effects of FLNA cleavage on TIF-IA–Pol I binding, pre-rRNA synthesis, and cell survival in primary AML cells. Fourteen samples from AML patients were divided into low and high FNLA cleavage expression groups (each group, n = 7), and the level of FLNA cleavage was measured based on the densitometry results from panel A. (D) TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody (left); effects of knockdown of FLNA on TIF-IA and Pol I binding in primary AML cells (right). A mixture of AML cells (n = 7) that have FLNA cleavage levels above the average value was transfected with siSCR or siFLNA for 36 hours. TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody. (E) The 5′ETS pre-rRNA was measured by qPCR (left), ChIP assay (middle), and MTT assay (right).
TIF-IA binds to FLNA in primary AML cells (Figure 4A, left), and FLNA interacts more strongly with TIF-90 than with TIF-FL (Figure 4A, right). To determine whether this interaction is direct or occurs through a Pol I complex intermediate, we used the Pol I–depleted lysate of FLNA-transfected cells to carry out a binding assay with GST-tagged TIF-90 protein. Supplemental Figure 5D shows that in the absence of Pol I, GST-TIF-90 still strongly interacts with FLNA, suggesting a direct interaction. We then mapped the binding domain of FLNA using sequentially deleted constructs. Only FLNA-wild type (WT) and the C-terminal (16-24) domain corresponding to the 90-kDa fragment interacted with TIF-90 (Figure 4B). Deletion of the N-terminal actin binding domain had no effect on the interaction, and the actin binding domain alone did not bind to TIF-90 (Figure 4B). The greater the binding of TIF-90 to FLNA, the less TIF-90 interacts with Pol I, whereas decreased FLNA expression increases the interaction of TIF-90 with Pol I (supplemental Figure 5E). Overexpression of FLNA strongly inhibits the TIF-90–Pol I interaction and TIF-90–mediated binding of Pol I to rDNA, as well as cell proliferation (Figure 4C; supplemental Figure 5F). In addition, with depletion of FLNA, a reduction in TIF-90 abolishes Pol I binding to rDNA and also reduces pre-rRNA synthesis and cell proliferation (supplemental Figure 5G-H). Thus, FLNA negatively regulates rRNA synthesis predominantly through TIF-90. Furthermore, only the C-terminal fragment inhibits the interaction of TIF-90 with Pol I and TIF-90–mediated enhancement of rRNA synthesis and cell proliferation (Figure 4D; supplemental Figure 6A). Both FLNA-WT and the C-terminal (16-24) fragment also inhibit basal levels of pre-rRNA synthesis and cell proliferation (supplemental Figure 6B). These data strongly suggest that the C-terminal 90-kDa fragment of FLNA regulates rRNA synthesis through its interaction with TIF-90. In summary, the interaction between the C-terminal FLNA fragment and TIF-90 prevents the recruitment of Pol I to rDNA through TIF-90 and inhibits both rRNA synthesis and cell proliferation (supplemental Figure 6C).
FLNA interacts with TIF-90 and prevents TIF-90 enhancement of rRNA synthesis. (A) Interaction of FLNA and TIF-IA in primary AML cells (left). AML lysate (500 μg) combined from 10 samples was used for immunoprecipitation experiments. Cell lysate was immunoprecipitated with anti-IgG or anti-FLNA antibody and immunoblotted with anti-TIF-IA antibody (left). Interaction of TIF-FL and TIF-90 with FLNA (right). The 293T cells were cotransfected with green fluorescent protein (GFP)-FLNA or vector control, Myc-TIF-FL, or Myc-TIF-90. FLNA protein was immunoprecipitated with anti-GFP antibody and immunoblotted with anti-Myc antibody. (B) Mapping the interaction domain of FLNA for TIF-90. Schematic structure of FLNA full length and fragments (left); interaction of GST-tagged TIF-90 with FLNA fragments in cell lysate (right). (C) Effects of FLNA expression on TIF-FL and TIF-90 enhancement of rRNA synthesis. The 293T cells were cotransfected with Myc-tagged TIF-FL or TIF-90 and vector control or GFP-FLNA. Myc-TIF protein was immunoprecipitated with anti-Myc antibody and immunoblotted with anti-Pol I antibody (left); 5′ETS pre-rRNA and RNA labeling (middle); MTT and colony-forming assay (right). (D) Effects of FLNA full length and fragments on TIF-90 enhancement of rRNA synthesis. The 293T cells were cotransfected with Myc-TIF-90 and the indicated FLNA constructs. FLNA protein was immunoprecipitated with anti-GFP antibody and immunoblotted with anti-Myc antibody (left, top row); TIF-90 protein was immunoprecipitated with anti-Myc antibody and blotted for Pol I binding (left, second row); 5′ETS pre-rRNA and RNA labeling (middle); MTT and colony-forming assays (right).
FLNA interacts with TIF-90 and prevents TIF-90 enhancement of rRNA synthesis. (A) Interaction of FLNA and TIF-IA in primary AML cells (left). AML lysate (500 μg) combined from 10 samples was used for immunoprecipitation experiments. Cell lysate was immunoprecipitated with anti-IgG or anti-FLNA antibody and immunoblotted with anti-TIF-IA antibody (left). Interaction of TIF-FL and TIF-90 with FLNA (right). The 293T cells were cotransfected with green fluorescent protein (GFP)-FLNA or vector control, Myc-TIF-FL, or Myc-TIF-90. FLNA protein was immunoprecipitated with anti-GFP antibody and immunoblotted with anti-Myc antibody. (B) Mapping the interaction domain of FLNA for TIF-90. Schematic structure of FLNA full length and fragments (left); interaction of GST-tagged TIF-90 with FLNA fragments in cell lysate (right). (C) Effects of FLNA expression on TIF-FL and TIF-90 enhancement of rRNA synthesis. The 293T cells were cotransfected with Myc-tagged TIF-FL or TIF-90 and vector control or GFP-FLNA. Myc-TIF protein was immunoprecipitated with anti-Myc antibody and immunoblotted with anti-Pol I antibody (left); 5′ETS pre-rRNA and RNA labeling (middle); MTT and colony-forming assay (right). (D) Effects of FLNA full length and fragments on TIF-90 enhancement of rRNA synthesis. The 293T cells were cotransfected with Myc-TIF-90 and the indicated FLNA constructs. FLNA protein was immunoprecipitated with anti-GFP antibody and immunoblotted with anti-Myc antibody (left, top row); TIF-90 protein was immunoprecipitated with anti-Myc antibody and blotted for Pol I binding (left, second row); 5′ETS pre-rRNA and RNA labeling (middle); MTT and colony-forming assays (right).
Akt enhances rRNA synthesis through TIF-90
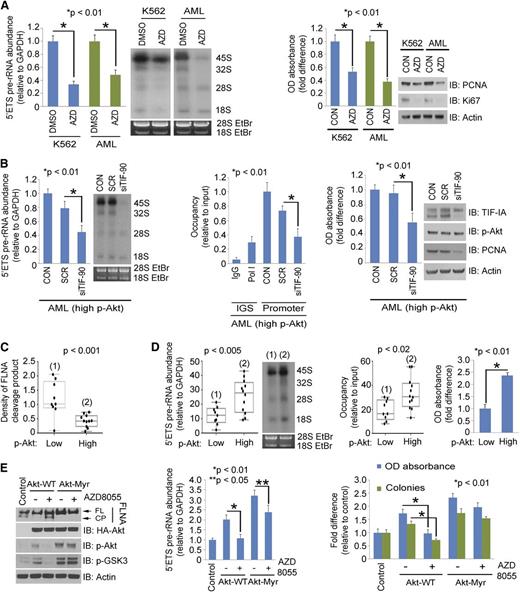
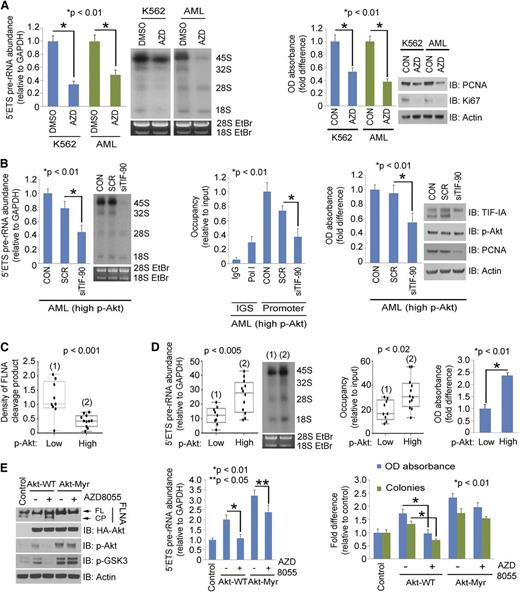
We have recently found that activated Akt directly regulates rRNA synthesis through TIF-IA.27 We therefore asked the role of activated Akt in regulation of TIF-90 functions. Overexpression of myristoylated Akt (Akt-Myr) significantly enhanced rRNA synthesis with both TIF proteins, although to a greater extent in TIF-90 cotransfected cells (supplemental Figure 7A). Akt-Myr also strongly increased both TIF-90 binding to Pol I and the recruitment of Pol I to rDNA (supplemental Figure 7B). Although overexpression of TIF-90 alone does not strongly increase cell proliferation, proliferation is markedly enhanced by the coexpression of Akt-Myr and TIF-90 (supplemental Figure 7C) and to a lesser extent by the coexpression of Akt-Myr and TIF-FL (supplemental Figure 7C). Depletion of endogenous Akt or inhibition of Akt activity disrupted the enhancement of rRNA synthesis by TIF-90 (supplemental Figure 7D). These results suggest that the coexpression of TIF-90 and p-Akt are important components of the regulation of rRNA synthesis and cell proliferation. Indeed, treatment of both K562 cells and AML cells with the Akt inhibitor AZD8055 decreased pre-rRNA synthesis and cell proliferation (Figure 5A; supplemental Figure 7E). Depletion of TIF-90 in Akt-transfected 293T cells strongly reduced pre-rRNA synthesis and cell proliferation, whereas depletion of TIF-FL had less effect (supplemental Figure 7F-G). Similar results were obtained in primary AML cells expressing high levels of p-Akt (Figure 5B).
Akt regulates rRNA synthesis through TIF-90 and FLNA cleavage. (A) Effects of Akt inhibition on rRNA synthesis and cell proliferation in AML cells. K562 and a mixture of primary AML cells (n = 10) were treated with AZD8055 for 3 hours (left). The 5′ETS pre-rRNA and RNA labeling were performed as shown. Values for qPCR represent the mean ± SD of triplicate determinations (n = 3). Western blot demonstrating Akt inhibition is shown in supplemental Figure 7E. K562 and a mixture of primary AML cells (n = 10) were treated with AZD8055, and cell survival and proliferation were determined by MTT assay and western blot using the proliferation markers proliferating cell nuclear antigen (PCNA) and antigen Ki-67 (Ki67) (right). (B) Effects of TIF-90 depletion on rRNA synthesis, promoter occupancy, and cell proliferation in primary AML cells. A mixture of AML cells (n = 7) with p-Akt expression levels above the mean value (as determined by densitometry on western blot; Figure 3A) was transfected with siSCR or siTIF-90 for 24 hours. The 5′ETS pre-rRNA (left), ChIP assay (middle), and MTT assay and western blot (right). (C and D) Correlation of p-Akt with cleavage of FLNA, pre-rRNA synthesis, and cell survival in primary AML cells. Twenty-one samples from AML patients were divided into low and high p-Akt expression groups, and the level of FLNA cleavage was measured based on the densitometry results from Figure 3A. (C) Correlation of p-Akt with FLNA cleavage. (D) Correlation of p-Akt with pre-rRNA synthesis (left); correlation of p-Akt and Pol I binding to rDNA (middle); MTT assay with a mixture of AML cells (low p-Akt, n = 9; high p-Akt, n = 12) (right). (E) Effects of AZD8055 on FLNA cleavage in the presence of Akt or Akt-Myr. K562 cells were transfected with the indicated Akt constructs or treated with AZD8055 or no drug. Western blots were performed as indicated (left); 5′ETS pre-rRNA was measured by qPCR (middle) and MTT and colony-forming assay (right).
Akt regulates rRNA synthesis through TIF-90 and FLNA cleavage. (A) Effects of Akt inhibition on rRNA synthesis and cell proliferation in AML cells. K562 and a mixture of primary AML cells (n = 10) were treated with AZD8055 for 3 hours (left). The 5′ETS pre-rRNA and RNA labeling were performed as shown. Values for qPCR represent the mean ± SD of triplicate determinations (n = 3). Western blot demonstrating Akt inhibition is shown in supplemental Figure 7E. K562 and a mixture of primary AML cells (n = 10) were treated with AZD8055, and cell survival and proliferation were determined by MTT assay and western blot using the proliferation markers proliferating cell nuclear antigen (PCNA) and antigen Ki-67 (Ki67) (right). (B) Effects of TIF-90 depletion on rRNA synthesis, promoter occupancy, and cell proliferation in primary AML cells. A mixture of AML cells (n = 7) with p-Akt expression levels above the mean value (as determined by densitometry on western blot; Figure 3A) was transfected with siSCR or siTIF-90 for 24 hours. The 5′ETS pre-rRNA (left), ChIP assay (middle), and MTT assay and western blot (right). (C and D) Correlation of p-Akt with cleavage of FLNA, pre-rRNA synthesis, and cell survival in primary AML cells. Twenty-one samples from AML patients were divided into low and high p-Akt expression groups, and the level of FLNA cleavage was measured based on the densitometry results from Figure 3A. (C) Correlation of p-Akt with FLNA cleavage. (D) Correlation of p-Akt with pre-rRNA synthesis (left); correlation of p-Akt and Pol I binding to rDNA (middle); MTT assay with a mixture of AML cells (low p-Akt, n = 9; high p-Akt, n = 12) (right). (E) Effects of AZD8055 on FLNA cleavage in the presence of Akt or Akt-Myr. K562 cells were transfected with the indicated Akt constructs or treated with AZD8055 or no drug. Western blots were performed as indicated (left); 5′ETS pre-rRNA was measured by qPCR (middle) and MTT and colony-forming assay (right).
Akt activation enhances rRNA synthesis by preventing the cleavage of FLNA
Western blot and densitometry analysis indicated that the expression of p-Akt correlates inversely with the expression of the FLNA cleavage product in 21 AML samples (Figures 3A and 5C). We therefore examined the relationship between p-Akt expression, FLNA cleavage, and pre-rRNA synthesis. Inhibition of Akt using a PI3K/mTOR inhibitor (LY294002) but not of mTORC1 (rapamycin) or mitogen-activated protein kinase (PD98059) induced FLNA cleavage (supplemental Figure 8A, right). The induction of the FLNA cleavage product reduced Pol I binding to rDNA and decreased pre-rRNA synthesis, as has been shown by Deng et al28 (supplemental Figure 8A, left and middle). In primary AML cells, the presence of the cleavage fragment is indicative of decreased Pol I binding to rDNA and a decrease in both pre-rRNA synthesis and cell survival (Figure 5C-D). To confirm the effect of Akt activation on these interactions, we first treated K562 cells with AZD8055, an inhibitor of mTORC1 and 2. Figure 6E confirms that Akt is inhibited by this compound, and supplemental Figure 10A shows that the inhibition occurs after 1 hour of exposure and that rRNA synthesis continues to decrease over 24 hours. Supplemental Figure 8B demonstrates that AZD8055 induces FLNA cleavage in a dose-dependent manner. The decrease in pre-rRNA synthesis and Pol I promoter occupancy that occurs with AZD8055 treatment correlates with the induction of FLNA cleavage (supplemental Figure 8C). AZD8055 treatment also inhibits the proliferation of K562 cells as well as their ability to form colonies (supplemental Figure 8D-E), whereas overexpression of Akt-Myr both prevents FLNA cleavage and enhances pre-rRNA synthesis and cell proliferation (Figure 5E). AZD8055 also reverses the effect of Akt-WT in preventing FLNA cleavage but has less effect on Akt-Myr (Figure 5E). Treatment with 2 more specific Akt inhibitors (MK-2206 and Akt inhibitor viii) also induces FLNA cleavage (supplemental Figure 8F) and decreases pre-RNA synthesis and cell proliferation in K562 and AML cells (supplemental Figure 8G-H). These data support the conclusion that activation of Akt inhibits FLNA cleavage and thereby enhances pre-rRNA synthesis and cell proliferation.
Effects of TIF-90 expression on FLNA C-terminal (FLNA-C) (16-24) regulated rRNA synthesis. (A) Correlation of FLNA cleavage and TIF-90 expression with rRNA synthesis. AML samples with high levels of FLNA cleavage as determined by western blot were divided into low and high TIF-90 expression groups. The level of rRNA synthesis (left) and the recruitment of Pol I to the rDNA promoter (right) were determined for each set of samples. (B) Effects of TIF-90 depletion and FLNA cleavage on rRNA synthesis and cell survival in primary AML cells. AML samples with high levels of both FLNA cleavage and TIF-90 (n = 5) were transfected with siSCR or siTIF-90 (20 nM). The 5′ETS pre-rRNA (left); ChIP assay and western blot (middle); MTT assay (right). (C and D) Effects of TIF-90 overexpression on the FLNA-C (16-24)–mediated decrease in rRNA synthesis, Pol I binding to rDNA, and cell proliferation. K562 cells were transfected with vector control of FLNA-C (16-24) before transfection with vector control or Myc-TIF-90. (C) The 5′ETS pre-rRNA was measured by qPCR (left); ChIP assay with anti-Pol I antibody (right). Values for qPCR represent the mean ± SD of triplicate determinations. (D) MTT and colony-forming assay (left); western blot (right). (E and F) Effects of TIF-90 overexpression on AZD8055-mediated decrease in rRNA synthesis, Pol I binding to rDNA, and cell proliferation. K562 cells were treated with AZD8055 before transfection with vector control or Myc-TIF-90. (E) Western blot with indicated antibodies. (F) ChIP assay with anti-Pol I antibody (left); 5′ETS pre-rRNA was measured by qPCR (middle); MTT and colony-forming assay (right).
Effects of TIF-90 expression on FLNA C-terminal (FLNA-C) (16-24) regulated rRNA synthesis. (A) Correlation of FLNA cleavage and TIF-90 expression with rRNA synthesis. AML samples with high levels of FLNA cleavage as determined by western blot were divided into low and high TIF-90 expression groups. The level of rRNA synthesis (left) and the recruitment of Pol I to the rDNA promoter (right) were determined for each set of samples. (B) Effects of TIF-90 depletion and FLNA cleavage on rRNA synthesis and cell survival in primary AML cells. AML samples with high levels of both FLNA cleavage and TIF-90 (n = 5) were transfected with siSCR or siTIF-90 (20 nM). The 5′ETS pre-rRNA (left); ChIP assay and western blot (middle); MTT assay (right). (C and D) Effects of TIF-90 overexpression on the FLNA-C (16-24)–mediated decrease in rRNA synthesis, Pol I binding to rDNA, and cell proliferation. K562 cells were transfected with vector control of FLNA-C (16-24) before transfection with vector control or Myc-TIF-90. (C) The 5′ETS pre-rRNA was measured by qPCR (left); ChIP assay with anti-Pol I antibody (right). Values for qPCR represent the mean ± SD of triplicate determinations. (D) MTT and colony-forming assay (left); western blot (right). (E and F) Effects of TIF-90 overexpression on AZD8055-mediated decrease in rRNA synthesis, Pol I binding to rDNA, and cell proliferation. K562 cells were treated with AZD8055 before transfection with vector control or Myc-TIF-90. (E) Western blot with indicated antibodies. (F) ChIP assay with anti-Pol I antibody (left); 5′ETS pre-rRNA was measured by qPCR (middle); MTT and colony-forming assay (right).
Activated Akt promotes rRNA synthesis by preventing the interaction of TIF-90 with the FLNA cleavage product
Overexpression of constitutively activated Akt-Myr prevents the inhibition of the TIF-90–Pol I interaction by the FLNA 16-24 fragment and increases Pol I recruitment to rDNA, rRNA synthesis, and proliferation (supplemental Figure 9A). Co-overexpression of Akt and TIF-90 in the presence of FLNA 1-15 enhances pre-rRNA synthesis and cell proliferation, confirming that only the C-terminal fragment is required for the inhibitory effect (supplemental Figure 9A). Similar effects of Akt on FLNA proteins and endogenous TIF-IA were observed in K562 cells (supplemental Figure 9B-C). Overexpression of both Akt-WT and Akt-Myr enhances TIF-90 binding to Pol I, Pol I–rDNA recruitment, rRNA synthesis, and proliferation, while preventing FLNA cleavage (supplemental Figure 9D). Although treatment with AZD8055 disrupted the downstream effects of Akt-WT, it had only minimal effects on Akt-Myr (supplemental Figure 9D).
Higher levels of TIF-90 expression are associated with increased pre-rRNA synthesis in primary AML cells (Figure 6A), whereas decreased TIF-90 expression is associated with a decrease in pre-rRNA synthesis and cell survival (Figure 6B). We therefore asked whether enhanced expression of TIF-90 reverses the FLNA-mediated repression of rRNA synthesis. Figure 6C-D shows that overexpression of TIF-90 partially increases Pol I recruitment to rDNA, rRNA synthesis, and cell proliferation, each of which is inhibited by FLNA 16-24. Although overexpression of TIF-90 does not affect the induction of FLNA cleavage by Akt inhibitors, TIF-90 expression does promote Pol I–rDNA binding and increase rRNA synthesis and proliferation in the presence of Akt inhibition (Figure 6E-F; supplemental Figure 9E). These data indicate that higher levels of TIF-90 expression enhance rRNA synthesis by reversing the inhibitory effects of FLNA 16-24.
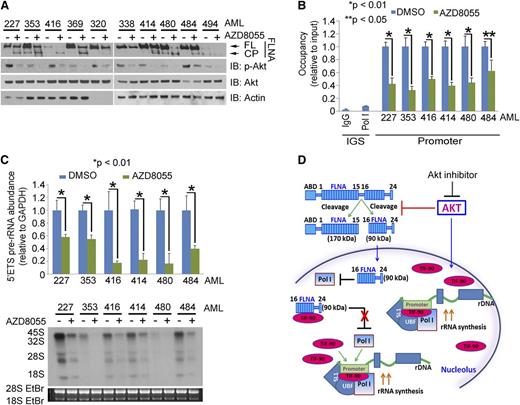
Finally, we examined the effects of AZD8055 treatment on 10 AML primary samples. AZD8055 decreased the phosphorylation of Akt and increased FLNA cleavage (Figure 7A), while inhibiting pre-rRNA synthesis and cell survival (Figure 7B-C; supplemental Figure 10B). Similar results were observed with MK-2206 and Akti-viii inhibitors (supplemental Figure 10C-D). In summary, these results support the conclusion that the p-Akt/FLNA/TIF-90 signaling pathway is important in maintaining the viability and growth potential of AML cells.
Akt inhibition induces FLNA cleavage and a decrease in rRNA synthesis and cell proliferation in AML cells. (A) Effects of AZD8055 on p-Akt expression and FLNA cleavage in primary AML cells. Ten AML patient samples were treated with AZD8055 (20 nM) or vehicle for 3 hours, and 30 μg lysate of protein was used for western blot and immunoblotted with the indicated antibodies. (B and C) Effects of AZD8055 on Pol I binding to rDNA, pre-rRNA synthesis, and cell survival in AML primary cells. Six AML samples were treated as in panel A. (B) ChIP assay was performed with anti-Pol I antibody. (C) The 5′ETS pre-rRNA was measured by qPCR (top); RNA was labeled with 32P (bottom). Values for qPCR represent the mean ± SD of triplicate determinations. (D) Schematic model of the regulation of rRNA synthesis by activated Akt as mediated through both the inhibition of FLNA cleavage and TIF-90 activity.
Akt inhibition induces FLNA cleavage and a decrease in rRNA synthesis and cell proliferation in AML cells. (A) Effects of AZD8055 on p-Akt expression and FLNA cleavage in primary AML cells. Ten AML patient samples were treated with AZD8055 (20 nM) or vehicle for 3 hours, and 30 μg lysate of protein was used for western blot and immunoblotted with the indicated antibodies. (B and C) Effects of AZD8055 on Pol I binding to rDNA, pre-rRNA synthesis, and cell survival in AML primary cells. Six AML samples were treated as in panel A. (B) ChIP assay was performed with anti-Pol I antibody. (C) The 5′ETS pre-rRNA was measured by qPCR (top); RNA was labeled with 32P (bottom). Values for qPCR represent the mean ± SD of triplicate determinations. (D) Schematic model of the regulation of rRNA synthesis by activated Akt as mediated through both the inhibition of FLNA cleavage and TIF-90 activity.
Discussion
We have found that TIF-90, a splice variant of TIF-IA, is coexpressed with TIF-IA and is increased in expression at both the mRNA and protein levels in proliferating cells and in the cells of patients with acute leukemia. Exogenously expressed TIF-90 localizes within both the nucleus and the nucleolus, whereas expression of TIF-FL is predominantly nuclear. In contrast, when actively growing cells are stained for endogenous TIF with an antibody that recognizes both forms, TIF is located predominantly in the nucleolus, shifting to the nucleus in quiescent cells that have been deprived of serum. To further validate the role of TIF-90, we demonstrated that exogenous expression leads to a greater increase in overall levels of 5′ETS pre-rRNA than does expression of TIF-FL in both the presence and absence of endogenous TIF expression. Moreover, specific depletion of endogenous TIF-90 in primary AML cells results in a significant decrease in pre-rRNA synthesis and proliferation. From these results, it is reasonable to conclude that TIF-90 plays a major role in the regulation of rRNA synthesis in AML cells.
In order to define more specifically the upstream signaling pathways that affect the activity of TIF-90, we examined the role of Akt activation. Akt has been shown to regulate ribosome biogenesis at multiple levels and to interact with both mTORC1 and c-Myc to stimulate 5′ETS pre-rRNA transcription.17 Although a number of studies had attested to the role of the mTOR pathway as the major link between nutrient availability, cell growth, and rRNA synthesis,16,20,38,39 recent work by ourselves and others has revealed that inhibition of mTORC1 by rapamycin does not ablate rRNA synthesis, whereas inhibition of Akt results in a much more pronounced decrease in 5′ETS pre-RNA levels.17 Our data support a central role for activated Akt in markedly enhancing the effects of TIF-90 on pre-rRNA synthesis. This evidence is based on the following observations: coexpression of p-Akt and TIF-90 has a greater effect on rRNA synthesis than does the coexpression of p-Akt and TIF-FL; the level of p-Akt correlates with TIF-90 expression and the level of pre-rRNA synthesis in primary AML cells; and selective TIF-90 depletion inhibits rRNA synthesis in AML cells expressing high levels of p-Akt.
FLNA plays a major intermediary role in this pathway. It interacts with more than 45 functionally diverse proteins including nuclear proteins and serves as a scaffold in a number of signaling networks.40,41 Studies had previously shown that Akt activation induces FLNA cleavage to generate a 90-kDa C-terminal product.29 These data suggested to us that TIF-90 might interact directly with FLNA. We therefore determined that FLNA interacts with TIF-90 through its C-terminal domain and that the expression of TIF-90 reverses the inhibition of rRNA synthesis that results from FLNA cleavage, while depletion of endogenous FLNA enhances the TIF-90–Pol I interaction. These results support 2 additional pathways involving TIF-90 that regulate rRNA synthesis: one in which FLNA-C suppresses rRNA synthesis when TIF-90 expression is limiting and a second pathway through which activated Akt increases rRNA synthesis by both increasing TIF-90 levels and inhibiting the cleavage of FLNA to FLNA-C (Figure 7D). This conclusion is borne out in both in vitro experiments in cell lines and in vivo correlative observations on primary AML cells (Figure 6A). TIF-90, and not TIF-FL, is thus the key intermediate in this pathway.
AML is associated with poor long-term survival, and new therapeutic approaches are urgently needed. The activation of the PI3K/Akt pathway is relatively common in AML leukemic cells and is associated with a poorer outcome.5,6 Recent research has shown that the compound AZD8055, an inhibitor of both Akt and mTORC1, may have more efficacy in inhibiting the growth of leukemic cells in vitro and in vivo than the mTORC1-specific inhibitor rapamycin and its analogs.42-45 AZD8055 decreases cell proliferation and cell cycle progression and induces autophagy, resulting in a significant decrease of AML cell survival without affecting the fate of normal CD34+ hematopoietic progenitors. Treatment with AZD8055 also markedly represses the growth of AML xenografts in mouse models at well-tolerated doses.43 We now show that AZD8055 inhibits the effect of activated Akt by inducing the cleavage of FLNA, thereby repressing rRNA synthesis in AML cells. Consistent with its inhibition of activated Akt, AZD8055 also significantly decreases AML cell proliferation and colony formation. These results, together with our previous findings, provide a comprehensive overview of the mechanisms by which activated Akt directly enhances rRNA synthesis (Figure 7D). These mechanisms provide support for the direct targeting of Akt as a therapeutic strategy in AML.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Fumihiko Nakamura for FLNA constructs.
This work was supported by a translational research grant and by a SCOR award from the Leukemia and Lymphoma Society.
Authorship
Contribution: L.X.T.N. and B.S.M. designed the research; L.X.T.N., S.M.C., T.D.N., and A.R. performed the research; L.X.T.N., K.K.K., R.M., and B.S.M. analyzed the data; and L.X.T.N. and B.S.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Beverly S. Mitchell, Lorry Lokey Building, 265 Campus Dr, Suite G2167, Stanford, CA 94305-5456; e-mail: bmitchell@stanford.edu.

![Figure 2. TIF-90 regulates pre-rRNA synthesis. (A) Colocalization of TIF-FL and TIF-90 with Pol I and rDNA using immunostaining and fluorescence in situ hybridization assays. The 293T cells were transfected with Myc-TIF-FL or Myc-TIF-90 and costained with anti-Myc and anti-Pol I antibodies (left). rDNA was labeled with an rDNA probe as described in “Materials and methods” (right). Fluorescence intensity was measured along the line through three-dimensional pictures on the left. (B) Interaction of TIF-FL and TIF-90 with Pol I. Interaction of glutathione S-transferase (GST)-tagged TIF-FL and TIF-90 with Pol I in cell lysate (left). Coimmunoprecipitation of Pol I and Myc-TIF-FL or Myc-TIF-90 in transfected 293T cells (right). (C and D) Effects of TIF-FL and TIF-90 expression on rRNA synthesis in primary AML cells. Primary AML cell samples (n = 10) were combined and transfected with siSCR or siRNAs specific for TIF-FL or TIF-90 for 24 hours. (C) Levels of pre-rRNA expression relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample as determined by qPCR and RNA synthesis using [32P] labeling. (D) Effect of TIF depletion on rDNA promoter occupancy by Pol I. ChIP assays were performed as described in “Materials and methods” using anti-Pol I antibody. Values represent the mean ± standard deviation (SD) of triplicate determinations (n = 3) (left). The levels of expression of TIF-IA in corresponding samples in panel C are shown by western blot (right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2013-12-544726/4/m_579f2.jpeg?Expires=1770049370&Signature=fgVdzLTWiV1YPHau8R7ZEmOBv~gCe3MYZNdAAZN1CaDCqB1yBUlnFptdJbWb4thEKqwrK~FzRsnVAKaQ8LL1m6oF0B40sVDD-V6ViIXm5uYzBMVyY16YfFkHulte95TPxLpyIENsF9yT~j4Uf1rTUqhII5PzQou34zHKu8LeO7mBQjqb8TNm3oYbTLwtWfRrotONIUuuEG8D1ByUWSq23sKUJhGMha6GgUzP8R4DGwc-5Dh6CKI4HRNukyTq1nk4ex3OI6FdaIjyYz0-jod~sLMzJcbbgFn7AajTOfKhPKbsUlf4zsoObtLefP58oaSplXoU2~vEG7sCKDEPUh8T1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Relationship of FLNA cleavage and TIF-90 expression to pre-rRNA synthesis in AML cells. (A) Relative expression of phosphorylated Akt (p-Akt) and FLNA in primary AML cells. Cell lysate (30 μg) from 4 normal bone marrows and 21 AML samples was separated on sodium dodecyl sulfate gels and immunoblotted with the indicated antibodies (FL, full length; CP, cleavage product). (B and C) Correlation of FLNA cleavage and TIF-90 expression with rRNA synthesis and cell survival. Twenty-one AML patient samples were divided into 4 groups based on the expression of TIF-90 and presence of the FLNA cleavage product. Measurement of TIF-90 was performed by qPCR (supplemental Figure 2D), and measurements of FLNA cleavage were carried out using densitometry on western blots (Figure 3A). Samples are divided into low and high expression relative to the average value for all samples. The value of 5′ETS pre-rRNA in supplemental Figure 2E and Pol I recruited to rDNA promoter in supplemental Figure 2F for each sample was used for graphic representation. (B) Level of pre-rRNA synthesis and RNA labeling with [32P] was performed with the mixture of AML cells from each group; level of Pol I recruited to rDNA (C, left). The bar indicates the average value and the ends of the whiskers represent minimum and maximum values. Significance was determined using the Student t test; 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay (C, right). (D and E) Effects of FLNA cleavage on TIF-IA–Pol I binding, pre-rRNA synthesis, and cell survival in primary AML cells. Fourteen samples from AML patients were divided into low and high FNLA cleavage expression groups (each group, n = 7), and the level of FLNA cleavage was measured based on the densitometry results from panel A. (D) TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody (left); effects of knockdown of FLNA on TIF-IA and Pol I binding in primary AML cells (right). A mixture of AML cells (n = 7) that have FLNA cleavage levels above the average value was transfected with siSCR or siFLNA for 36 hours. TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody. (E) The 5′ETS pre-rRNA was measured by qPCR (left), ChIP assay (middle), and MTT assay (right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2013-12-544726/4/m_579f3.jpeg?Expires=1770049370&Signature=hEGoOUrFfEpcbMXijK3Cjofqu5a4bKVRXLqXYKlHTeBoTe6iLMzvhrLAKscnfQAlLphoJtI2sA0S4L47tNRUPtsetTo5nc-OaDNpOa~91-Im5XG~4dGHH~WfaTEP8xbUeqy5u2lERd6nJScOsOeFvbyaKq5h2GRsOcD8QSkKVNAawAivD9-l4qOlWHv7WqlQ5W9txs9Y~vp0noXw8G8G4vZT-0sgV0cdFs0yHJHAPRpJhfo9F6GJR-H~wjnbFWAUa0UIyegpA3aiIisYPGxAZCX9dQG9BsCMMY6DqH3DBJxqYwfNiEBzb8OqSLZdMYcYH7X3AWioiiPUoCs0APUetg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. TIF-90 regulates pre-rRNA synthesis. (A) Colocalization of TIF-FL and TIF-90 with Pol I and rDNA using immunostaining and fluorescence in situ hybridization assays. The 293T cells were transfected with Myc-TIF-FL or Myc-TIF-90 and costained with anti-Myc and anti-Pol I antibodies (left). rDNA was labeled with an rDNA probe as described in “Materials and methods” (right). Fluorescence intensity was measured along the line through three-dimensional pictures on the left. (B) Interaction of TIF-FL and TIF-90 with Pol I. Interaction of glutathione S-transferase (GST)-tagged TIF-FL and TIF-90 with Pol I in cell lysate (left). Coimmunoprecipitation of Pol I and Myc-TIF-FL or Myc-TIF-90 in transfected 293T cells (right). (C and D) Effects of TIF-FL and TIF-90 expression on rRNA synthesis in primary AML cells. Primary AML cell samples (n = 10) were combined and transfected with siSCR or siRNAs specific for TIF-FL or TIF-90 for 24 hours. (C) Levels of pre-rRNA expression relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample as determined by qPCR and RNA synthesis using [32P] labeling. (D) Effect of TIF depletion on rDNA promoter occupancy by Pol I. ChIP assays were performed as described in “Materials and methods” using anti-Pol I antibody. Values represent the mean ± standard deviation (SD) of triplicate determinations (n = 3) (left). The levels of expression of TIF-IA in corresponding samples in panel C are shown by western blot (right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2013-12-544726/4/m_579f2.jpeg?Expires=1770049371&Signature=zYonIxnq9rmqPqvsDHOHWO39CUcSj63areCBDxe1WXaFDzn7zuCgk2quZP209JkOlRI~OLfEmMHiGwVXmw9Z9ZK7kfGNK9YaXecdTGXQpOU9nP~rDrGnyW7xD~zJlqbJIR59eqIRFhK~FwRgeoJ6vIOlgDzvgE9938CWb0teQBojZbYe3NEFSpY6IQYsLVMYPiMCRaO~nY2O23gqVCrKJBBDwpoPMLn6kBHBPjwBuDn~vbesdJubVERE~DhYIOh2MfHMU-HNs7Llf7gl9ByOy1ztpYPUPoc7~B3EDsq58pwqocLGGVV56EA9uOkJAQ3n4q3tFqiZ13TH1cAOLBcemg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Relationship of FLNA cleavage and TIF-90 expression to pre-rRNA synthesis in AML cells. (A) Relative expression of phosphorylated Akt (p-Akt) and FLNA in primary AML cells. Cell lysate (30 μg) from 4 normal bone marrows and 21 AML samples was separated on sodium dodecyl sulfate gels and immunoblotted with the indicated antibodies (FL, full length; CP, cleavage product). (B and C) Correlation of FLNA cleavage and TIF-90 expression with rRNA synthesis and cell survival. Twenty-one AML patient samples were divided into 4 groups based on the expression of TIF-90 and presence of the FLNA cleavage product. Measurement of TIF-90 was performed by qPCR (supplemental Figure 2D), and measurements of FLNA cleavage were carried out using densitometry on western blots (Figure 3A). Samples are divided into low and high expression relative to the average value for all samples. The value of 5′ETS pre-rRNA in supplemental Figure 2E and Pol I recruited to rDNA promoter in supplemental Figure 2F for each sample was used for graphic representation. (B) Level of pre-rRNA synthesis and RNA labeling with [32P] was performed with the mixture of AML cells from each group; level of Pol I recruited to rDNA (C, left). The bar indicates the average value and the ends of the whiskers represent minimum and maximum values. Significance was determined using the Student t test; 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assay (C, right). (D and E) Effects of FLNA cleavage on TIF-IA–Pol I binding, pre-rRNA synthesis, and cell survival in primary AML cells. Fourteen samples from AML patients were divided into low and high FNLA cleavage expression groups (each group, n = 7), and the level of FLNA cleavage was measured based on the densitometry results from panel A. (D) TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody (left); effects of knockdown of FLNA on TIF-IA and Pol I binding in primary AML cells (right). A mixture of AML cells (n = 7) that have FLNA cleavage levels above the average value was transfected with siSCR or siFLNA for 36 hours. TIF-IA protein was immunoprecipitated and immunoblotted with anti-Pol I antibody. (E) The 5′ETS pre-rRNA was measured by qPCR (left), ChIP assay (middle), and MTT assay (right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/4/10.1182_blood-2013-12-544726/4/m_579f3.jpeg?Expires=1770049371&Signature=afNMP7Q78cCf4h~tuw8LCAFiQabL86~Kg1cyYNqyDBfYZsvP6-CUGQyXHZghZsssN1sZ9peqlD3uFypZwpJPdwdOJVd8IiXI6BhUPUHF0MDVlhga4~fAEN-9flkarfDjFBLmTrZ4~kHiI9c3nWSlGWs6gGKrjzVp7YG4fWnMzV0hkhJr1HAhkcMEq0oZBYSi5IF~tAsNBcN4Pta4S4nDaeEG33deVFt30GxONqbDlFBTeRi~PWIwkKsyGqIBOhPi3Pde5QaKCnQBiOb302OUg1pB2c1egIZ6UgvX0WhhbbumckmC966FumROKZB2T70fbmtjyHDAOoH8YOxkTD-Gng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)