Key Points
Microdeletions represent an additional inactivation mechanism for PTEN in human T-cell acute lymphoblastic leukemia.
PTEN microdeletions are RAG-mediated aberrations.
Abstract
Phosphatase and tensin homolog (PTEN)-inactivating mutations and/or deletions are an independent risk factor for relapse of T-cell acute lymphoblastic leukemia (T-ALL) patients treated on Dutch Childhood Oncology Group or German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia protocols. Some monoallelic mutated or PTEN wild-type patients lack PTEN protein, implying that additional PTEN inactivation mechanisms exist. We show that PTEN is inactivated by small deletions affecting a few exons in 8% of pediatric T-ALL patients. These microdeletions were clonal in 3% and subclonal in 5% of patients. Conserved deletion breakpoints are flanked by cryptic recombination signal sequences (cRSSs) and frequently have non-template-derived nucleotides inserted in between breakpoints, pointing to an illegitimate RAG recombination-driven activity. Identified cRSSs drive RAG-dependent recombination in a reporter system as efficiently as bona fide RSSs that flank gene segments of the T-cell receptor locus. Remarkably, equivalent microdeletions were detected in thymocytes of healthy individuals. Microdeletions strongly associate with the TALLMO subtype characterized by TAL1 or LMO2 rearrangements. Primary and secondary xenotransplantation of TAL1-rearranged leukemia allowed development of leukemic subclones with newly acquired PTEN microdeletions. Ongoing RAG activity may therefore actively contribute to the acquisition of preleukemic hits, clonal diversification, and disease progression.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) represents 10% to 15% of pediatric acute leukemias. Despite major therapeutic improvements due to treatment intensification and refined risk-adapted stratification during the past decade, ∼30% of T-ALL cases relapse with very poor prognosis.1
T-cell transformation is characterized by aberrant expression of oncogenic transcription factors combined with inactivation of tumor suppressor genes (eg, phosphatase and tensin homolog [PTEN], CDKN2A) and/or activation of the NOTCH1 pathway.2 The ectopic expression of oncogenes is typically caused by chromosomal rearrangements, the so-called type A hits, that place oncogenes under the control of T-cell–specific promoters or enhancer elements.3,4 The analysis of translocation breakpoints revealed frequent involvement of illegitimate V(D)J recombination in these translocations by binding of recombination-activating gene 1/2 (RAG1/2) proteins to sequences that resemble authentic recombination signal sequences (RSSs).5 These recurrent chromosomal rearrangements activate several oncogenes, such as TAL1, LMO2, TLX3, TLX1, or NKX2-1/NKX2-2, which are believed to represent the clonal disease drivers.2,6
Besides near mutually exclusive type A mutations, recurrent genetic aberrations that affect cell viability and/or proliferation, the so-called type B hits, are found in nearly all T-ALL genetic subgroups. Type B mutations include NOTCH1-activating mutations affecting NOTCH1 and FBXW7 that are found in over 60% of pediatric T-ALL patients7-11 (reviewed in Ferrando12 ), as well as less frequent events such as IL7R mutations in ∼10% of T-ALL cases.13,14 In addition, mutations in the PTEN tumor suppressor gene have been associated with poor prognosis,15-18 resulting in overactive phosphatidylinositol 3-kinase (PI3K)–AKT signaling that drives enhanced cell proliferation and cell metabolism, and impairs apoptosis.16,19,20 PTEN is considered to be a haploinsufficient tumor suppressor gene because PTEN dose determines cancer susceptibility.21-23 The majority of PTEN aberrations in T-ALL are deletions affecting the entire PTEN locus or mutations that truncate the membrane-binding C2 domain.15,18
In our previous studies, we detected PTEN aberrations in 13% to 20% of T-ALL patients18,24 and revealed that those mutations are especially associated with TAL or LMO rearrangements and nearly absent in TLX3-rearranged T-ALL.18 In general, PTEN-mutated T-ALL appears to be devoid of NOTCH1-activating mutations.18 Interestingly, we did not observe differential AKT activation when comparing PTEN mutant/deleted with wild-type patient samples, indicating that other mechanisms may influence the PI3K-AKT pathway. In this respect, nondeletional posttranslational inactivation of PTEN,24 rare mutations in PIK3CA (encoding PI3K) and AKT themselves,16 or PI3K-AKT pathway activation downstream of activated NOTCH1 have been described.15 However, none of these mechanisms explain the absence of PTEN protein in some T-ALL patient samples that have retained at least 1 PTEN wild-type allele.18
In this study, we have used multiplex ligation-dependent probe amplification (MLPA) to investigate copy-number variations among PTEN exons to detect potential additional PTEN deletions. We identified PTEN microdeletions in T-ALL patient samples and we provide evidence that these are driven by illegitimate RAG-mediated recombination events.
Materials and methods
Patients
A total of 146 primary pediatric T-ALL patient samples enrolled in the Dutch Childhood Oncology Group (DCOG) protocols (n = 72)25-27 or the German Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia study (COALL-97) (n = 74) were included in this study.28 Normal thymocytes were isolated from thymic tissue obtained from children undergoing cardiac surgery.24,29 Informed consents were in accordance with the institutional review boards of the Erasmus MC (Rotterdam, The Netherlands), the ethics committee of the City of Hamburg, Germany, the Hospital Sta. Cruz, Centro Hospitalar de Lisboa Ocidental (Lisboa, Portugal), and the Declaration of Helsinki.
Computational detection of putative RAG RSSs
The human PTEN gene (ENSG00000171862) was screened for the presence of cryptic RAG RSSs (cRSSs) using the PERL software algorithms developed by Cowell et al.30
Generation of GFPi-PTEN cRSS reporter constructs and recombination assay
To measure efficiencies of predicted cRSSs in mediating recombination of inverted green fluorescent protein (GFPi)-monomer red fluorescent protein (mRFP) reporter constructs, polymerase chain reaction (PCR)-amplified PTEN cRSS1-4 or defined RSS control sequences were cloned into this reporter construct and recombination assays were carried out as described.29
Statistics
Statistics was performed using IBM SPSS Statistics 21 software. The Pearson χ2 was used for nominal distributed data; the Fisher exact test was alternatively used in case the number of patients in individual groups was lower than 5. The Mann-Whitney U test was used for continuously distributed data. Differences in relapse-free survival (RFS) were tested using the log-rank test. Proportional risk for relapse was done by univariate and multivariate Cox regression analyses. The recombination efficiencies of cRSSs were compared using a 1-way analysis of variance with the Bonferroni multiple comparison posttest. Data were considered significant when P ≤ .05 (2-sided).
See supplemental Methods (available at the Blood Web site) for further experimental details.
Results
PTEN microdeletions in T-ALL patients
In our previous study, we identified various T-ALL primary patient samples that lack PTEN protein expression and seemed PTEN wild-type or that contained an inactivating mutation or PTEN deletion in only 1 allele (summarized in Table 1).18 To identify additional PTEN-inactivating mechanisms, we performed MLPA analysis to screen for potential microdeletions affecting single or a few PTEN exons that had been missed by array comparative genomic hybridization (array-CGH) and fluorescence in situ hybridization (FISH) analyses. We analyzed 146 T-ALL patient samples for copy-number alterations in any of all 9 coding exons. Heterozygous microdeletions were detected in 3 T-ALL patients (Figure 1A), encompassing exons 2 and 3 in 2 patients (no. 21, no. 11) and exons 4 and 5 in another patient (no. 20). Accordingly, 2 of the 3 patients (no. 11, no. 20) demonstrated defective PTEN splicing with previously unknown underlying genetic aberrancies.18 A fourth patient (no. 12) was identified with a homozygous deletion of exons 2 and 3 that was confirmed by high-resolution array-CGH analysis (Figure 1A-B).
PTEN genetics of T-ALL patients
| Patient . | PTEN mutation . | PTEN deletion (FISH/ array-CGH) . | PTEN deletion (MLPA) . | Genomic breakpoint PCR . | Aberrant transcript . | PTEN protein . | Cytogenetic aberration . | NOTCH/ FBXW7 mutation . | |
|---|---|---|---|---|---|---|---|---|---|
| Allele A . | Allele B . | ||||||||
| Clonal inactivation of 2 alleles | |||||||||
| 1 | R129G | T231fsX24 | — | — | — | ND | Mutant | LMO3 | — |
| 2 | F144fsX37 | R232fsX23 | Subcl Del | — | — | ND | Absent | LMO2 | PEST |
| 3 | P246fsX11 | — | Het Del | Het Del exons 2-9 | — | ND | Absent | SIL-TAL1 | — |
| 4 | D235fsX9 | P245fsX12 | ND | ND | — | ND | Absent | CALM-AF10 | PEST |
| 5 | R232fsX13 | Q244fsX8 | — | — | — | ND | Absent | Unknown | — |
| 6 | R233fsX10 | P245fsX9 | — | — | — | ND | ND | Unknown | FBXW7 |
| 7 | R232fsX10 | P243fsX18 | — | — | — | ND | ND | TLX1 | — |
| 8 | R232fsX10 | P245fsX14 | — | — | — | ND | Absent | LMO2 | FBXW7 |
| 9 | L180fsX2 | I305fsX7 | — | — | Intron 1-3 Del (type II; Subcl) | ND | Absent | NKX2-5 | HD |
| 10 | C104fsX2 | K236fsX5 | — | — | — | ND | Absent | SIL-TAL1 | — |
| 11 | P245fsX3 | — | ND | Het Del exons 2-3 | Intron 1-3 Del (type II, clonal) | in1/2-ex4 | ND | MYC | — |
| 12 | — | — | Hom Del/FISH negative | Hom Del exons 2-3 | 2 Intron 1-3 Del variants (type I, clonal) | in1/2-ex4 | Absent | SIL-TAL1 | — |
| Clonal inactivation of 1 allele | |||||||||
| 13 | R129fsX4/ P245fsX3 | — | Subcl Del | Het Del exons 1-9 | — | ND | ND | Unknown | — |
| 14 | R232 (STOP) | — | — | — | — | — | ND | SIL-TAL1 | — |
| 15 | C249fsX10 | — | — | — | Intron 1-3 Del (type I; Subcl) | — | Absent | Unknown | — |
| 16 | T231fsX14 | — | — | — | — | — | Absent | NKX2-1 | HD/FBXW7 |
| 17 | T276A | — | — | — | Intron 1-3 Del (type I; Subcl) | — | Absent | Unknown | — |
| 18 | — | — | Het Del | Het Del exons 1-9 | — | WT | Absent | Unknown | PEST |
| 19 | — | — | Het Del | Het Del exons 3-9 | Intron 1-3 Del (type I; Subcl) | Altered | Absent | SIL-TAL1 | — |
| 20 | — | — | — | Het Del exons 4-5 | Intron 3-5 Del (type III, clonal) /Intron 1-3 Del (type I; Subcl) | ex3-ex6 splice | Absent | SIL-TAL1 | — |
| 21 | — | — | ND | Het Del exons 1-3 | Intron 1-3 Del (type I, clonal) | in1/2-ex4 | ND | SIL-TAL1 | HD |
| 22 | — | — | Het Del | Het Del exons 2-9 | — | ND | ND | Unknown | — |
| Subclonal-inactivating event only | |||||||||
| 23 | R232fsX (Subcl) | — | — | — | — | — | ND | SIL-TAL1 | — |
| 24 | — | — | — | — | Intron 1-3 Del (type I; Subcl) | ND | ND | SIL-TAL1 | HD/PEST |
| 25 | — | — | ND | — | Intron 1-3 Del (type I; Subcl) | ND | ND | SIL-TAL1 | — |
| 26 | — | — | — | — | Intron 1-3 Del (type I; Subcl) | ND | ND | Unknown | — |
| Others | |||||||||
| 27 | — | — | — | — | — | — | Absent | TAL2 | FBXW7 |
| 28 | — | — | — | — | — | — | Absent | TAL1 | HD |
| Patient . | PTEN mutation . | PTEN deletion (FISH/ array-CGH) . | PTEN deletion (MLPA) . | Genomic breakpoint PCR . | Aberrant transcript . | PTEN protein . | Cytogenetic aberration . | NOTCH/ FBXW7 mutation . | |
|---|---|---|---|---|---|---|---|---|---|
| Allele A . | Allele B . | ||||||||
| Clonal inactivation of 2 alleles | |||||||||
| 1 | R129G | T231fsX24 | — | — | — | ND | Mutant | LMO3 | — |
| 2 | F144fsX37 | R232fsX23 | Subcl Del | — | — | ND | Absent | LMO2 | PEST |
| 3 | P246fsX11 | — | Het Del | Het Del exons 2-9 | — | ND | Absent | SIL-TAL1 | — |
| 4 | D235fsX9 | P245fsX12 | ND | ND | — | ND | Absent | CALM-AF10 | PEST |
| 5 | R232fsX13 | Q244fsX8 | — | — | — | ND | Absent | Unknown | — |
| 6 | R233fsX10 | P245fsX9 | — | — | — | ND | ND | Unknown | FBXW7 |
| 7 | R232fsX10 | P243fsX18 | — | — | — | ND | ND | TLX1 | — |
| 8 | R232fsX10 | P245fsX14 | — | — | — | ND | Absent | LMO2 | FBXW7 |
| 9 | L180fsX2 | I305fsX7 | — | — | Intron 1-3 Del (type II; Subcl) | ND | Absent | NKX2-5 | HD |
| 10 | C104fsX2 | K236fsX5 | — | — | — | ND | Absent | SIL-TAL1 | — |
| 11 | P245fsX3 | — | ND | Het Del exons 2-3 | Intron 1-3 Del (type II, clonal) | in1/2-ex4 | ND | MYC | — |
| 12 | — | — | Hom Del/FISH negative | Hom Del exons 2-3 | 2 Intron 1-3 Del variants (type I, clonal) | in1/2-ex4 | Absent | SIL-TAL1 | — |
| Clonal inactivation of 1 allele | |||||||||
| 13 | R129fsX4/ P245fsX3 | — | Subcl Del | Het Del exons 1-9 | — | ND | ND | Unknown | — |
| 14 | R232 (STOP) | — | — | — | — | — | ND | SIL-TAL1 | — |
| 15 | C249fsX10 | — | — | — | Intron 1-3 Del (type I; Subcl) | — | Absent | Unknown | — |
| 16 | T231fsX14 | — | — | — | — | — | Absent | NKX2-1 | HD/FBXW7 |
| 17 | T276A | — | — | — | Intron 1-3 Del (type I; Subcl) | — | Absent | Unknown | — |
| 18 | — | — | Het Del | Het Del exons 1-9 | — | WT | Absent | Unknown | PEST |
| 19 | — | — | Het Del | Het Del exons 3-9 | Intron 1-3 Del (type I; Subcl) | Altered | Absent | SIL-TAL1 | — |
| 20 | — | — | — | Het Del exons 4-5 | Intron 3-5 Del (type III, clonal) /Intron 1-3 Del (type I; Subcl) | ex3-ex6 splice | Absent | SIL-TAL1 | — |
| 21 | — | — | ND | Het Del exons 1-3 | Intron 1-3 Del (type I, clonal) | in1/2-ex4 | ND | SIL-TAL1 | HD |
| 22 | — | — | Het Del | Het Del exons 2-9 | — | ND | ND | Unknown | — |
| Subclonal-inactivating event only | |||||||||
| 23 | R232fsX (Subcl) | — | — | — | — | — | ND | SIL-TAL1 | — |
| 24 | — | — | — | — | Intron 1-3 Del (type I; Subcl) | ND | ND | SIL-TAL1 | HD/PEST |
| 25 | — | — | ND | — | Intron 1-3 Del (type I; Subcl) | ND | ND | SIL-TAL1 | — |
| 26 | — | — | — | — | Intron 1-3 Del (type I; Subcl) | ND | ND | Unknown | — |
| Others | |||||||||
| 27 | — | — | — | — | — | — | Absent | TAL2 | FBXW7 |
| 28 | — | — | — | — | — | — | Absent | TAL1 | HD |
PTEN frameshift mutations are indicated with the number of encoded amino acids in the alternative reading frame. PTEN deletion status was determined by FISH, array-CGH, and/or MLPA. Introns harboring the genomic breakpoints of PTEN deletions and/or microdeletions have been indicated. Exons for alternative spliced PTEN transcripts are indicated.
Del, deletion; FS, frameshift; HD, NOTCH1 heterodimerization domain mutation; Het, heterozygous; Hom, homozygous; PEST, NOTCH1 mutation in the proline, glutamine, serine and threonine-rich C-terminal region; Subcl, subclonal; WT, wild-type.
Identification of PTEN microdeletions in T-ALL patients. (A) MLPA electropherograms of normal reference DNA and representative examples of T-ALL patients with heterozygous or homozygous PTEN microdeletions affecting exons 2-3 or a heterozygous deletion of exons 4-5. Fluorescence intensities of amplified PCR products for specific PTEN exons are shown. PCR product sizes are shown at the top. Each arrow points to a homo- or heterozygously deleted exon. (B) Array-CGH plot exhibiting the homozygous PTEN exon 2-3 microdeletion in one T-ALL patient sample.
Identification of PTEN microdeletions in T-ALL patients. (A) MLPA electropherograms of normal reference DNA and representative examples of T-ALL patients with heterozygous or homozygous PTEN microdeletions affecting exons 2-3 or a heterozygous deletion of exons 4-5. Fluorescence intensities of amplified PCR products for specific PTEN exons are shown. PCR product sizes are shown at the top. Each arrow points to a homo- or heterozygously deleted exon. (B) Array-CGH plot exhibiting the homozygous PTEN exon 2-3 microdeletion in one T-ALL patient sample.
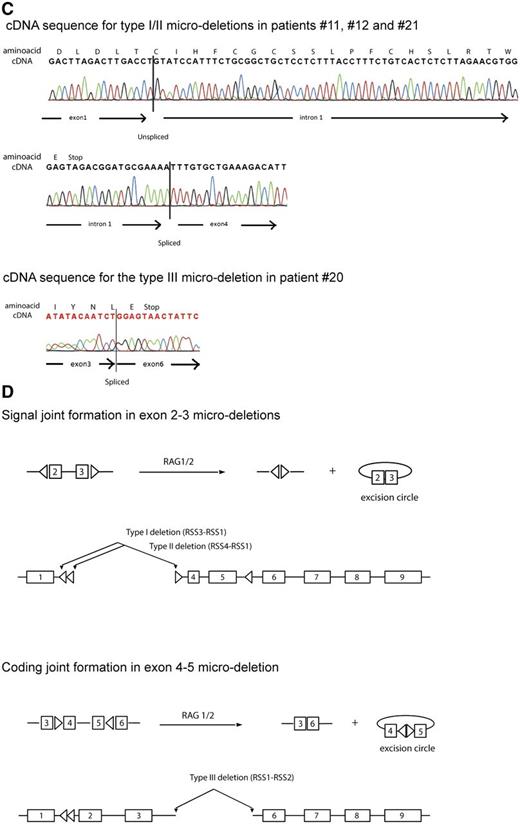
To clone the breakpoint regions of these microdeletions, a PCR-based strategy was designed for introns 1 and 3 (patient no. 21, patient no. 11, and patient no. 12) and intron 3 and 5 (patient no. 20) (Figure 2A-B), and resulting positive reactions were cloned and sequenced. These analyses predicted microdeletions of ∼65 kb that encompassed exons 2 and 3 and of ∼11 kb that encompassed exons 4 and 5 (Figure 2B). The homozygously deleted patient (no. 12) revealed different breakpoints that point to independent deletion events for each allele, with insertion of random bases in between breakpoints for 1 allele (Figure 2B). Breakpoints for the exon 2 and 3 microdeletion in patient no. 21 were identical to breakpoints of 1 allele of patient no. 12 and also lacked insertion of random bases. T-ALL patient no. 11 had a similar exon 2-3 deletion that shared the identical breakpoint in intron 3 but had an alternative breakpoint in intron 1 (Figure 2A-B). Breakpoints for the fourth T-ALL patient (no. 20) with a microdeletion that affected exons 4 and 5 were located in introns 3 and 5 (Figure 2A-B). All 3 types of microdeletions result in out-of-frame PTEN transcripts (Figure 2C).
Breakpoints of PTEN microdeletions. (A) Schematic representation of the PTEN gene. Missense mutations are represented by open triangles above the exons, whereas a silent mutation is presented as a filled gray triangle as shown before.18 Nonsense insertion/deletion mutations are indicated by a filled black triangle. Left- or right-pointing open triangles in introns 1, 3, and 5 represent cRSSs. (B) Sequences of cloned intron 1-3 type I and type II breakpoints and the intron 3-5 type III breakpoint for T-ALL patients with PTEN microdeletions. cRSSs are indicated by a box with the canonic CAC trinucleotide sequences or the corresponding GTG nucleotides in heptamer sequences indicated in bold and underlined. Insertion of non-template, random nucleotides are shown in bold. (C) Examples of sequence traces of cDNA resulting from type I, II, and III microdeletions. (D) Involvement of specific cRSSs in illegitimate RAG-mediated recombination events resulting in types I and II microdeletions (signal joint) and aberrant PTEN splice variant or the type III with the aberrant exon 4-5 microdeletion PTEN transcript (coding joint).
Breakpoints of PTEN microdeletions. (A) Schematic representation of the PTEN gene. Missense mutations are represented by open triangles above the exons, whereas a silent mutation is presented as a filled gray triangle as shown before.18 Nonsense insertion/deletion mutations are indicated by a filled black triangle. Left- or right-pointing open triangles in introns 1, 3, and 5 represent cRSSs. (B) Sequences of cloned intron 1-3 type I and type II breakpoints and the intron 3-5 type III breakpoint for T-ALL patients with PTEN microdeletions. cRSSs are indicated by a box with the canonic CAC trinucleotide sequences or the corresponding GTG nucleotides in heptamer sequences indicated in bold and underlined. Insertion of non-template, random nucleotides are shown in bold. (C) Examples of sequence traces of cDNA resulting from type I, II, and III microdeletions. (D) Involvement of specific cRSSs in illegitimate RAG-mediated recombination events resulting in types I and II microdeletions (signal joint) and aberrant PTEN splice variant or the type III with the aberrant exon 4-5 microdeletion PTEN transcript (coding joint).
As MLPA does not allow for sensitive detection of subclones with microdeletions, we performed PCR analysis to screen the T-ALL cohort for similar microdeletions. Seven additional patients were identified with deletions affecting exons 2-3 that were similar to the breakpoints as observed in patient no. 21 and patient no. 12 (Figure 2B). Based on the conservation of breakpoints, this microdeletion was denoted as a type I microdeletion. One additional patient was identified with breakpoints similar to patient no. 11, therefore this deletion was denoted as a type II microdeletion. The deletion affecting exons 4 and 5 as identified in patient no. 20 was accordingly denoted as a type III microdeletion. These deletions had not been detected before by array-CGH, FISH, or MLPA analyses. One patient sample (no. 20) had already been identified by MLPA as having a clonal microdeletion affecting exons 4 and 5, but also contained a subclonal type I microdeletion in exons 2 and 3 as detected by PCR (Figure 2B, Table 1). In another case (no. 19), array-CGH had revealed a heterozygous PTEN deletion of exons 3-9, but now PCR also revealed a subclonal type I exon 2-3 microdeletion (Table 1). Sequencing of the breakpoints in these additional T-ALL cases revealed that 5 of the 7 type I deletions and the type II deletion involved the insertion of unique, random nucleotide sequences, thereby excluding false-positives due to PCR contamination. Notably, the PCR product for these patient samples as visualized by gel electrophoresis was much weaker than those for the 4 patient samples with clonal microdeletions (data not shown). This strongly indicates that these deletions must be present on the subclonal level and therefore only detectable by specific PCRs. Overall, we have identified PTEN microdeletions in 11 of 146 T-ALL patients (8%), comprising a total of 13 deletional events. Only 4 patients presented these mutations at the clonal level.
Microdeletion breakpoints are flanked by cRSSs
The conservation of breakpoints among patient samples and the inclusion of non-template-derived nucleotides by terminal deoxynucleotidyltransferase (TdT)31 in most breakpoint regions pointed to a RAG-mediated deletion mechanism. We then searched for the presence of cRSSs that could function as putative RAG-mediated recombination signals, such as the RSS involved in T- or B-cell receptor gene segment rearrangements.32 Analysis of sequences directly flanking the breakpoints immediately revealed typical CAC canonic trinucleotides (Figure 2B, supplemental Table 5), which is a hallmark of the heptamer sequences of RSSs. The search for nearby nonamer sequences with A-nucleotide enrichment revealed a putative 12-spacer cRSS in intron 3 with a 5′ to 3′ orientation (cRSS1). A 23-spacer RSS was identified that directly flanks the breakpoint in intron 5 (cRSS2), and 2 others were identified flanking both breakpoints in intron 1 (cRSS3 and cRSS4; Figure 2A-B, supplemental Table 5). All 23-spacer cRSSs (cRSS2, cRSS3, and cRSS4) are present in a 3′ to 5′ orientation with respect to the PTEN reading frame orientation, and are therefore correctly positioned to allow illegitimate RAG-mediated recombinations with cRSS1 (Figure 2A,C). In this scenario, RAG1/2 molecules bind a pair of 12 and 23 RSSs resulting in 2 DNA double-strand breaks adjacent to each heptamer. Most microdeletion breakpoints are the consequence of heptamer-to-heptamer sequence fusions resembling signal joints of excision circles that are generated during normal T- or B-cell receptor gene segment rearrangements (Figure 2D, top): type I and II microdeletions result from cleaved DNA sequences 3′ of cRSS3 or cRSS4, respectively, that are fused to sequences 5′ of cRSS1. This retains both cRSSs in the genomic sequences that flank the deletion breakpoints as depicted in Figure 2D. The type III deletion resembles a typical coding joint that results from cleaved DNA sequences 5′ of cRSS1 that are fused to sequences 3′ of cRSS2 resulting in the loss of cRSSs from the genomic sequence (Figure 2D, bottom). T- or B-cell receptor coding joints give rise to fused gene segments with potential exonuclease processing of both ends and incorporation of random nucleotides whereby directly flanking RSSs and intervening DNA sequences are lost as excision circles. For the type III deletion of patient no. 20 (Figure 2B), this led to the fusion of sequences 5′ of cRSS1 to sequences 3′ of cRSS2 with loss of 14 nucleotides and incorporation of 17 guanine cytosine-rich N-nucleotides.
Prediction of cRSSs
To further characterize these cRSSs and estimate their recombination potential, we calculated RSS information content (RIC) scores.30 Cryptic RSSs with RIC scores close to the threshold levels that discriminate bona fide functional RSSs from cRSSs (ie, −38.81 for 12-spacer RSSs and −58.45 for 23-spacer RSSs) were further investigated.30 This search in the PTEN locus predicted a 12-spacer cRSS1 with a strong RIC score of −34.23 as well as 23-spacer cRSS2 (−55.59) and cRSS3 (−59.78) with RIC scores that were close to the threshold levels separating RSSs from cRSSs. A 23-spacer cRSS4 was predicted with a RIC score of −75.59 that is barely above the mean background RIC score value for 39-nucleotide non-RSS DNA sequences (−77.76). Thus, the obtained RIC scores for cRSS1, cRSS2, and cRSS3 strongly support PTEN microdeletions as RAG-mediated recombination events with similar recombination potential to that of bona fide RSSs flanking immunoglobulin V(D)J gene segments.
Cryptic RSS1-4 support RAG-mediated recombination
We then tested whether the predicted cRSS1-4 could functionally mediate RAG recombinations. We used the GFPi-mRFP RAG reporter construct29 (Figure 3A), in which the 12-spacer cRSS1 was cloned in combination with a consensus 23-spacer RSS. Also, the 23-spacer RSSs (cRRS2, cRSS3, and cRRS4) were cloned in combination with a consensus 12-spacer RSS. Recombination efficiency of each variant GFPi-cRSS-mRFP construct was measured by flow cytometry as the frequency of GFP-positive (recombination-positive) HEK293T cells within the population of RFP-positive (transfected) cells (Figure 3A). Indeed, all 4 PTEN cRSSs were able to mediate RAG-dependent recombination of the GFPi substrate (Figure 3B). Recombination efficiencies were 7.5% ± 0.19% for cRSS1 (Figure 3B, left panel), 2.2% ± 0.15% for cRSS2, 4.1% ± 0.21% for cRSS3, and 2.1% ± 0.16% for cRSS4 (right panel). For comparison, the putative 12-spacer cRSS SCL(12)29 or the 23-spacer cRSS SCL(23)33 from the human SCL gene yielded 1.3% ± 0.09% and 1.1% ± 0.13% of GFP-positive cells, respectively. Both of these SCL cRSSs were used as references for the lower limit of detection in the GFPi-mRFP RAG reporter assay, as these do not give rise to distinct GFP-positive cell populations in the reporter assay. In contrast, the 12-spacer RSS that flanks the Jβ2-2 gene segment of the mouse TCRβ locus yielded 8.0% ± 0.31% of GFP-positive cells. Also, the 12-spacer cRSS that is involved in recurrent LMO2 translocations in T-ALL5 yielded 11.4% ± 0.30% of GFP-positive cells. These reporters highlight the capability of the recombination assay to measure low-efficiency RSS and cRSS activities. Despite the low frequencies of recombination, cRSS2-4 reporters give rise to distinct populations of GFP-positive cells (Figure 3B), in contrast to SCL(12) and SCL(23) cRSSs. Moreover, the efficiencies of recombination of PTEN cRSS1-4 differed significantly from the those of SCL(12) or SCL(23) cRSSs (Figure 3C). These results strongly support the involvement of predicted cRSS1-4 in illegitimate RAG-mediated recombination events causing PTEN microdeletions. Additionally, the recombination potential of these cRSSs are in line with the observed frequencies of type I microdeletions (cRSS3-cRSS1) vs type II (cRSS4-cRSS1) and type III (cRSS1-cRSS2) microdeletions in T-ALL patients (Figure 2B).
Intronic PTEN cRSS mediate RAG recombination events. (A) Top panel: Linear representation of the GFPi reporter construct that results in the inversion of GFP coding sequence during RAG-mediated recombination, and consequent GFP expression. The inverted GFP sequence (light green box) is flanked by a proximal 12-spacer RSS (light gray triangle) and a distal 23-spacer RSS (dark gray triangle) followed by the IRES-RFP as transfection control reporter (red box). GFP positivity is a measure for recombination potential. Bottom panel: Control in vitro RAG recombination assay; flow cytometry analysis of HEK293T cells transiently transfected with either an irrelevant, mock vector (in the absence of RAG1/2 expression vectors; negative control) or the GFPi-reporter construct containing the consensus 12- and 23-RSS in the presence of RAG1/2 expression vectors.29 The flow cytometry plots show the expression of GFP and RFP within gated live cells defined by FSC and SSC parameters (not shown) and the values represent the percentage of each cell population in the quadrants. The gate used to discriminate RFP-positive from RFP-negative cells is depicted by a red square and used for the contour plot analysis. The efficiency of recombination is indicated as the percentage of GFP-positive (recombination positive) cells within the RFP-positive (transfected) population. (B) Flow cytometry analysis of HEK293T cells transiently transfected with the GFPi variant constructs containing specific 12-spacer cRSS (LMO2, SCL/TAL1, PTEN-cRRS1, or the Jβ2.2-RSS) site combined with the consensus 23-spacer RSS (left panel). The GFPi variant constructs containing the consensus 12-spacer RSS29 were combined with 23-spacer PTEN cRSS2, cRSS3, cRSS4, or the control SCL/TAL1 23-spacer cRSS version (right panel). The human LMO2 12-spacer cRSS and the mouse Jβ2-2 bona fide RSS were used to establish the range of recombination activitites for low-efficiency RSSs as measured by the GFPi reporter assay. The 12- and 23-spacer versions of the human SCL/TAL1 cRSS were used to define the lower limit of detection of cRSS function in this reporter assay. Average percentage ± SD of GFP+ cells in the RFP+ population are derived from 4 to 5 independent experiments. (C) Recombination index was determined by normalizing the recombination efficiencies of each indicated reporter to that of GFPi Con12/23 and recombination efficiencies were calculated subtracting the GFP background of each respective unrecombined control. Values represent the mean ± SEM of 3 independent experiments with 3 replicates per condition; *P < .05; **P < .01; and ***P < .0001.
Intronic PTEN cRSS mediate RAG recombination events. (A) Top panel: Linear representation of the GFPi reporter construct that results in the inversion of GFP coding sequence during RAG-mediated recombination, and consequent GFP expression. The inverted GFP sequence (light green box) is flanked by a proximal 12-spacer RSS (light gray triangle) and a distal 23-spacer RSS (dark gray triangle) followed by the IRES-RFP as transfection control reporter (red box). GFP positivity is a measure for recombination potential. Bottom panel: Control in vitro RAG recombination assay; flow cytometry analysis of HEK293T cells transiently transfected with either an irrelevant, mock vector (in the absence of RAG1/2 expression vectors; negative control) or the GFPi-reporter construct containing the consensus 12- and 23-RSS in the presence of RAG1/2 expression vectors.29 The flow cytometry plots show the expression of GFP and RFP within gated live cells defined by FSC and SSC parameters (not shown) and the values represent the percentage of each cell population in the quadrants. The gate used to discriminate RFP-positive from RFP-negative cells is depicted by a red square and used for the contour plot analysis. The efficiency of recombination is indicated as the percentage of GFP-positive (recombination positive) cells within the RFP-positive (transfected) population. (B) Flow cytometry analysis of HEK293T cells transiently transfected with the GFPi variant constructs containing specific 12-spacer cRSS (LMO2, SCL/TAL1, PTEN-cRRS1, or the Jβ2.2-RSS) site combined with the consensus 23-spacer RSS (left panel). The GFPi variant constructs containing the consensus 12-spacer RSS29 were combined with 23-spacer PTEN cRSS2, cRSS3, cRSS4, or the control SCL/TAL1 23-spacer cRSS version (right panel). The human LMO2 12-spacer cRSS and the mouse Jβ2-2 bona fide RSS were used to establish the range of recombination activitites for low-efficiency RSSs as measured by the GFPi reporter assay. The 12- and 23-spacer versions of the human SCL/TAL1 cRSS were used to define the lower limit of detection of cRSS function in this reporter assay. Average percentage ± SD of GFP+ cells in the RFP+ population are derived from 4 to 5 independent experiments. (C) Recombination index was determined by normalizing the recombination efficiencies of each indicated reporter to that of GFPi Con12/23 and recombination efficiencies were calculated subtracting the GFP background of each respective unrecombined control. Values represent the mean ± SEM of 3 independent experiments with 3 replicates per condition; *P < .05; **P < .01; and ***P < .0001.
PTEN microdeletions in xenografted human T-ALL cells
Subclonal microdeletions in PTEN, even in patients that already had undergone clonal inactivating events affecting one allele, strongly imply that acquisition of microdeletions is an ongoing phenomenon in T-ALL leading to clonal diversity.34 To test this, we performed primary and secondary xenotransplantation experiments into NSG mice (Figure 4A) using TAL1-rearranged T-ALL blasts from patient no. 24 at diagnosis that had a subclonal microdeletion (Figure 4B). Several months posttransplantation,mice developed overt leukemia. Primary (×1) and secondary (×2) xenotransplanted material was then analyzed for the presence of PTEN microdeletions in bone marrow, thymus, spleen, and liver biopsies. Using MLPA analysis, no PTEN microdeletions were detected (data not shown), indicating that the subclonal PTEN microdeletion in the diagnostic patient material had not been clonally selected following xenotransplantation. Three distinct, subclonal PTEN microdeletions were detected by PCR in thymocyte and liver biopsies: 1 (X1-24 thymus-1) was identical to the microdeletion as originally identified in this patient (Figure 4B), whereas 2 novel microdeletions were detected, suggesting that these had occurred upon serial retransplantation.
PTEN microdeletions in xenotransplants of a T-ALL primary patient sample and in human thymocytes from healthy individuals. (A) Schematic representation of the xenotransplantation strategy. Several months posttransplant of patient no. 24’s leukemic cells into immunodeficient NSG mice (n = 9), cells from bone marrow, thymus, spleen, and liver were collected. Primary (×1) and secondary (×2) xenotransplanted material was then analyzed for the presence of any of the 3 different PTEN microdeletions. (B) Breakpoint sequences of PTEN microdeletions as detected in samples from primary (×1) and secondary (×2) xenotransplanted mice. Canonic CAC trinucleotide sequences or the corresponding GTG nucleotides in heptamer sequences are indicated in bold and underlined. (C) Sequences of the breakpoints for PTEN type I microdeletions as identified in thymocytes of healthy individuals (H-Thy1 – H-Thy3). BM, bone marrow; Spl, spleen; Thy, thymus.
PTEN microdeletions in xenotransplants of a T-ALL primary patient sample and in human thymocytes from healthy individuals. (A) Schematic representation of the xenotransplantation strategy. Several months posttransplant of patient no. 24’s leukemic cells into immunodeficient NSG mice (n = 9), cells from bone marrow, thymus, spleen, and liver were collected. Primary (×1) and secondary (×2) xenotransplanted material was then analyzed for the presence of any of the 3 different PTEN microdeletions. (B) Breakpoint sequences of PTEN microdeletions as detected in samples from primary (×1) and secondary (×2) xenotransplanted mice. Canonic CAC trinucleotide sequences or the corresponding GTG nucleotides in heptamer sequences are indicated in bold and underlined. (C) Sequences of the breakpoints for PTEN type I microdeletions as identified in thymocytes of healthy individuals (H-Thy1 – H-Thy3). BM, bone marrow; Spl, spleen; Thy, thymus.
PTEN aberrations are associated with TALLMO T-ALL patients
PTEN aberrations have been associated with a low incidence of NOTCH1-activating mutations, but with a high incidence of rearrangements in TAL1- and/or LMO2-related oncogenes.18 We now extend these findings, totaling 26 of 146 T-ALL patients (18%), which have PTEN aberrations including point, missense, or nonsense mutations, entire locus deletions, and/or microdeletions at the clonal or subclonal level as summarized in Table 1. Twelve patients had clonally inactivated PTEN on both alleles and 10 patients on 1 allele. Evidence for subclonal PTEN aberrations was found in 11 patients, 7 of whom also had clonally inactivated PTEN at least in 1 allele. The other 4 patients had either a subclonal missense mutation (patient no. 23) or subclonal microdeletions (3 patients) only. Still, for 8 T-ALL patients for whom protein data were available, absence of PTEN protein could not be solely explained by the genetic aberrations found, suggesting that additional PTEN-inactivating mechanisms await identification. Overall, our previously observed association with TAL- or LMO-rearranged leukemia18 became considerably more significant (P = .003; supplemental Table 6). Also, the significance levels for absence of these mutations in TLX3-rearranged T-ALL (P = .002; supplemental Table 6) and reduced overlap with NOTCH1-activating mutations were further strengthened (P = .001, supplemental Table 6).
PTEN aberrations and outcome
Our results do not support observations by others34 that PTEN-inactivated T-ALL subclones become selected during disease progression giving rise to relapse. Therefore, we regarded T-ALL patients with subclonal PTEN aberrations as wild-type patients in outcome analyses. PTEN/AKT aberrant T-ALL patients, including patients lacking PTEN protein expression, were not significantly associated with poor outcome in both treatment cohorts (5-year RFS for PTEN/AKT mutant patients is 64% ± 15% vs 70% ± 6% for wild-type patients on DCOG protocols and 57% ± 15% vs 76% ± 6% for patients on COALL protocols). This is due to the fact that PTEN/AKT mutations and NOTCH-activating mutations predominantly behave as mutually exclusive mutations. In addition, NOTCH-activating mutations have a strong trend toward poor outcome (5-year RFS for NOTCH-activated patients is 62% ± 8% vs 82% ± 8% for wild-type patients on DCOG protocols and 68% ± 8% vs 80% ± 9% for patients on COALL protocols, P = .06 [stratified for protocol]; supplemental Table 7).35 However, if NOTCH-activated and PTEN/AKT-mutated T-ALL patients are being compared with wild-type patients, wild-type patients demonstrate significantly fewer relapses (stratified P = .04; Figure 5), albeit having more frequent events including toxic deaths and secondary malignancies.18 Using the Cox regression proportional hazard method, NOTCH-activating and PTEN/AKT mutations were investigated along with male gender and the presence of TLX3 rearrangements, which negatively relate with poor outcome (supplemental Table 7, Table 2). NOTCH1-activating mutations and PTEN/AKT mutations did not significantly predict for increased risk for relapse in univariate analyses, but both were identified as strong, independent risk factors along with male gender in multivariate analysis (Table 2).
T-ALL patients lacking PTEN/AKT mutations and NOTCH-activating mutations have a good outcome. RFS curves for T-ALL patients treated on (A) Dutch DCOG ALL7/8 or 9 protocols or (B) German COALL-97/03 protocols. Green line: NOTCH-activating mutations including mutations in NOTCH and FBXW7; red line: PTEN-inactivating or AKT-activating mutations; blue line: NOTCH-activating mutations and PTEN-inactivating or AKT-activating mutations combined; black line: wild type for NOTCH/FBXW7 and PTEN/AKT. Tick marks in figures refer to patients that are lost from further follow-up. The numbers of patients included at various time points in these studies are shown.
T-ALL patients lacking PTEN/AKT mutations and NOTCH-activating mutations have a good outcome. RFS curves for T-ALL patients treated on (A) Dutch DCOG ALL7/8 or 9 protocols or (B) German COALL-97/03 protocols. Green line: NOTCH-activating mutations including mutations in NOTCH and FBXW7; red line: PTEN-inactivating or AKT-activating mutations; blue line: NOTCH-activating mutations and PTEN-inactivating or AKT-activating mutations combined; black line: wild type for NOTCH/FBXW7 and PTEN/AKT. Tick marks in figures refer to patients that are lost from further follow-up. The numbers of patients included at various time points in these studies are shown.
NOTCH1-activating and PTEN/AKT mutations predict for poor outcome in pediatric T-ALL treated on DCOG ALL7/8/9 or COALL-97/03 protocols
| Analyses using Cox regression model . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| Univariate | ||||
| Male gender | 146 | 3.278 | 1.267-8.486 | .014* |
| TLX3 | 146 | 2.044 | 1-4.175 | .05* |
| NOTCH1/FBXW7 | 141 | 2.077 | 0.946-4.560 | .068 |
| PTEN/AKT aberrations† | 142 | 1.675 | 0.787-3.567 | .18 |
| Multivariate | ||||
| Male gender | 141 | 2.910 | 1.117-7.577 | .029* |
| TLX3 | 141 | 2.018 | 0.921-4.424 | .079 |
| NOTCH-activating | 141 | 2.588 | 1.083-6.183 | .032* |
| PTEN/AKT aberrations† | 141 | 3.407 | 1.254-7.400 | .014* |
| Analyses using Cox regression model . | n . | HR . | 95% CI . | P . |
|---|---|---|---|---|
| Univariate | ||||
| Male gender | 146 | 3.278 | 1.267-8.486 | .014* |
| TLX3 | 146 | 2.044 | 1-4.175 | .05* |
| NOTCH1/FBXW7 | 141 | 2.077 | 0.946-4.560 | .068 |
| PTEN/AKT aberrations† | 142 | 1.675 | 0.787-3.567 | .18 |
| Multivariate | ||||
| Male gender | 141 | 2.910 | 1.117-7.577 | .029* |
| TLX3 | 141 | 2.018 | 0.921-4.424 | .079 |
| NOTCH-activating | 141 | 2.588 | 1.083-6.183 | .032* |
| PTEN/AKT aberrations† | 141 | 3.407 | 1.254-7.400 | .014* |
Univariate and multivariate Cox regression analyses stratified for DCOG or COALL treatment protocols using RFS for various parameters that were significantly associated with poor RFS (see supplemental Table 7).
CI, confidence interval; HR, hazard ratio.
P value represents P < .05.
Includes T-ALL patients who do not express PTEN protein while lacking PTEN aberrations, but does not include patient samples with PTEN aberrations only on the subclonal level.
PTEN microdeletions in healthy human thymocytes
The presence of recombination-prone cRSSs in PTEN intron sequences led us to speculate that microdeletions may occur in healthy thymocytes. Screening DNAs that were isolated from thymocytes of nonleukemic children for the presence of PTEN microdeletions by PCR revealed evidence for subclonal type I deletions in 3 of 11 (27%) thymocyte biopsies. Two of those microdeletions had unique random nucleotide sequences inserted in between the breakpoints (Figure 4C), ruling out false PCR positivity due to contamination. Thus, RAG-mediated PTEN microdeletions are not exclusive to T-ALL; they also occur during normal T-cell development.
Discussion
PTEN has been identified as a haploinsufficient tumor suppressor gene,21-23 for which gene mutations and/or deletions have been associated with poor outcome in T-ALL in various17,18,36,37 but not all studies.16,38 For T-ALL patients treated on DCOG ALL-7/8/9 or COALL97/03 treatment protocols, we demonstrate that clonal PTEN-inactivating aberrations or loss of PTEN protein are an independent factor that predicts for relapse just like NOTCH-activating mutations and male gender. About half of the T-ALL patients that retained 1 wild-type allele do not express PTEN protein. This indicates that additional genetic, epigenetic, or posttranslational-inactivating events are expected. We have identified recurrent inactivating PTEN microdeletions in T-ALL patients due to illegitimate RAG-mediated recombination events, mediated through cRSSs. Taking into account all point, missense, or nonsense mutations as well as deletions including microdeletions, 18% of T-ALL patients in our patient cohort harbor PTEN-inactivating aberrations.
Increasing evidence suggests that cRSSs can participate in oncogenic mechanisms, including chromosomal translocations in lymphoma, B-cell acute lymphoblastic leukemia39,40 and T-ALL,4,41,42 such as SIL-TAL1 gene fusions and HPRT deletions. Different chromosomal translocation mechanisms have been described as a consequence of erroneous rearrangements between cRSS that flank oncogenes with RSS sequences of T-cell receptor gene segments. Alternatively, broken DNA strands near oncogenes become mistakenly fused through a non-cRSS mechanism to T-cell receptor gene during V(D)J assembly (reviewed in Radich and Sala43 ). Other illegitimate, cRSS-driven coding-joint recombination events may cause intrachromosomal deletions such as described for IKZF1,44,45 ERG1,46 BTG1,47 and CDKN2A/B48-50 in humans and Notch151,52 and Bcl11b53 in mice.
PTEN microdeletions occur as a consequence of aberrant RAG-mediated recombination events, and breakpoints are flanked by cRSSs containing heptamer and nonamer sequences separated by 12- or 23-nucleotide spacers.54 These cRSSs facilitate recombinations in in vitro recombination assays with efficiencies that match the frequencies of different types of microdeletions in T-ALL patients. Approximately one-third of all signal joint-related type I and II microdeletions have perfect heptamer-to-heptamer fusions that lack incorporation of random nucleotides just like immunoglobulin H (IgH) or T-cell receptor excision circle signal joints. Two-thirds of microdeletion signal junctions represent atypical joints that incorporated guanine cytosine-rich N-nucleotides, phenomena which is also observed in V(D)J-associated signal junctions in mouse lymphocytes.55 The T-cell receptor–mediated translocation in T-ALL line SUP-T1 is a comparable atypical signal junction.56 Some of these microdeletion atypical signal joints (patient no. 12, patient no. 26, and patient no. 11) had undergone exonuclease processing of signal ends (Figure 2B). Noncanonic heptamer sequence variations may destabilize the RAG complex, allowing alternative joining mechanisms of coding ends and signal ends.57 This may include open-and-shut joint recombinations,58,59 resulting from a single RAG cleavage adjacent to cRSS heptamers, exonuclease processing, and insertion of random nucleotides before re-ligation of the DNA ends. This phenomenon may explain 1 rare T-ALL case24 with a mutation that replaces 13 nucleotides including the start codon by 15 random nucleotides. The mutation is flanked by a 12-spacer cRSS with a strong RIC score of −45.48 that allows RAG-mediated recombinations equal to the efficiency (2.5% ± 0.13%) of the mouse IgH locus VH/87 RSS (L.M.S., unpublished data and Davila et al60 ). However, no other T-ALL patient in our current series had an equivalent mutation at this position, indicating that these events are rare.
The identification of subclonal PTEN microdeletions, as well as entire PTEN locus deletions,18 indicates that RAG activity may be ongoing in (at least part of) the leukemic cell population. This may explain clonal diversity and selection that results in disease progression and relapse. This latter is also supported by in vitro recombination assays using T-ALL cell lines, demonstrating that about 1% of leukemic cells or less will undergo recombinations of the reporter construct within a 1-week time frame.29 Because intraclonal heterogeneity at diagnosis and clonal evolution at relapse are known to occur in ALL,46,61-64 we checked whether PTEN microdeletions in minor leukemic clones at presentation of disease become clonally selected following xenograft transplantation just like lentiviral PTEN-silenced T-ALL blasts.34 However, we did not observe preferential selection of leukemic cells with PTEN microdeletions to near clonal levels following xenotransplantation. This could be explained by preferential outgrowth of leukemic subclones having other mutations that were advantageous for engraftment in mice over subclones having PTEN microdeletions. Furthermore, additional and new illegitimate RAG-mediated PTEN microdeletions possibly as consequence of ongoing RAG activity were detected that were not found in primary leukemic cells. We cannot formally rule out that subclonal selection of a leukemic subpopulation with a novel PTEN microdeletion occurred from a PCR-undetectable subclone that was already present at diagnosis. In addition, 1 patient (no. 23) had a subclonal missense mutation, indicating that there is an ongoing pressure on TALLMO-dysregulated leukemic cells to inactivate remaining wild-type PTEN alleles. RAG activity also results in PTEN microdeletions in developing thymocytes of healthy individuals. These rearrangements may facilitate a premalignant condition from which leukemia can develop. Likewise, Marculescu et al5 described 2 mechanisms of illegitimate V(D)J chromosomal rearrangement that were found in healthy children, that is, the Dδ2/LMO2 recombination in the t(11;14)(p13;q11) and the TAL2/TCRβ translocation t(7;9)(q34;q32),65 known as driving oncogenic lesions in T-ALL.
Overall, our discovery of PTEN microdeletions has reinforced the fact that PTEN aberrations are especially abundant in TAL- or LMO-rearranged leukemia but not in TLX3-rearranged patients,18 as also observed in adult T-ALL patient series.37 PTEN abnormalities seem to be associated with a reduced incidence of NOTCH1-activating mutations. The TALLMO subtype represents an immunophenotypically mature subtype of arrested leukemic cells in T-ALL, in which ongoing RAG activity creates an opportunistic and extended time window for cRSS-mediated illegitimate recombination events. These may provoke disease progression and relapse in leukemia patients, adding a new level of complexity that should be addressed in the development of future antileukemic strategies for ALL. Taking into account all currently known PTEN inactivation mechanisms (PTEN mutations, entire locus deletions, and PTEN microdeletions), some seemingly wild-type T-ALL patients still lack PTEN protein expression indicating that other PTEN inactivation mechanisms await identification.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Gustavo G. L. Costa and Izabella A. P. Neshich for help with computational detection of cRSSs.
R.M., K.C.-B., and L.Z. were financed by the Stichting Kinderen Kankervrij (grant nos. KiKa 2007-12, KiKa 2008-29, and KiKa 2013-116). L.M.S. received a postdoctoral fellowship from Fundação para a Ciência e a Tecnologia (F.T.C.). This work was supported by grants PTDC/SAU-ONC/113202/2009 from Fundação para a Ciência e a Tecnologia (J.T.B.) and Fundação de Amparo à Pesquisa do Estado de São Paulo 08/10034-1.
Authorship
Contribution: R.D.M., L.M.S., J.T.B., and J.P.P.M. designed the study and wrote the manuscript; R.D.M., W.K.S., L.Z., and J.G.C.A.M.B.-G. performed the MLPA analyses, breakpoint mapping, and PCR-based screening of PTEN microdeletions; J.A.Y. performed the computational detection of putative RAG RSSs; L.M.S. and V.P. were responsible for generation of GFPi-PTEN cRSS reporter constructs and recombination assays; K.C.-B. and W.K.S. performed the xenotransplantation experiments; J.P.P.M. performed the statistical analyses of the data; M.A., E.S., and M.A.H. collected and provided patient samples and their characteristics; R.D.M., L.M.S., K.C.-B., R.P., J.T.B., and J.P.P.M. analyzed and interpreted data; and all authors read, revised, and approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jules P. P. Meijerink, Department of Pediatric Oncology/Hematology, Room Na-1611, Erasmus MC–Sophia Children’s Hospital, Wytemaweg 80, 3015 CN Rotterdam, The Netherlands; e-mail: j.meijerink@erasmusmc.nl.
References
Author notes
R.D.M. and L.M.S. are cofirst authors.
J.T.B. and J.P.P.M. contributed equally to this work.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal