Key Points
Uridine diphospho glucuronosyltransferase 2B17 (UGT2B17) is overexpressed in poor prognostic chronic lymphocytic leukemia.
Abstract
Uridine diphospho glucuronosyltransferase 2B17 (UGT2B17) glucuronidates androgens and xenobiotics including certain drugs. The UGT2B17 gene shows a remarkable copy number variation (CNV), which predisposes for solid tumors and influences drug response. Here, we identify a yet undescribed UGT2B17 mRNA overexpression in poor-risk chronic lymphocytic leukemia (CLL). In total, 320 CLL patients and 449 healthy donors were analyzed. High (above median) UGT2B17 expression was associated with established CLL poor prognostic factors and resulted in shorter treatment-free and overall survival (hazard ratio ([death] 2.18; 95% CI 1.18-4.01; P = .013). The prognostic impact of mRNA expression was more significant than that of UGT2B17 CNV. UGT2B17 mRNA levels in primary CLL samples directly correlated with functional glucuronidation activity toward androgens and the anticancer drug vorinostat (R > 0.9, P < .001). After treatment with fludarabine containing regimens UGT2B17 was up-regulated particularly in poor responders (P = .030). We observed an exclusive involvement of the 2B17 isoform within the UGT protein family. Gene expression profiling of a stable UGT2B17 knockdown in the CLL cell line MEC-1 demonstrated a significant involvement in key cellular processes. These findings establish a relevant role of UGT2B17 in CLL with functional consequences and potential therapeutic implications.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by a considerable heterogeneity regarding clinical presentation, need for treatment, and outcome. Many prognostic markers have been identified.1 Although most of them provide information about risk of progression and survival, the functional role of these markers is often unclear and therapeutic consequences are therefore lacking. Apart from the clinical Rai and Binet staging systems and cytogenetics,2-4 molecular markers, such as immunoglobulin heavy chain variable (IGHV) gene mutational status5,6 and lipoprotein lipase (LPL) mRNA expression have strong prognostic value.7,8 In a pilot gene expression study with 20 CLL patients, we identified a significant association of uridine diphospho (UDP) glucuronosyltransferase 2B17 (UGT2B17) with these prognostic factors.9
Metabolizing phase 2 enzymes of the UGT2B super-family conjugate various endogenous compounds, in particular steroid hormones as well as several pharmaceutical drugs.10,11 The UGT2B genes and pseudogenes are clustered on chromosome 4q13 and display up to 95% sequence homology among each other, which is reflected in some overlap in substrate specificity but often distinct expression profile. Isoform UGT2B17 is a major androgen inactivating enzyme playing a role in local tissue-specific regulation of it is substrates.12 Importantly, antileukemic drugs, such as anthraquinones or the histone deacetylase (HDAC) inhibitor vorinostat, are also subject to glucuronidation by this enzyme.13,14 An influence of UGT2B17 on clinical outcome after vorinostat therapy in Asian women with breast cancer was recently reported.15
UGT2B17 is affected by a remarkable copy number variation (CNV) spanning a 117-kb region encompassing the entire gene.16,17 The frequency of copy numbers shows exceptional differences between populations from Africa, Europe, or Asia.17-19 Interindividual variability in UGT2B17 allele-frequency is accompanied by pronounced differences in gene expression characterized by more than 29 times higher mRNA levels in whites compared with Japanese.20 This is by far the greatest difference in gene expression observed between these 2 ethnic groups.
Several reports point to a role of UGT2B17 in cancer susceptibility. UGT2B17 null genotype is more frequent in adenocarcinoma of the lung in women.21 For an association between the CNV and prostate cancer risk contradictory results have been obtained.22-24 However, the presence of the UGT2B17 deletion was recently associated with cancer recurrence and sex-steroid hormone levels suggesting a prognostic role of this gene in prostate cancer patients after initial treatment.25 Studies on graft versus host disease identified UGT2B17 as a minor histocompatibility antigen presented by different HLA-ABC molecules, which acts as a potential trigger of antibody response.26,27 So far, no association with leukemia has been described.
Here we report a detailed analysis of the role of UGT2B17 in CLL. We investigated (1) UGT2B17 mRNA expression as a prognostic marker; (2) the impact of the UGT2B17 copy number on CLL susceptibility and prognosis; (3) an association between UGT2B17 mRNA and specific enzymatic glucuronidation activity, as well as (4) potential functional consequences of modulating UGT2B17 expression in CLL.
Methods
Patients and healthy donors
Peripheral blood samples from 320 patients with CLL diagnosed between 1973 and 2011 at the Vienna General Hospital and from 449 healthy donors were analyzed. Of the 320 patients, 13 were diagnosed before 1990, 72 from 1990 to 1999, 88 between 2000 and 2003, 123 in the period from 2004 to 2007, and 24 between 2008 and 2011. Median observation time of patients was 73.7 months (range: 0.4-382.3 months). The majority of patients were untreated at time of blood collection. CLL characterization and treatment response were defined according to National Cancer Institute Working Group guidelines.28 Treatment-free survival (TFS) was defined from date of diagnosis until first day of CLL specific therapy or death. Overall survival (OS) was counted from date of CLL diagnosis until death from any cause. Patient samples were investigated for cytogenetic aberrations and IGHV mutational status as previously described.8 The control subjects were unrelated white men and women, who were participants in a Viennese health care program, 45 years or older, free of vascular disease and any other severe disease at the time of sample collection between 1998 and 2000. Informed consent was obtained from the study participants according to the Declaration of Helsinki and the study protocol was approved by the institutional review board (ethical approvals at the Medical University of Vienna, EK-No. 385/2007, 025/2009).
Cell lines
MEC-1 were obtained from Leibniz Institute DSMZ-German Collection of Microorganisms and cultured in RPMI without phenolic red (Gibco, Life Technologies) containing 10% of FBS Gold (PAA). HEK-293T cells were obtained from ATCC and cultured in fetal bovine serum albumin from Sigma-Aldrich Canada. Cells were maintained in the incubator at 37°C, 5% CO2. Additional human cell lines (n = 28) were obtained only for analysis of RNA expression as listed in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Drugs and chemicals
Vorinostat was obtained from Merck. Drugs were diluted in DMSO. Androsterone (ADT), dihydrotestosterone (DHT), androstane-3α-diol (3α-diol) were purchased from Steraloids.
Quantitative real-time PCR
DNA and RNA were prepared from unsorted frozen PBMCs. Total RNA from 240 CLL patients (143 men, 97 women; 195 Binet stage A) and from cell lines was analyzed for UGT2B17 mRNA expression by quantitative real-time PCR (qPCR) analysis using Applied Biosystems assay-on-demand and Taqman universal master mix without AmpErasesUNG. Samples were run on the ABI Prism 7000 Sequence Detector (Applied Biosystems) according to the manufacturer's instructions. Beta actin was used as a housekeeping gene and expression was calculated relative to the mean of 10 unrelated healthy donor samples.
UGT family mRNA expression profiling for UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A8, UGT1A9, UGT1A10, UGT2B7, UGT2B15, UGT2B17, and UGT2B28 was performed with cDNA from MEC-1 using SYBR green. Nineteen CLL patient samples were also analyzed for UGT2B7 and UGT2B15, isoforms showing the highest sequence homology and largely overlapping substrate specificity with UGT2B17. Reactions were run on a StepOnePlus real-time PCR system (Applied Biosystems). Primers and protocols were used as listed in supplemental Table 1.
Conventional PCR
DNA samples of 277 Austrian CLL patients (167 men, 110 women; 223 stage Binet A) and 449 healthy donors (307 men, 142 women) were compared regarding the distribution of UGT2B17 genotype. PCR was carried out with primers for marker E (in UGT2B17 exon 6) and marker J (flanking UGT2B17 deletion) as well as reaction conditions according to Wilson et al and run on Biometra T gradient or MWG Biotech Primus 96.17
UGT2B17 silencing in MEC-1 cells
For knockdown experiments 5 pLKO.1 clones containing different shRNAs targeting human UGT2B17 and a control vector were obtained from Sigma-Aldrich and lentiviral particles were produced according to standard protocols. In brief, HEK-293T cells were cotransfected with pLKO.1 plasmids, ΔR 8.91, and pVSV-G using lipofectamine2000 (Invitrogen, Life Technologies) to produce recombinant VSV-G pseudotyped lentiviruses. MEC-1 cells were transduced by spin infection (800g, 90 minutes, 32°C) in the presence of polybrene (7 μg/mL, Sigma-Aldrich) and selected with puromycin (PAA). Knockdown of UGT2B17 was assessed by quantitative real-time PCR as previously described. For subsequent experiments, the clone inducing the best knockdown was applied. After 2 weeks of culture, RNA for microarray analysis of 3 independent stably transfected MEC-1 cell lines was isolated using Trizol (Invitrogen) and a subsequent clean-up step with RNAeasy columns (QIAGEN).
Enzymatic assays and detection of glucuronides by mass spectrometry
To assess enzymatic glucuronidation activity of UGT2B17 in MEC-1 and CLL patient samples, microsomes were isolated as previously described.29 Detection of dihydrotestosterone (DHT)-glucuronide (-G), androsterone-G (ADT-G), androstane-3α, 17β-diol 17-glucuronide (3α-diol-17-G), and androstane-3α, 17β-diol 3-glucuronide (3α-diol-3-G) was performed by liquid chromatography coupled to tandem mass spectrometry as previously described.25
Gene expression profiling
Microarray analysis of CLL patient samples was reported previously.9,30 Briefly, CD19-sorted PBMCs from 10 patients treated with fludarabine and cyclophosphamide (FC) and from 10 patients treated with FC in combination with rituximab (FCR) were analyzed before and after initiation of treatment. To investigate the significance of our initial findings using unsorted PBMCs,9 we compared UGT2B17 expression with this dataset using CD19+ selected cells from untreated patients.30
The samples from 3 independent UGT2B17 shRNA-induced knockdown experiments in MEC-1 were run on GeneChip Human Gene 1.0 ST Array (Affymetrix). GeneChip datasets are available online as Gene Expression Omnibus entry GSE15490 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE15490) and GSE38367 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token = djobjcoeykgksro&acc = GSE38367), respectively.
Statistical and bioinformatical analysis
Gene expression was given as median, quartiles (q1, q3), and range. The influence of UGT2B17 expression on TFS and OS was illustrated by Kaplan-Meier plots (including P values from log-rank tests) and quantified by hazard-ratios (HR) from univariate and multivariable Cox regression models. In these models, UGT2B17 expression was dichotomized at the sample median. As a recommendation for further studies a cut-off level for UGT2B17 expression optimal for predicting TFS was exploratively determined.31 This recommended cut-off was not used for dichotomization in the models of the present study to avoid over-fit (ie, to avoid fitting a model that is overly adapted to the concrete sample data at hand and thus lacks external validity). Prognostic subgroups were investigated with respect to TFS by testing the interaction of UGT2B17 expression with each of IGHV mutation, CD38, sex, and time of diagnosis before or after 2004. Only in the case of a significant interaction, subgroups were considered separately.
As an external in silico validation, previously published Affymetrix gene expression array datasets on CLL prognostic markers from the French Cooperative Group on CLL using the U133A array as in our first analysis,32 and U95A array expression data from Haslinger et al were analyzed for UGT2B17-expression.33 Although Vasconcelos et al analyzed CD19+ selected samples from 18 CLL patients, unsorted PBMCs of 100 patients were analyzed in the German study.32
Genotype frequencies and prognostic markers were compared between groups using chi-square tests. The influence of high UGT2B17 on treatment response was quantified by an odds ratio (OR) from a logistic regression model. Means and confidence intervals of relative changes in expression levels were computed on the log-scale and then back-transformed. Spearman correlation coefficients were used to quantify the association between UGT2B17 mRNA expression and enzymatic activity. The reported P values are the results of 2-sided tests. P values of ≤ .05 were considered to be statistically significant. Computations were performed using SAS Version 9.2 (SAS Institute) and PASW Statistics Version 18.0.3 (SPSS) software.
For data import handling and all further calculations of gene expression microarrays in MEC-1 we used the tools provided by the Bioconductor project in R (www.bioconductor.org, cran.r-project.org). Boxplots, histograms, and a correlation plot were drawn for quality assessment. The function “RMA” implemented in the “Affy” package was used for background adjustment, normalization and summarization of the data into expression sets. A filtering step was performed where genes with a small variation across groups were deleted. A heatmap of the dataset is depicted with hierarchical clustering of samples and probe sets. To compare the genes of the 2 groups (2-sample t tests) the function lmFit provided by the “limma” package was applied on the data which results in one P value for each probe set. NTK Stat gives the test statistic that tests the hypothesis if the genes in a gene set show the same pattern of association with the phenotype compared with the rest of the genes, NTK q-value gives the minimum false discovery rate at which this test is considered significant. To adjust for multiplicity the Benjamini-Hochberg method was applied to control the false discovery rate.34 For pathway analysis the method by Tian et al was used.35 These computations were done using the statistical computing environment R 2.12.0.
Results
Association of UGT2B17 with other prognostic markers in CLL
Compared with the mean of 10 normal healthy donor PBMC samples, the relative UGT2B17 gene expression in 240 patients ranged from 0 to 264.12 (median: 1.77). High UGT2B17 above median (≥ 1.77-fold versus healthy donors) was significantly associated with established poor prognostic factors such as unmutated IGHV, high LPL and CD38 expression, absence of deletion 13q−, and trisomy 12 (Table 1). Interestingly, we found a trend toward a negative association with deletion 17p (odds ratio = 0.61, 95% CI 0.24-1.56).
The in silico comparison of our data with previously published datasets confirmed differential expression of UGT2B17 between IGHV mutated and unmutated CLL in other studies. Although UGT2B17 was the number top 12 significantly differentially expressed gene (fold change = 3.4, P = .023) in our results derived from unsorted PBMCs, it was top 11 (fold change = 3.9, P = .031) in the dataset using CD19+ selected cells from 18 CLL patients. In the work analyzing unsorted cells from 100 CLL patients, a different UGT2B17 probe set exhibited a fold change of 1.9 (P < .001). Comparing UGT2B17 expression in CD19-sorted and unsorted PBMCa in our own 2 unrelated datasets, differential expression between prognostic groups was even more pronounced using CD19+ cells (Table 2).
UGT2B17 mRNA expression predicts treatment-free and overall survival in CLL
High UGT2B17 expression levels above the median were significantly associated with a shorter treatment-free (HR = 2.25, 95% CI 1.58-3.21, P < .001) and overall survival (HR = 2.18, 95% CI 1.18-4.01, P = .013). TFS was significantly shorter in patients with high UGT2B17 expression (median TFS 62.6 months versus 154.1 month; P < .001). Furthermore, median OS was doubled in patients with low UGT2B17 compared with those with high expression (median OS 152.4 versus 299.5 months; P = .011; Figure 1A).
Prognostic significance of UGT2B17 mRNA expression and genotype in CLL. (A) Impact of high UGT2B17 mRNA expression (> 1.77 fold of healthy donors) on TFS and OS. (B) Illustration of UGT2B17 expression and TFS in IGHV mutated subgroup only. (C) Lack of gene dosage effect of UGT2B17 copy number and (D) Impact of UGT2B17 CNV on TFS and OS.
Prognostic significance of UGT2B17 mRNA expression and genotype in CLL. (A) Impact of high UGT2B17 mRNA expression (> 1.77 fold of healthy donors) on TFS and OS. (B) Illustration of UGT2B17 expression and TFS in IGHV mutated subgroup only. (C) Lack of gene dosage effect of UGT2B17 copy number and (D) Impact of UGT2B17 CNV on TFS and OS.
In multivariable models the influence of UGT2B17 on TFS was independent from each of sex, cytogenetics, LPL and CD38 expression, treatment period (before and in the rituximab era), and Binet stage, but not independent from IGHV mutational status. The effect of UGT2B17 adjusted for sex, LPL and CD38 expression, Binet stage, and IGHV mutational status is quantified by a hazard ratio (HR) of 1.68 (95% CI 0.89-3.16, P = .110). However, analysis of different prognostic subgroups revealed a pronounced impact of UGT2B17 among patients with mutated IGHV genes (HR = 2.40; P = .014 for the interaction; Figure 1B, supplemental Figure 2).
These data indicate that high UGT2B17 gene expression is suitable to identify poor-risk CLL patients at early stages of the disease. The explorative search for a cutoff level of UGT2B17 mRNA expression, which optimally discriminates with respect to TFS, yielded a relative level of 4.11-fold in relation to healthy donors to be recommended for use in further studies.
The role of UGT2B17 genotype in development and course of CLL
Practically no gene dosage effect for UGT2B17 CNV was observed although average mRNA-expression was slightly higher in homozygous carriers of the UGT2B17 gene (median 6.3; q1 = 0.5; q3 = 31.3) compared with heterozygous (median 4.3; q1 = 0.2; q3 = 19.7), and remained undetectable in patients with the null genotype (Figure 1C).
There was also a trend for association of longer TFS and OS with the null genotype (median TFS 113.9 months versus 83.9 months; HR = 0.60; 95% CI 0.35-1.03; P = .066) but the effect of genotype was weak compared with mRNA-expression (Figure 1D). Trends of association between CNV and prognostic markers were in line with results obtained for mRNA expression, but far from statistical significance and with a much weaker impact. Of note, no patient with the UGT2B17 null genotype (−/−) had a positive Coombs test.
Genotype frequencies in 277 CLL patients were 41.2% for carriers of 2 copies of the UGT2B17 gene (+/+), 45.8% for heterozygosity (+/−), and 13.0% with no copies of UGT2B17 or the null genotype (−/−), respectively. In 449 healthy donors we found 43.0% +/+, 46.3% +/−, and 10.7% −/− (Figure 2A). These results are consistent with other investigations on UGT2B17 copy number variations in whites.17,21-23 The frequency of the deleted allele did not differ significantly between CLL patients and healthy subjects or between men and women. This indicates that there was no decisive influence of UGT2B17 CNV on the development of CLL in our cohort.
Role of UGT2B17 in CLL susceptibility and drug response. (A) UGT2B17 genotype and CLL susceptibility and (B) change in UGT2B17 mRNA expression in CLL cells during treatment with fludarabine and cyclophosphamide.
Role of UGT2B17 in CLL susceptibility and drug response. (A) UGT2B17 genotype and CLL susceptibility and (B) change in UGT2B17 mRNA expression in CLL cells during treatment with fludarabine and cyclophosphamide.
UGT2B17 mRNA expression and response to therapy
Among 72 patients treated with fludarabine-based therapy, there was no association of single pretreatment UGT2B17 mRNA-level with final response or progression free survival on these regimens. However, sequential microarray analysis performed in 20 patients before and 3 days after initiation of therapy with FC or FCR showed a substantial up-regulation of UGT2B17 mRNA within 48 hours of treatment only in patients with stable or progressive disease (mean relative change = 134.1%, CI 106.0%-169.7%) compared with patients with partial and complete response to therapy (mean relative change = 99.1%; CI 84.2%-116.7%; P = .030; Figure 2B).
UGT family profiling in MEC-1 and primary CLL cells
RNA from 28 different human cell lines originating from various types of solid tumors and leukemias was obtained and analyzed for UGT2B17 (supplemental Figure 1). Among all samples tested, the CLL cell line MEC-1 showed by far the highest level of UGT2B17 mRNA and was selected as a model for further functional studies of UGT2B17 in CLL.
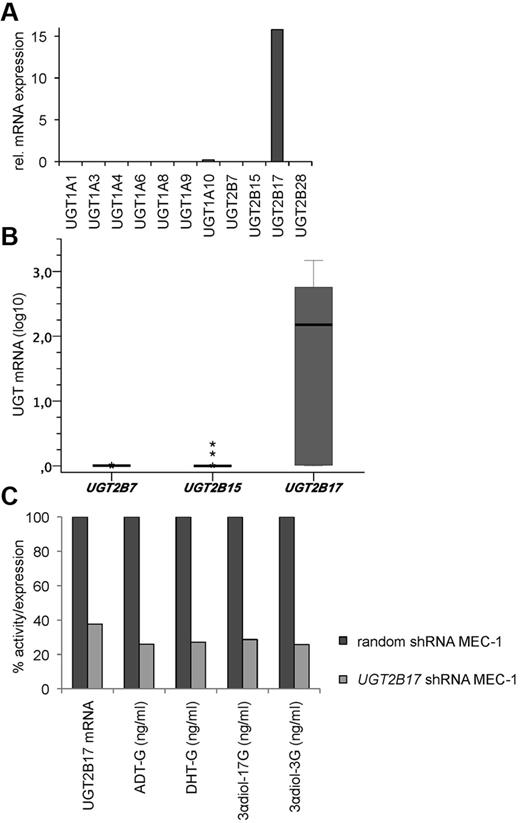
Expression of 11 functional UGTs (UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A8, UGT1A9, UGT1A10, UGT2B7, UGT2B15, UGT2B17, and UGT2B28) was then investigated in MEC-1. UGT2B17 was the only one showing a significant expression (Figure 3A). In addition, in all 19 patient samples tested the expression of UGT2B17-homolog isoforms UGT2B7 and UGT2B15 was also very low (Figure 3B). This makes a significant involvement of other enzymes of the UGT family with similar substrate specificity or regulation unlikely.
CLL-specific expression and enzymatic activity of UGT2B17. (A) UGT family expression profiling in MEC-1, (B) UGT2B isoform expression in CLL patients (n = 19), and (C) functional UGT2B17 shRNA knockdown in MEC-1 (mean of 3 replicates).
CLL-specific expression and enzymatic activity of UGT2B17. (A) UGT family expression profiling in MEC-1, (B) UGT2B isoform expression in CLL patients (n = 19), and (C) functional UGT2B17 shRNA knockdown in MEC-1 (mean of 3 replicates).
Correlation between UGT2B17 mRNA expression and enzymatic activity in CLL samples
In the 19 CLL patient samples tested, area under the curve (AUC) for vorinostat glucuronidation ranged from 0.00 to 0.73, and formation of DHT-G was between 0.0 and 58.1 pg/mL/mg protein. Glucuronidation of these substrates strongly correlated with UGT2B17 mRNA expression (Spearman correlation coefficient > 0.9, P < .001; supplemental Figure 3).
Gene expression profiling of functional UGT2B17 knockdown in MEC-1
Using 5 UGT2B17-targeting shRNA sequences, between 30% and 90% knockdown of UGT2B17 mRNA compared with the random shRNA control were achieved in the cell line model MEC-1. The clone inducing the best knockdown (90%) was NM_001077.1-1554s1c1, shRNA sequence 5′-CCG GCG TGG CAA CTA TGA TAT TTA TCT CGA GAT AAA TAT CAT AGT TGC CAC GTT TTT G-3′. Stable repression of UGT2B17 expression was still achieved after 2 weeks of culture posttransfection. In long-term culture over several months, a knockdown of approximately 70% was maintained. This shRNA-induced suppression of UGT2B17 mRNA was directly proportional to a decrease of UGT2B17-specific enzymatic functions (Figure 3C).
Microarray gene expression profiling of 3 independent UGT2B17 knockdown MEC-1 cell lines revealed significant changes in expression of 864 genes compared with the negative controls (supplemental Figure 4). The genes with reduced expression on UGT2B17 silencing were involved in pathways related to chromatin and nucleosome organization, regulation of proliferation and apoptosis, protein and cytokine synthesis and transport as well as inflammation and immunologic defense (supplemental Table 2). In contrast, a significant up-regulation of genes involved in intracellular transport and secretion including the endoplasmic reticulum and Golgi compartments was found (supplemental Table 3). Among the top differentially expressed probe sets, we also observed several genes known to be involved in CLL susceptibility and progression, such as CASP1,36 CD38,37 GRAMD1B,38 CCND2 (cyclin D2),39 ITGAL (LFA-1; CD11a),40 GZMK,41 and FCRL342 (Figure 4). These data indicate that UGT2B17 knockdown leads to perturbations in gene expression, which may influence CLL cell behavior.
Top 50 differentially expressed genes after UGT2B17 gene expression knockdown in MEC-1. The rows are scaled to have a mean of zero and standard deviation of one (z-score).
Top 50 differentially expressed genes after UGT2B17 gene expression knockdown in MEC-1. The rows are scaled to have a mean of zero and standard deviation of one (z-score).
Discussion
This is the first report describing a role of a UDP glucuronosyltransferase metabolic enzyme in leukemia. We identified a strong association of UGT2B17 mRNA expression with other prognostic markers in CLL translating into a significantly shorter treatment-free and overall survival among patients with high UGT2B17 levels. The impact of UGT2B17 on TFS was independent from each of sex, cytogenetics, LPL mRNA levels, CD38 expression, and Binet stage, respectively. A particular discriminating power was observed within favorable prognostic CLL subgroups, such as patients with mutated IGHV. Thus, UGT2B17 gene expression provides novel information for risk assessment in CLL in addition to previously described markers. In silico analysis of large datasets from other groups analyzing IGHV mutated and unmutated CLL patients also identified significant differences in UGT2B17 expression. This supplies an external validation of our findings. Comparison with mRNA expression values derived from CD19-sorted cells confirmed the significance of UGT2B17 using unsorted PBMCs.
Our data also point to a functional role of UGT2B17 in CLL. We demonstrate that the highly variable UGT2B17 mRNA levels found in quantitative real-time PCR linearly correlate with specific enzymatic protein function. Among known substrates of the enzyme are androgens and drugs being used for leukemia treatment, such as chlorambucil (unpublished data), anthraquinones,13 and the HDAC-inhibitor vorinostat.14,43 Of note, we also observed a significant up-regulation of UGT2B17 during treatment with standard regimens. Fludarabine and cylophosphamide are not directly subject to significant glucuronidation. However, constitutive overexpression or up-regulation of UGT2B17 during treatment with these drugs may lead to an increased elimination of other substrates. Such inducible and highly variable glucuronidation rates of drugs in CLL cells could be of therapeutic relevance because drug inactivation might also occur directly in leukemic cells in addition to classic metabolic tissues, such as liver, intestine, and kidney. Thus, the presence of UGT2B17 may have an impact on antineoplastic drug metabolism in cancer cells with the potential to affect overall drug response.
Interestingly, expression profiling of various UGT genes in patient samples and in the CLL cell line model MEC-1 revealed no significant mRNA expression or activity of any other UGT2B enzyme with a similar substrate spectrum in CLL cells. These results point to an exclusive role of the UGT2B17 isoform and suggest that its overexpression in CLL cells plays a role probably beyond glucuronidation of androgens and drugs. Indeed, UGT2B17 knockdown in MEC-1 affected predominantly genes involved in intracellular trafficking, regulation of proliferation and apoptosis, as well as immunologic pathways, including genes known for an association with CLL. The decrease of CD38 expression found on down-regulation of UGT2B17 is in agreement with our data observed in CLL patients. Interestingly, in contrast to the very high expression in the p53–deficient MEC-1 cell line, UGT2B17 mRNA levels were below median in our primary CLL samples with 17p deletion. However, larger sample series would be required to determine whether UGT2B17 is independent from p53. Because MEC-1 cells were obtained from a patient who was already in prolymphoblastic transformation, results derived from this cell line are to be interpreted with caution. Thus, factors inducing UGT2B17 overexpression, related functional consequences, and the influence of the microenvironment remain to be elucidated and are currently subject to further projects.
We also studied the role of UGT2B17 copy number on CLL susceptibility and prognosis. Genotype distribution did not differ significantly between CLL patients and healthy donors in our central European study population. This indicates that the UGT2B17 polymorphic deletion status by itself does not have a major impact on CLL development. The UGT2B17 expression was more relevant for CLL prognosis than the gene copy number alone. Therefore, the role of UGT2B17 in CLL susceptibility remains unclear. A comparative genetic study of Asian and white patients has recently been initiated to address the impact of UGT2B17 on the different incidence of CLL in the Asian and white populations.44
In summary, the findings of this study suggest a relevant role of the UGT2B17 pathway in progressive CLL and provide novel prognostic information. Linear correlation of UGT2B17 mRNA levels with specific glucuronidation activity demonstrated for DHT and vorinostat may probably apply for further substrates and could result in significant functional and therapeutic implications.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and donors who provided their cells for this research project. They also thank Martin and Bernadette Hilgarth, Elisabeth Reiter, Ulrike Zeman, and Particia Weiss for technical support on CLL patient sample collection and experiments; Lyne Villeneuve and Mario Harvey for technical support on UGT2B related experiments; Patrick Caron for mass spectrometry–based analyses of sex steroids; and Markus Jeitler for microarray experiments. The authors are also grateful to Christine Einberger for support with clinical data collection, Peter Valent for provision of vorinostat and leukemia cell lines, as well as Thomas Grunt, Gerald Prager, and Martin Filipits for provision of carcinoma cell lines. The authors also thank Robin Riestl for uploading our microarray data to the Gene Expression Omnibus platform.
This work was supported in part by the Austrian Genome Program Gen-AU C.h.i.l.d. II (GZ 200.136/1-VI/1/2005 to K.V.), Initiative Krebsforschung of the Medical University of Vienna (K.V.), the Canadian Institutes of Health Research (MOP-42 392 to C.G.), and the Canada Research Chair Program (C.G.). M.G. was a recipient of a Translational Research Training in Hematology (TRTH) grant from the European Hematology Association and the American Society of Hematology and was supported by a Marietta Blau Fellowship for a research stay in Québec, by the Hans und Blanca Moser Stiftung, and by a Förderungsstipendium from the Austrian Ministry of Science. J.B. was supported by a Canadian Institutes of Health Research (CIHR) Frederick Banting and Charles Best studentship award. E.L. holds a CIHR clinician-scientist award. C.G. holds a tier II Canada research chair in pharmacogenomics.
Authorship
Contribution: M.G. designed the research, performed experiments, collected clinical data, analyzed data, and wrote the paper; J.B. performed experiments and writing on UGT2B functional experiments; G.H. did experiments and writing on lentiviral transfections; M.B. performed microarray analysis; A.Gl. and S.Z. provided expert statistical analysis; E.P. and T.L. performed experiments; C.M., A.Ga., and M.S. provided samples and molecular data; K.F. and C.S. provided patient clinical information; E.L. and C.G. supervised and wrote UGT2B family functional and glucuronidation experiments; K.V. supervised UGT2B17 PCR experiments on CLL patients and healthy donors; U.J. designed the research, supervised the study, and wrote the paper; and all authors critically revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ulrich Jaeger, Medical University of Vienna, Department of Internal Medicine I, Division of Hematology, Waehringer Guertel 18-20, 1090 Vienna, Austria; e-mail: ulrich.jaeger@meduniwien.ac.at.