We have demonstrated by a detailed statistical analysis of proteome and transcriptome data of human platelets and human cell lines that protein and transcript abundance in platelets, if at all, are only weakly correlated.1 This analysis appears to be in contradiction to previous claims made inter alia by Rowley and Weyrich,2 who again advanced their opinion that transcript numbers would indeed reflect the extent of protein expression in human platelets.3 However, we do not agree that clear evidence about close transcriptome and proteome correlation is provided by previous publications, and from our perspective, the publications Rowley and Weyrich allude to4-6 do not convey clear proof for their hypothesis. None of the publications deal with the problem of comprehensive and comparative analysis of the transcriptome and proteome of human platelets, but rather are primarily focused either on the transcriptome6 or on a small number of individual proteins.4,5 In the publication by Gnatenko et al, the authors clearly state that: “the molecular analysis of the platelet transcriptome may be confounded by the constant decay of m[essenger] RNAs in the absence of new gene transcription”.5
We have carefully analyzed their response in order to understand the reason for the apparently contradicting view. Some of the remarks in the letter by Rowley and Weyrich are undeniably correct. For instance, in a few cases, we have missed transcripts and assumed that, although the protein being present as evident from mass spectrometry, the corresponding transcript would be absent. Indeed the transcript was present (eg, for GPIbα), but because of inconsistencies in the annotation systems, the refseq identifier could not be mapped to the correct protein. Unfortunately, this is a rather common problem with large data sets, so that some of the refseq identifiers provided by the authors2 were actually deleted or superseded in the meantime and in certain cases could even not be mapped on the sequence data level. Splicing variants and multiple identifiers assigned to the same protein further complicate the alignment of proteome and transcriptome data. Using the gene names that were listed along with the refseq identifiers did not appear recommendable to us, because they cannot be expected to be unique. Inspired by the letter from Rowley et al, we once more revised the data using alternative approaches for mapping transcripts to protein identifiers. Again identifiers were mapped exclusively to stable identifiers, and pseudogenes, hypothetical proteins, and so on were omitted. However, unexpectedly, this extended strategy yielded only 24 additional transcript-protein pairs for reads per kilobase per million (RPKM) >1 (Table 1); except for GPIbα, only 5 additional transcripts contributed significantly (RPKM > 100).
A major challenge for studies dealing with native material, particularly when isolated from blood, is posed by the high demands on sample purity. Because the protein content of platelets is comparable to other cells, contaminations have a comparatively small and predictable effect on the quality of proteome analysis. For instance, plasma proteins cannot be entirely removed from platelet preparations because of the “sponge-like” platelet surface formed by the open canalicular system, which is virtually inaccessible to purification techniques. In contrast, RNA content in platelets is ∼4 orders of magnitude lower than in leukocytes7 ; consequently, contaminations have a much stronger impact on data quality in platelet transcriptome analysis. Platelet RNA content is governed by exogenous and endogenous conditions as well as intrinsic factors. Because, to our knowledge, platelets have no transcription machinery, the RNA found apparently might be a relic of megakaryocyte RNA from proplatelet formation, rendering it difficult to deduce which of the transcripts contribute to the actual platelet proteome. Moreover, the amount of platelet RNA is affected by aging and most probably by platelet activating mechanisms.8,9 Apart from contamination by other cells or material, platelets may also incorporate foreign RNA, as demonstrated for tumor biomarkers,10 and may also transfer their RNA to other cells, as described recently.11
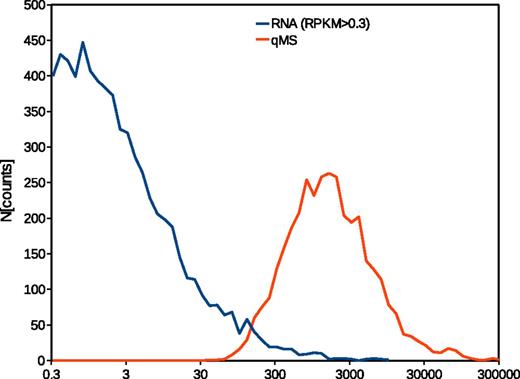
Considering the constraints and technical limitations of both techniques, we decided to choose a statistical approach rather than a straightforward comparison of the data. Quantitative proteomic data reflect normal distributions for protein frequency densities, as to be expected. In contrast, the transcriptome data provided by the authors show an almost exponential distribution, indicating a strong increase in the number of transcripts with decreasing transcript frequency (Figure 1). A recent publication by the Mann group12 provided evidence that transcriptome data may yield an almost identical frequency density distribution as proteomics data. However, in this analysis, a bimodal distribution was observed when the threshold for detection was set below 1 FPKM—the authors hypothesized that the low-frequency peak results from transcripts indeed not expressed as proteins. In our opinion, lowering the threshold for comparing the data, as proposed by Rowley et al, will thus certainly increase the coverage of the proteome, however, at the expense of validity, because the number of false-positive transcripts will concurrently increase. Because the frequency density distributions of the transcriptome data by Rowley et al and our proteome data do not share any similarities, we chose to rank each data set. In addition, we stratified the data to enable a direct comparison of high, medium, and low expression/transcription. Neither the rank correlation for the whole data set nor the correlation of the stratified data resulted in a correlation coefficient greater than 0.3. By including more low-rank data, the correlation can be improved, as Rowley et al demonstrated in their letter,3 but even then does not exceed 0.5, which would suggest a systematic rather than a purely random relation.
Frequency density distribution of RNAseq and quantified by mass spectometry (qMS) data from human platelets. The traces represent logarithmized original data from the publications.1,2 The lower threshold for RNAseq has been set to RPKM > 0.3 as proposed elsewhere.3
With respect to the articles cited and the reasoning of Rowley et al, we presume that the present discussion may partly result from a misunderstanding of the term “correlation.” Whereas there is no doubt that the presence of a transcript may on the whole serve as an indication for the expression of the related protein and vice versa, any definite or even quantitative claim is only possible by careful, direct observation. The numerous factors affecting the kind and number of transcripts in anucleated cells such as platelets, most of which are concealed from analysis, prohibit a valid quantitative assertion. In contrast, proteomic studies are suited to provide quantitative data on protein expression as we and others could unquestionably show,1 yet not to appraise the actual presence or absence of a particular protein. In consequence, it seems that neither of the 2 methods on its own is sufficient to meet the requirements of current and, most probably future, systems biology research on human platelets, though most probably nucleated cells may be investigated by both methods with comparable quality of results, as suggested by Nagaray et al.12
Authorship
Contribution: J.G. analyzed the statistical data and wrote the manuscript; J.M.B. collected and analyzed the data and edited the manuscript; S.G. provided study material and critically reviewed and edited the manuscript; U.W. and A.S. designed the study and critically reviewed and edited the manuscript; and R.P.Z. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joerg Geiger, Interdisciplinary Bank of Biomaterials and Data, Straubmuehlweg 2a/Bldg A9, 97078 Wuerzburg, Germany; e-mail: joerg.geiger@uni-wuerzburg.de.