Abstract
Mounting evidence indicates that grouping of chronic lymphocytic leukemia (CLL) into distinct subsets with stereotyped BCRs is functionally and prognostically relevant. However, several issues need revisiting, including the criteria for identification of BCR stereotypy and its actual frequency as well as the identification of “CLL-biased” features in BCR Ig stereotypes. To this end, we examined 7596 Ig VH (IGHV-IGHD-IGHJ) sequences from 7424 CLL patients, 3 times the size of the largest published series, with an updated version of our purpose-built clustering algorithm. We document that CLL may be subdivided into 2 distinct categories: one with stereotyped and the other with nonstereotyped BCRs, at an approximate ratio of 1:2, and provide evidence suggesting a different ontogeny for these 2 categories. We also show that subset-defining sequence patterns in CLL differ from those underlying BCR stereotypy in other B-cell malignancies. Notably, 19 major subsets contained from 20 to 213 sequences each, collectively accounting for 943 sequences or one-eighth of the cohort. Hence, this compartmentalized examination of VH sequences may pave the way toward a molecular classification of CLL with implications for targeted therapeutic interventions, applicable to a significant number of patients assigned to the same subset.
Introduction
The analysis of the Ig genes in chronic lymphocytic leukemia (CLL) has contributed significantly toward deciphering the molecular pathogenesis of the disease. Studies from the 1990s provided the first indications for a possible role of Ag(s) in selecting the CLL progenitor cells, through the discovery of a biased Ig heavy variable (IGHV) gene repertoire, different from that of normal B cells, as well as distinctive Ag-binding sites among unrelated cases.1-5
By the late 1990s, it emerged that the mutational status of the rearranged IGHV genes directly correlated with patient survival. In particular, patients with unmutated IGHV genes were found to follow a more aggressive clinical course and have significantly shorter survival than patients carrying mutated IGHV genes.6,7 Yet, there were exceptions to this rule: cases using the IGHV3-21 gene, although mostly expressing mutated Ig, had a survival similar to that of unmutated cases.8 Intriguingly, approximately half of the IGHV3-21 cases were found to display restricted and, in some instances, essentially identical variable heavy complementarity determining region 3 (VH CDR3) sequences and identical light chains, strongly suggesting recognition of a common antigenic determinant.9
Soon thereafter, the study of Ig sequences in CLL by groups in both Europe and the United States led to the identification of several other subsets of cases carrying highly similar BCR Igs among both mutated and unmutated cases (stereotyped BCR).10-14 The identification of stereotypy among unrelated and geographically distant cases was widely accepted as evidence for the recognition of individual, discrete Ags or classes of structurally similar epitopes, likely selecting the leukemic clones.10-13
Subsequently, the study of stereotypy in large cohorts revealed that a significant fraction of CLL cases (20%-28%) carried stereotyped VH CDR3 sequences within their BCR Ig,14-17 and, more importantly, that stereotypy may extend from restricted sequence patterns within the Ig to shared biologic and clinical characteristics and, perhaps, outcome.10,14,16 In addition, it was conclusively demonstrated that the IGHV gene repertoire restrictions typical of CLL were in essence a property confined to stereotyped cases, clearly segregating them from the “heterogeneous” (nonstereotyped) cases.17 Hence, it was proposed that ontogenies of the 2 CLL groups (stereotyped vs heterogeneous) as well as the selective forces shaping the Ig repertoire of the CLL precursor cell population(s) might differ.17
The nature of the selecting Ags and the functional consequences of their recognition cannot be directly deduced from primary Ig rearrangement features and have so far remained largely unknown. However, recent studies on BCR reactivity using CLL clones from stereotyped cases suggest that the expression of a stereotyped BCR may be linked to the reactivity profile of the CLL clone,18-21 and, eventually, to disease outcome.22
Against this background, several issues remain to be further addressed: (1) the criteria for the discovery of stereotypy and subset assignment; (2) the question as to whether stereotypy may be found for each BCR Ig provided enough cases are analyzed; (3) the number and size of different subsets; and (4) the identification of “CLL-biased” features among BCR Ig stereotypes with potential implications for disease ontogeny.
To address these issues, we systematically explored stereotypy based on VH CDR3 in a series of > 7000 VH (IGHV-IGHD-IGHJ) sequences from patients with CLL, 3 times the size of the largest published study. The results reported herein provide definitive evidence to our previous hypothesis17 that not all CLL cases will end up being part of stereotyped subsets or, in other words, that CLL indeed comprises 2 distinct categories: one with stereotyped and the other with heterogeneous Ig, in an approximate ratio of 1:2. The major stereotyped subsets collectively represent a substantial proportion of the respective category, with “CLL-biased” and often highly distinctive molecular features. Consequently, this deeper and compartmentalized view of BCR Ig primary structures in conjunction with biologic and clinical information may not only lead to a better basic understanding of the disease, but also potentially pave the way for tailored treatment strategies applicable to each major stereotyped subset.
Methods
Patient group
A total of 7424 patients diagnosed with CLL from collaborating institutions in Europe and the United States were included in the present study. All cases met the recently revised diagnostic criteria of the National Cancer Institute Working Group.23 The study was approved by the local ethics review committee of each institution.
PCR amplification of IGHV-IGHD-IGHJ rearrangements
Sequence analysis and data mining
PCR amplicons were subjected to direct sequencing on both strands. Sequence data were analyzed using the IMGT databases25 and the IMGT/V-QUEST tool (http://www.imgt.org).26 Codons and amino acid positions are according to the IMGT unique numbering for V domain.27 Only productive rearrangements were evaluated.
Output data from IMGT/V-QUEST for all productive IGHV-IGHD-IGHJ rearrangements were parsed, reorganized, and exported to a spreadsheet through the use of computer programming. Information was extracted regarding Ig gene repertoires, VH CDR3 length and amino acid sequence and somatic hypermutation (SHM), as previously described.15
Identification and clustering of IGHV-IGHD-IGHJ rearrangements based on stereotypy within VH CDR3 sequences
To identify and cluster stereotyped rearrangements, we used a purpose-built bioinformatics method, which has been previously described and applied with efficacy.17 As shown previously,17 VH CDR3 sequences were connected if sharing at least 50% amino acid identity and 70% similarity calculated through common sequence patterns. For the present study though, a novel parameter and more stringent criteria were introduced.
In particular, the application of the novel parameter was based on evidence that IGHV genes are phylogenetically related,28 that is, they originate from 3 ancestral phylogenetic clans: IGHV1/5/7 subgroup genes from clan I, IGHV2/4/6 from clan II, and, finally, IGHV3 genes from clan III.28 Consequently, we required that only sequences carrying IGHV genes of the same clan could be assigned to the same cluster, thus taking into account the role of the IGHV region in Ag recognition.
Furthermore, more stringent criteria were related to the 3-dimensional structure of the BCR and included the requirement for identical VH CDR3 lengths and identical offsets (ie, exact locations within the VH CDR3 region) of shared patterns between connected sequences.
The clustering process starts by building clusters on a first (ground) level, called level 0. Members of each level 0 cluster are guaranteed to meet all the aforementioned criteria for cluster assignment and can appear in more than one level 0 cluster, highlighting complex relationships. Finally, the existence of common amino acid patterns between sequences belonging to different level 0 clusters leads to their grouping in clusters at progressively higher levels of hierarchy describing more distant, and thus relaxed, sequence relationships with more widely shared sequence patterns. A schematic example of the hierarchical clustering process is given in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Data visualization tools
VH CDR3 amino acid patterns of different subsets were visualized using WebLogo (http://weblogo.berkeley.edu/). Each logo consists of stacks of symbols, one stack for each position of the sequence. CDR3 are shown from IMGT positions 105 to 117.27
Results
A synopsis of IGH gene repertoires, somatic hypermutation status, and VH CDR3 features
A total of 7596 VH (IGHV-IGHD-IGHJ) productive rearrangements from 7424 cases with CLL were evaluated; 172 cases appeared to carry double productive rearrangements. The IGHV, IGHD, and IGHJ subgroup and gene repertoires were generally similar to those reported in previous studies5,11-17 (listed in detail in supplemental Tables 1-3). Following the 98% germline identity (GI) cutoff value, 4171 (55%) of 7596 rearrangements were assigned to the mutated subgroup, whereas the remainder (3425 [45%] of 7596,) were classified as unmutated, of which 2516 (73.5%) of 3425 displayed 100% sequence identity against the corresponding germline IGHV gene sequences.
Focusing on rearrangements of the predominant IGHV genes, specific associations with certain IGHD and IGHJ genes were identified, often accounting for sizeable fractions of cases. This is best depicted by rearrangements using the IGHV1-69 and IGHV1-2 genes, which were strongly biased toward the usage of the IGHD3-3 gene (298 [30.6%] of 973) and the IGHD6-19 gene (80 [23%] of 348), respectively. On the contrary, less pronounced or no biases were noted for rearrangements using other frequent IGHV genes, namely IGHV4-34, IGHV3-7, and IGHV3-23. Specific combinations of IGHV and IGHJ genes were also identified: for example, IGHV1-69 and IGHV3-21 with IGHJ6 (542 [55.7%] of 973, and 248 [70.3%] of 353 cases, respectively). VH CDR3 median length was 17 aa (range, 5-37).
One-third of CLL cases were assigned to subsets with stereotyped VH CDR3
Through the use of our recently established bioinformatics method based on shared VH CDR3 amino acid sequence patterns17 herein updated with more stringent and novel criteria, 2308 (30.4%) of 7596 CLL VH CDR3 sequences were placed in 952 subsets at the ground level (level 0) that included 2-56 sequences each, that is, considered as stereotyped.
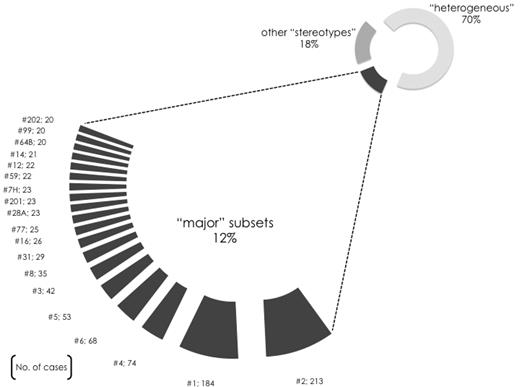
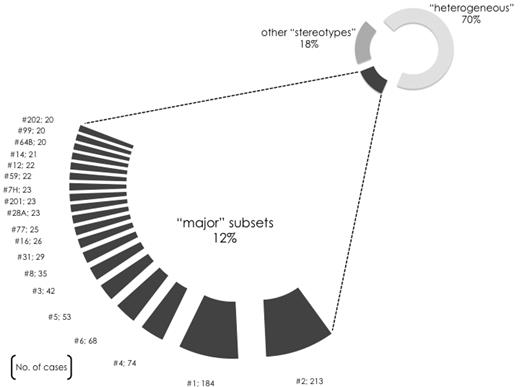
VH CDR3 sequences may contain multiple shared patterns and this allowed their concurrent assignment to different ground-level subsets. This overlap enabled the discovery of subsets at 3 successive hierarchically higher levels (supplemental Figure 1), characterized by more broadly shared sequence patterns and, hence, greater size. At the highest level, 19 different subsets contained 20 or more (up to 213) sequences and were defined as major (supplemental Tables 4-5): collectively, major subsets included 943 cases and, thus, accounted for 41% of the stereotyped cases and 12% of the cohort, respectively. In other words, 1 in 8 of all CLL patients belonged to a major subset (Figure 1).
A limited number of major subsets accounts for a sizeable proportion of the CLL Ig repertoire. Nineteen different subsets were identified in the present study containing 20 or more cases and defined as major. The relative size of each major subset (no. 1, 2, etc) is indicated in the graph, while their actual member sequences are listed in supplemental Table 5. Altogether, the 19 major subsets comprised 943 rearrangements in total and accounted for ∼ 41% of the stereotypes and for ∼ 12% of the cohort sequences, hence indicating that an important fraction of CLL cases can be represented by only few VH CDR3 stereotypes.
A limited number of major subsets accounts for a sizeable proportion of the CLL Ig repertoire. Nineteen different subsets were identified in the present study containing 20 or more cases and defined as major. The relative size of each major subset (no. 1, 2, etc) is indicated in the graph, while their actual member sequences are listed in supplemental Table 5. Altogether, the 19 major subsets comprised 943 rearrangements in total and accounted for ∼ 41% of the stereotypes and for ∼ 12% of the cohort sequences, hence indicating that an important fraction of CLL cases can be represented by only few VH CDR3 stereotypes.
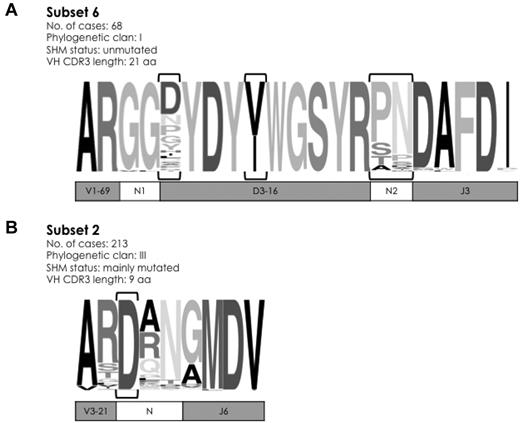
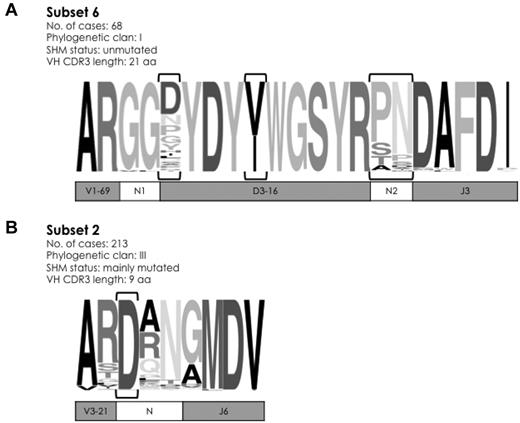
Aside from unique lengths, the widely shared sequence patterns characteristic of major subsets could comprise the entire VH CDR3 or a few, even a single, strategically positioned residue. A prime example of the former case is subset 6 that comprises cases with stereotyped, unmutated IGHV1-69/IGHD3-16/IGHJ3 rearrangements.12,21,22 In the present series, subset 6 included 68 rearrangements, thus accounting for 0.9% of the cohort, in keeping with the literature.12 Except for 4 CDR3 positions, which were characterized by variability, all other 17 positions were extremely, if not entirely, conserved. Notably, the first 2 aa (Gly G107, Gly G108) of the N1 region at the IGHV-IGHD junction were identical in 67 of 68 subset 6 cases (Figure 2A).
Sequence logos of selected major subsets in CLL. (A) Subset 6 comprises 68 unmutated IGHV1-69/IGHD3-16/IGHJ3 rearrangements, characterized by pronounced overall similarity. In fact, except for 4 VH CDR3 positions (encircled by brackets), which were characterized by variability, all other 17 positions were extremely, if not entirely, conserved. (B) Subset 2 is the largest high-level subset in the present study. Rearrangements belonging to this subset can be simply identified by a 9-aa long VH CDR3 with an acidic residue (aspartic acid D) at position 107 (encircled by brackets). The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27
Sequence logos of selected major subsets in CLL. (A) Subset 6 comprises 68 unmutated IGHV1-69/IGHD3-16/IGHJ3 rearrangements, characterized by pronounced overall similarity. In fact, except for 4 VH CDR3 positions (encircled by brackets), which were characterized by variability, all other 17 positions were extremely, if not entirely, conserved. (B) Subset 2 is the largest high-level subset in the present study. Rearrangements belonging to this subset can be simply identified by a 9-aa long VH CDR3 with an acidic residue (aspartic acid D) at position 107 (encircled by brackets). The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27
At the opposite end stands subset 29,14 with 213 IGHV3-21–expressing CLL cases (2.8% of the cohort) characterized by a very short VH CDR3 sequence (9 aa) involving the IGHJ6 gene and no identifiable IGHD gene. Interestingly, at the center of the short VH CDR3, virtually all subset 2 cases (211 [99%] of 213) carried a landmark amino acid (aspartic acid, D) at position 107, between the amino acids qualified as IGHV- or IGHJ-encoded; 2 cases lacking an aspartic acid were found to carry a glutamic acid (Glu E) at the same position, that is, an amino acid of the same IMGT physicochemical class29 (both acidic; Figure 2B). On these grounds, CLL IGHV3-21 rearrangements can be assigned to subset 2 if meeting only 2 criteria: (1) restricted VH CDR3 length of 9 aa, and (2) a “landmark” acidic amino acid (D or E) at VH CDR3 position 107.
Two types of CDR3 sequence patterns define CLL subsets
Shared sequence patterns defining subsets were broadly divided into 2 types: (1) “mainly combinatorial,” that is, largely encoded by unmutated sequences of the D region and 5′J-region of specific combinations of IGHD-IGHJ genes, and (2) “combinatorial + junctional,” that is, encoded in part by the N-diversity regions (N1 and N2) generated by junctional diversity mechanisms (as a result of P nucleotide addition, exonuclease trimming of the 3′ IGHV, 5′ and 3′ IGHD, and 5′ IGHJ gene ends and N nucleotide addition) leading to restricted motifs at the IGHV-IGHD and/or IGHD-IGHJ gene junctions.
Subset 810,14 epitomizes the mainly combinatorial type of VH CDR3 sequence patterns. The stereotyped unmutated heavy chains are encoded by the specific combination of the IGHV4-39/IGHD6-13/IGHJ5 genes (IGHD gene in reading frame 1) and carry VH CDR3s with a length of 19 aa (Figure 3A). The junctional N1 (IGHV-IGHD) and N2 (IGHD-IGHJ) regions were heterogeneous, yet this restricted combination of IGHV, IGHD, and IGHJ genes proved to be “subset 8–specific” and “CLL-specific, as revealed by the extensive examination of IGH rearrangements deposited in public generalist and specialist (IMGT/LIGM-DB30 ) databases.
Two types of subset-defining VH CDR3 sequence patterns. (A) Mainly combinatorial. The pattern typical of subset 8 is exclusively composed of amino acids encoded by the unmutated D region of the IGHD6-13 and 5′J region of the IGHJ5 genes, whereas the junctional N-diversity regions (N1 and N2) are diverse. (B) Combinatorial+junctional. The pattern defining subset 4 consists of the junctional N2 amino acids [KR]R at positions 112.4 (tip of the CDR3 loop) and 112.3 and of the IGHJ6-encoded motif YYYYG. The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27
Two types of subset-defining VH CDR3 sequence patterns. (A) Mainly combinatorial. The pattern typical of subset 8 is exclusively composed of amino acids encoded by the unmutated D region of the IGHD6-13 and 5′J region of the IGHJ5 genes, whereas the junctional N-diversity regions (N1 and N2) are diverse. (B) Combinatorial+junctional. The pattern defining subset 4 consists of the junctional N2 amino acids [KR]R at positions 112.4 (tip of the CDR3 loop) and 112.3 and of the IGHJ6-encoded motif YYYYG. The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27
Exemplifying the combinatorial + junctional type of VH CDR3 patterns is subset 4,11,14 represented by 74 cases (0.97% of the cohort sequences). The stereotyped mutated IGH chains are generated through the combination of the IGHV4-34 and IGHJ6 genes (reliable IGHD gene assignment was not possible because of heterogeneity at the tip of the VH CDR3 loop, possibly because of SHM). Interestingly, in subset 4, conserved amino acids characterize the N regions. Thus, the N1 region comprises glycine G107 and tyrosine Y108 (or the other aromatic amino acids, phenylalanine F or tryptophan W) whereas the N2 region has 2 arginine R at positions 112.4 and 112.3 (in 13 of 74 sequences, the arginine R112.4 located at the tip of the CDR3 is replaced by a lysine K, another basic amino acid; Figure 3B). Public database examinations revealed the [RK]RYYYY pattern that reflects the junctional N2 conserved amino acids and the IGHJ6 use to be “subset 4–specific” and “CLL-specific.” Other examples of major subsets defined by mainly combinatorial and combinatorial + junctional patterns are given in supplemental Table 6.
Subset-defining sequence patterns in CLL differ from those underlying VH CDR3 stereotypy in other B-cell malignancies
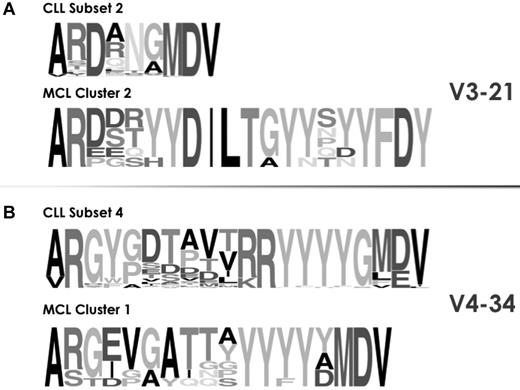
Recent studies by us and others have documented the existence of stereotyped BCR Ig in mantle cell lymphoma (MCL)31 and splenic marginal-zone lymphoma (SMZL),32,33 albeit at significantly lower frequencies than CLL. We therefore sought to investigate whether the stereotypes observed in CLL are disease-biased or common by performing cross-entity comparisons.
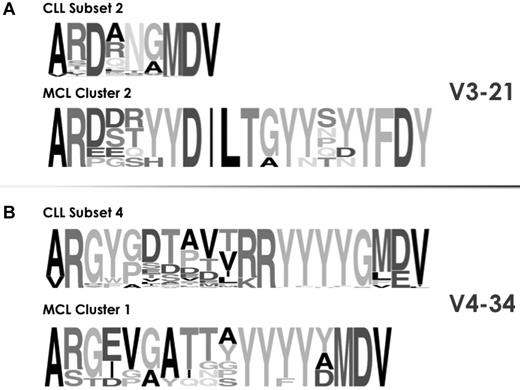
In the case of MCL, stereotyped IGHV-IGHD-IGHJ rearrangements collectively represent ∼ 10% of the cohort31 and display extreme restrictions in IGHV gene utilization, with the IGHV4-34 and IGHV3-21 genes accounting for 67% of all such cases. Stereotyped MCL subsets were compared with major CLL subsets using the same IGHV genes, namely subsets 4, 16 (both IGHV4-34), and 2 (IGHV3-21). In all comparisons, significant differences were identified in terms of IGHD and IGHJ gene usage, VH CDR3 sequence length and amino acid composition (Figure 4).
Stereotypes in CLL are disease-biased. As an example, the cross-entity comparison of VH CDR3 sequences among rearrangements from CLL and MCL using the same IGHV genes showed clear differences in a series of molecular features: IGHD and IGHJ gene utilization and also VH CDR3 length and amino acid composition. The height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. Amino acid position is according to the IMGT numbering for V domain.27
Stereotypes in CLL are disease-biased. As an example, the cross-entity comparison of VH CDR3 sequences among rearrangements from CLL and MCL using the same IGHV genes showed clear differences in a series of molecular features: IGHD and IGHJ gene utilization and also VH CDR3 length and amino acid composition. The height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. Amino acid position is according to the IMGT numbering for V domain.27
Differences were even more fundamental when CLL stereotypes were compared with those identified in SMZL, in this case comparing stereotypes using the IGHV1-2 gene, which by far predominates in SMZL.32,33 Hence, BCR Ig VH stereotypes in CLL were significantly different from “related” ones in MCL or SMZL, even though they expressed the same IGHV gene, and can thus be considered as disease-biased (supplemental Table 7).
Even if numbers increase significantly, not all CLL cases can be classified as stereotyped
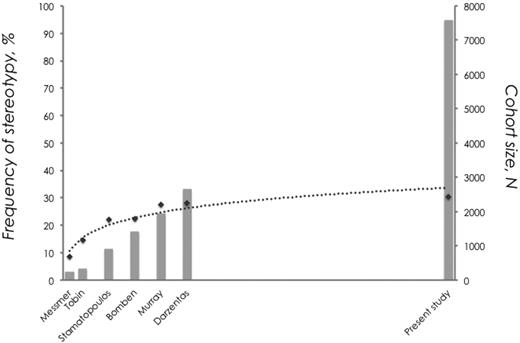
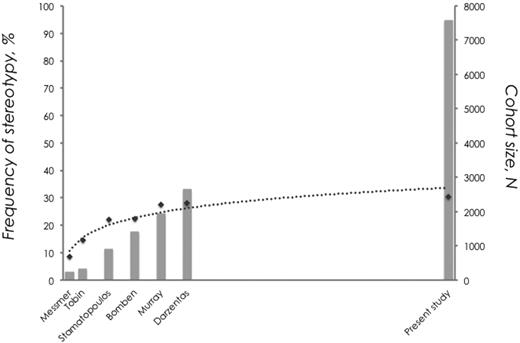
From this large study involving > 7000 sequences, it became evident that the frequency of stereotyped rearrangements in CLL is not directly correlated to cohort size. Despite a significant increase in sample size between the present series and previous studies (including up to 2662 sequences),11,13-17 the increase in the frequency of stereotypy is only marginal (30.4% vs 28%, respectively). In other words, although the proportion of stereotyped BCR Ig VH in CLL seems to continuously increase with larger cohort sizes, the rate of this increase starts to drop after a critical value of cohort size until the frequency of stereotypy plateaus at ∼ 30% of the cohort, independent of cohort size (Figure 5).
CLL Ig repertoire: one-third stereotyped, two-thirds heterogeneous. A continuous increase in cohort size results in a nonproportional increase in the frequency of VH CDR3 stereotypy. This is best depicted when considering the fact that despite a significant increase in sample size between the present series and the largest published series (almost 5000 additional cases), the increase in the frequency of stereotypy was only 2.4% (data are shown using a logarithmic trendline).
CLL Ig repertoire: one-third stereotyped, two-thirds heterogeneous. A continuous increase in cohort size results in a nonproportional increase in the frequency of VH CDR3 stereotypy. This is best depicted when considering the fact that despite a significant increase in sample size between the present series and the largest published series (almost 5000 additional cases), the increase in the frequency of stereotypy was only 2.4% (data are shown using a logarithmic trendline).
To corroborate this finding, while avoiding potential bias because of slightly different criteria to define stereotypy across the published studies, we performed random set simulations using the latest version of the algorithm and the current cohort of VH CDR3 sequences. Indeed, we found that above a critical value of cohort size of 2000 sequences, each addition of a thousand sequences resulted in an increase in the frequency of stereotypy by a steadily decreasing and disproportional rate (supplemental Table 8).
Distinctive immunogenetic characteristics and CLL ontogenetic implications
The present analysis led to the discovery of several noteworthy immunogenetic features, with implications for CLL ontogeny and Ag selection.
Subset 10: virtually identical VH CDR3 sequences.
VH CDR3 sequence homogeneity reached its maximum potential for cases assigned to subset 10. This subset included 18 sequences expressing stereotyped, unmutated IGHV4-39/IGHD2-2/IGHJ6 Ig and is quite remarkable for the fact that the defining sequence pattern covered all 22 aa of the VH CDR3. In fact, only 10 amino acid differences were identified in a total of 396 VH CDR3 amino acid residues (2.5%) of subset 10 cases, rendering it the most homogeneous high-level subset thus far described (supplemental Figure 2).
Subsets with different IGHV genes of the same clan.
Subset 59 comprised 21 unmutated cases with stereotyped VH CDR3 using one of the IGHV1-2, IGHV1-58, or IGHV1-69 genes (2, 8 and 12 sequences, respectively), which are all members of the IGHV1 subgroup. Further exemplifying and extending this principle, subset 12 consisted of 22 cases with stereotyped VH CDR3 using one of the IGHV1-2, IGHV1-46, or IGHV5-a genes (11, 10, and 1 sequences, respectively), which, though belonging to different IGHV subgroups (namely, IGHV1 and IGHV5), are members of the same clan of IGHV genes (clan I), that is, they still exhibit significant sequence similarity. This may indicate that amino acids of the CDR1 and/or CDR2 may contribute, in addition to the CDR3, to the Ig specificity. Subsets 59 and 12, along with additional examples, are presented in supplemental Table 9.
Closely related subsets using different IGHV genes.
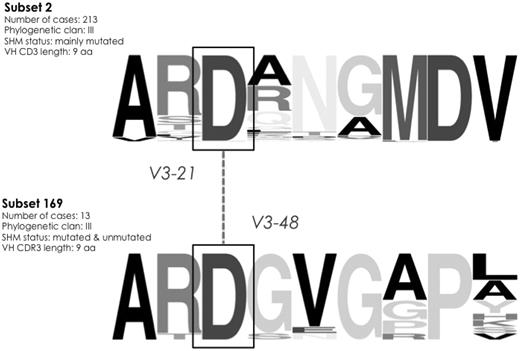
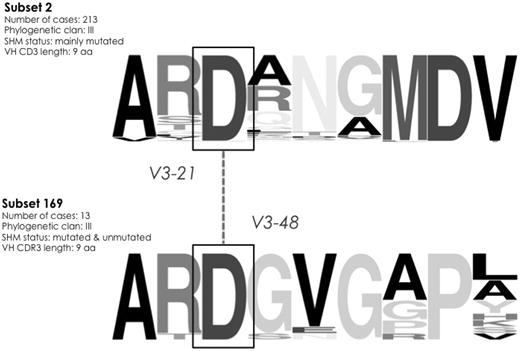
The IGHV3-48 gene is very similar to the IGHV3-21 gene (overall amino acid identity 97%). Thirteen rearrangements of the cohort using the IGHV3-48 gene were assigned to a novel subset (169) with a short, 9-aa long VH CDR3, that is, identical in size to that of subset 2, which includes stereotyped rearrangements of the IGHV3-21 gene. Intriguingly, as in subset 2, all subset 169 rearrangements carried an aspartic acid D107. The fact that sequences from these 2 subsets were not finally grouped together indicates that they differ to an extent that did not satisfy the algorithmic criteria. However, the similarity of the IGHV3-21 and IGHV3-48 genes along with identical VH CDR3 length and the presence of D107, suggests that the 2 subsets 169 and 2 may be closely related (Figure 6).
Intriguing sequence similarities between different high-level subsets. VH CDR3 sequences grouped to subsets 2 and 169 share molecular characteristics: a VH CDR3 composed of 9 aa and an aspartic acid (D) residue at position 107. Furthermore, the IGHV3-48 gene (utilized by all 169 rearrangements), is highly similar to the IGHV3-21 gene. The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27
Intriguing sequence similarities between different high-level subsets. VH CDR3 sequences grouped to subsets 2 and 169 share molecular characteristics: a VH CDR3 composed of 9 aa and an aspartic acid (D) residue at position 107. Furthermore, the IGHV3-48 gene (utilized by all 169 rearrangements), is highly similar to the IGHV3-21 gene. The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27
Sequence convergence induced by somatic hypermutation.
We and others have previously shown that sequences assigned to subsets with stereotyped VH CDR3 can also exhibit identical amino acid changes (as a result of SHM across the IGHV region11,15 ; supplemental Table 10), clearly indicating that certain positions throughout the VH domain of cases belonging to the same subset seem to be under selective pressure for specific changes.
We here extend this notion showing that certain subsets with stereotyped VH CDR3 expressing different, yet related, IGHV genes can exhibit SHM-derived sequence convergence. This is illustrated by subset 77, which consists of 25 mutated stereotyped rearrangements using either the IGHV4-4 or the IGHV4-59 gene. In selected positions where the germline sequences of the above IGHV genes varied, we noted that the changes introduced by SHM could lead to higher sequence identity, hence eliminating germline-encoded variation: for instance, 4 of 13 IGHV4-59 cases carried a tyrosine (Y) to histidine (H) change at VH CDR2 codon 58 (Y58 > H), resembling the IGHV4-4*02 and IGHV4-4*07 alleles; moreover, 16 (67%) of 24 of all subset 77 cases carried an isoleucine (I) to methionine (Met M) change at VH FR3 codon 78 (I78 > M), regardless of whether they expressed the IGHV4-4 or the IGHV4-59 gene (supplemental Figure 3). These findings could not be attributed to ambiguous IGHV germline gene assignment because rearrangements using one or the other gene retained “gene-specific” features (supplemental Table 11).
Discussion
Emerging data suggest that grouping CLL cases into subsets based on BCR Ig stereotypy can reflect similar Ag-reactivity profiles22 and similar clinical outcomes,14,16 at least for certain subsets. Therefore, the study of stereotypy in CLL has important implications for both understanding the pathogenesis of the disease34 and, most likely, for improving patient stratification toward personalized therapeutic applications.
Here, we analyzed the Ig gene repertoire of 7424 patients with CLL to address several open issues related to: (1) the proper criteria for stereotypy detection and eventual subset assignment; (2) the exact magnitude of and future projections about BCR Ig stereotypy; (3) the identification of CLL-biased features in BCR Ig stereotypes. This latter issue is particularly timely given recent reports by several groups, including ours, regarding the existence of BCR Ig stereotypes in the normal repertoire35 as well as in other B-cell malignancies, namely MCL31 and SMZL.32,33
To answer these questions, we applied an updated version of our published clustering method,17 purpose-built for the identification of shared amino acid sequence patterns within VH CDR3 sequences and their subsequent clustering. Accumulating evidence from the recent literature,35,36 combined with our experience from similar analyses, led us to adopt more stringent subset-defining criteria than we did previously, so that the identified primary amino acid sequence relatedness may correspond to actual structural similarity.37,38 The revised criteria mainly related to (1) VH CDR3 length, known to be a critical determinant of the structure of the Ag recognition loop,39 and (2) the exact location of the shared pattern within the VH CDR3 region, given ample evidence that the positioning of certain amino acids may affect Ig structure stabilization.40,41 For these reasons and to ensure maximum accuracy, we followed a conservative approach and required for same-subset assignment at the ground level no less than total identity in VH CDR3 length and offset of shared patterns. Through this approach, groups of rearrangements using the same IGHV, IGHD, and IGHJ genes with highly similar VH CDR3 sequences, yet slightly dissimilar VH CDR3 lengths, were eventually clustered in related yet different subsets (supplemental Table 12). This maximizes homogeneity and is now our preferred strategy for subset assignment, at least until other types of evidence (functional, structural, clinical) become available to support that rearrangements should be placed in a single subset irrespective of slight VH CDR3 length variation, as has been the case so far for subset 1.11,14,16
The novel parameter introduced in our clustering algorithm required that only sequences carrying IGHV genes of the same clan could be placed in the same subset. This development takes into account the role of the CDR1 or CDR2 amino acids in classic Ag recognition as well as the binding of specific classes of Ags41 (ie, superantigens, bacterial polysaccharides) to amino acids of the framework regions. Its implementation highlighted several subsets of cases that express different yet phylogenetically related IGHV genes, including the well-established subset 111,14,16 (IGHV genes belonging to clan I) as well as other cases, for example, subsets 12 (IGHV1-2 and IGHV1-46), 59 (IGHV1-58 and IGHV1-69), and 77 (IGHV4-4 and IGHV4-59). The latter, including cases with mutated Ig, is also remarkable for the fact that distinct yet related IGHV genes, when rearranged, can eventually become more similar through functionally selected SHM. In other words, starting from different germline-encoded genes, the affinity maturation process can reduce germline sequence disparities leading to sequence convergence.
Altogether, it now becomes clear that the existence of CLL subsets with stereotyped VH CDR3s using different, yet related, IGHV genes is not a peculiarity but a rather general phenomenon. Potentially, these related genes might also be prefavored for selection in response to specific Ags and amenable to stereotypy. In addition, it may mean that a few critical IGHV-encoded positions are important for defining the specificity of certain CLL Ig and, hence, their ability to recognize Ag and therefore to be selected. This is similar to what has been described for recombinant mAbs using CDR shuffling approaches42 and, very recently, for potent broadly neutralizing CD4-binding site anti-HIV Abs that mimic binding to CD4.43 Intriguingly, in the latter case, despite extensive SHM, the anti-HIV Abs shared a consensus motif of 68 aa in the VH domain and arose independently from 2 distinct yet related IGHV genes (IGHV1-2 and IGHV1-46, also used in major CLL stereotyped subsets, eg, 1 and 12).
Reviewing previous studies of BCR Ig stereotypy in CLL reporting on incrementally larger series of patients (starting from 255 and peaking at 2662 sequences),11-17 a trend was apparent toward a rise in the percentage of stereotyped cases (from 8.6% up to 28%) along with the increase in the size of the series. Although not proportional to the increase of the cohort, in principle, this upward trend might continue to rise to an unknown figure if the series of cases available for analysis would continue to expand. The large size of our cohort—3 times the size of the largest published study17 —enabled us to reliably investigate this issue and ascertain the mathematical relationship of BCR Ig stereotypy with cohort size. Our findings support the notion that not all CLL cases will eventually be clustered: in fact, only a fraction of CLL—approximately one-third of cases—could be assigned to subsets defined by stereotyped VH CDR3 with the most recent criteria, a ratio also supported by random simulations. Hence, overall, the present study reinforces our earlier claim that CLL can be subdivided into 2 broad categories: one with stereotyped BCR Ig and the other displaying heterogeneity in terms of BCR Ig sequence features.17
Given that some of the participating institutions of the present study are secondary referral centers and, thus, have a larger proportion of cases at advanced stages, the relative sizes of the IGHV-mutated and IGHV-unmutated subgroups reported here appear to differ somewhat from those found in everyday clinical practice (community CLL). Therefore, the relative frequencies of individual subsets may be slightly different from those expected in community CLL. That notwithstanding, considering that similar frequencies have been reported previously by us and others for several major subsets,12,14-17 it is reasonable to claim that, collectively, just a few subsets account for a sizeable proportion of the CLL cohort.
The recent identification of BCR VH CDR3 stereotypes in other B-cell malignancies, namely MCL31 and SMZL,32,33 abrogated the notion that stereotypy by itself is CLL-unique. Furthermore, it raised questions regarding the possible existence of common immune-mediated mechanisms of lymphomagenesis among different entities, reflected in shared Ag-binding sites.44 To address these questions, we took advantage of our recently published, large Ig sequence datasets from MCL31 and SMZL33 and performed cross-entity comparisons to search for commonalities between their Ag-binding sites that might allude to common immune pathways to lymphoma development. Through this approach, it became apparent that differences were fundamental regarding both VH CDR3 length and composition. We interpret this finding as an indication that distinct functional and/or developmental processes act to shape the respective Ig repertoires. A corollary of these comparisons is that the characteristics of VH CDR3 stereotypes in CLL can be considered as truly disease-biased. Needless to say, these rearrangements are expected to be present within the repertoire of normal B cells, as reported in the literature,35 however it remains scientifically interesting and unique that few specific ones are expanded repeatedly and consistently in the same lymphoma subtype.
A major such characteristic is the VH CDR3 sequence itself. In certain instances, CLL-biased sequence patterns extended over the entire VH CDR3 sequence (eg, in subset 10); in contrast, in other instances, only a few VH CDR3 amino acids seemed to be sufficient for defining a subset. The most impressive example of the latter case is subset 2, where subset assignment is reliable provided the following 3 requirements are met: (1) IGHV3-21/IGHJ6 gene association; (2) 9-aa long VH CDR3; and, most intriguingly (3) an acidic residue at VH CDR3 position 107. In other words, the defining features of subset 2 IGHV3-21 BCR are VH CDR3 length restriction and sequence restriction at a single landmark position. In keeping with previous reports for certain mAbs with defined specificities,45 these 2 features might be linked, in that VH CDR3 length restriction could ensure proper positioning of a specificity determinant within the VH CDR3 itself or of critical contacts between the heavy and light chains.
The finding of this and several other examples of CLL BCR stereotypes which are defined by constraints in VH CDR3 length and composition in a few critical positions, should in no way be taken to imply that the actual sequence of the entire VH CDR3 is less important for defining specificity toward the cognate Ag. However, it leaves open the possibility of selection of a paratope endowed with adequate plasticity for binding a range of diverse ligands, recalling what has been reported in the mouse for Abs against small haptens or carbohydrates, where a preferred length usually dominates for a given specificity.46
What could be the biologic and practical implications of these findings? From an ontogenetic perspective, it is worth underscoring the molecular analogies of the Ag-binding sites in CLL with stereotyped BCR Ig with those of murine B-1 cells. These cells have been reported to exhibit a different and considerably more restricted Ig repertoire than conventional B cells, distinctive for the frequent occurrence of identical Ig heavy and light chain rearrangement.47 Their repertoire and reactivity pattern seems to be stable within each species and even between species, likely reflecting evolutionary selection for specificities which would afford protection against pathogen invasion and also serve housekeeping functions of recognizing and removing everyday apoptotic material.47 Interestingly, a similar reactivity profile has been identified for CLL mAbs that can be polyreactive18,19 (in particular when unmutated) and have been shown to react with molecular structures present on apoptotic cells and bacteria.19-21,48 Whether these observations and facts are relevant for CLL ontogeny, especially in view of a recent report describing human fundamental cells with functional properties of B-1 cells,49 is currently unknown and will require further multidisciplinary research.
From a practical perspective, the sheer size of the stereotyped CLL fraction has important potential clinical implications. This is supported by independent recent studies which have shown striking correlations between certain CLL stereotypes and disease presentation, course, or outcome, for example, young age at diagnosis and remarkably indolent course for subset 4,14-16 significantly increased risk for Richter transformation for subset 8,50 or adverse clinical course for subsets 1 and 2,14,16 even compared with cases with similar IGHV gene mutational status. Hence, it seems reasonable to propose that reliable recognition of stereotypy may soon be instrumental in patient risk stratification, perhaps even beyond IGHV gene mutational status, and, eventually, form the basis for the development and implementation of “subset-specific” therapeutic protocols.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the following members of the collaborating institutions for their help with sample processing: Vasiliki Douka, Hana Skuhrova Francova, Evangelia Stalika, Dennis Tielemans, Ashley van der Spek, Brenda Verhaaf, Joyce Vermeulen, and Ingrid L. M. Wolvers Tettero.
This work was supported in part by the ENosAI project (code 09SYN-13-880) co-funded by the European Union and the Hellenic General Secretariat for Research and Technology (N.D., K. Stamatopoulos); the Cariplo Foundation, Milan, Italy (P.G., K. Stamatopoulos); Program Molecular Clinical Oncology-5 per mille no. 9965 and Investigator grant, Associazione Italiana per la Ricerca sul Cancro (AIRC), Milano, Italy (P.G.); the National Cancer Institute/National Institutes of Health RO1 CA81554 (N. Chiorazzi); the Nordic Cancer Union, the Swedish Cancer Society, the Swedish Research Council, the Uppsala University Hospital, and the Lion's Cancer Research Foundations in Uppsala (R.R.) for the analysis of the Swedish cases; and grant IGA MZ CZ NS10439-3/2009 by the Ministry of Health of the Czech Republic (S.P.) for the analysis of the Czech cases. A. Agathangelidis is the recipient of a grant from the A. G. Leventis Foundation (http://www.leventisfoundation.org/).
National Institutes of Health
Authorship
Contribution: A. Agathangelidis performed research, analyzed data, and wrote the manuscript; N.D., A.W.L., D.G.O., N. Chiorazzi, C.B., F.D., R.R., and P.G. designed the study and supervised research; A.H. performed research and analyzed data; X.B. and V.G. were responsible for the curation of IGHV-IGHD-IGHJ sequences; F.M., X.-J.Y., Z.D., E.J.v.G.-M., C.T., C.C.C., N. Cahill, B.T., L.B.P., L.F., L.B., A.J., and P.F.d.C. performed research; K. Smedby, A.T., and M.-P.L. supervised research; A. Anagnostopoulos, H.M.-B., N.L., G.J., S.P., and J.J. provided samples and associated clinicopathologic data and supervised research; and K. Stamatopoulos designed the study, supervised research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Jesper Jurlander, one of the co-authors of the present contribution, unexpectedly passed away during the preparation of the manuscript. This work is dedicated to his memory.
Correspondence: Paolo Ghia, MD, PhD, Università Vita-Salute San Raffaele, Via Olgettina 58, 20132 Milano, Italy; e-mail: ghia.paolo@hsr.it.

![Figure 3. Two types of subset-defining VH CDR3 sequence patterns. (A) Mainly combinatorial. The pattern typical of subset 8 is exclusively composed of amino acids encoded by the unmutated D region of the IGHD6-13 and 5′J region of the IGHJ5 genes, whereas the junctional N-diversity regions (N1 and N2) are diverse. (B) Combinatorial+junctional. The pattern defining subset 4 consists of the junctional N2 amino acids [KR]R at positions 112.4 (tip of the CDR3 loop) and 112.3 and of the IGHJ6-encoded motif YYYYG. The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/19/10.1182_blood-2011-11-393694/4/m_zh89991290020003.jpeg?Expires=1768101498&Signature=rH63DGOXcAQYNtFESAC4fTWafa2IzOuSrKnqo~CjcHXe2M2HjtTcOloLbZ5q2yeR9onF4IbiS1tCV3pGYw-jdBqOPc7RLhMHHyjqMDryz7OqiU5ESKzyGCJzFMqiEwabeGRaZxJ~ldSU7H4NC6Zr8Ss5UllLIXafXHWuOgUIyL2hxwXmW0vbuPDyY7FGbKNf~JXrwbwLvZyfKLltR78A3OXmk3UvxBlKBuPwnTqJOrJl1Ue~ZqjsRZsU-4qDVjS-ct7GrO0b2YOdnBvJrwZY0Lb2e2O-heXXHGwyikrIlMF2lJxrbNIeuRukCau778VKYoFbAYAGBdYl2pCwhAaHlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Two types of subset-defining VH CDR3 sequence patterns. (A) Mainly combinatorial. The pattern typical of subset 8 is exclusively composed of amino acids encoded by the unmutated D region of the IGHD6-13 and 5′J region of the IGHJ5 genes, whereas the junctional N-diversity regions (N1 and N2) are diverse. (B) Combinatorial+junctional. The pattern defining subset 4 consists of the junctional N2 amino acids [KR]R at positions 112.4 (tip of the CDR3 loop) and 112.3 and of the IGHJ6-encoded motif YYYYG. The height of symbols within the stack indicates the relative frequency of each amino acid at that position. Amino acid position is according to the IMGT numbering for the V domain.27](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/19/10.1182_blood-2011-11-393694/4/m_zh89991290020003.jpeg?Expires=1768227996&Signature=on50~rshDnQy0I9zo2A6q5JWvlonCg1CiJygT3lzbONqxmHdvBn5kGLjX4ZQAAG7D4fP-p7dWJ6kVdZ7EQpe~gQ6-oq~3dTFOfb6ny9r1-PVJpt5Cy5DDyfTI5spk8Tgpv5r3Y0XLsoPQWyIhbLSHLck8oej6WpNIQvuYso4iCZyLeI5z-kZC-Vn3yug8fty7ulbYyjirEE9aRmhQU-lGmlmS1Docmd~3lrTgEeD5rEQHZydE0hipYm5lFOlwfwoej63dmWdzl79IjHMZCnRvrV9vPYlUlGTYkb47HryNgVCSsp4iJowI9b1dm~POc~fG9mfTtBHLADSFguf0-1ddQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)