Abstract
To identify rational therapeutic combinations with cytarabine (Ara-C), we developed a high-throughput, small-interference RNA (siRNA) platform for myeloid leukemia cells. Of 572 kinases individually silenced in combination with Ara-C, silencing of 10 (1.7%) and 8 (1.4%) kinases strongly increased Ara-C activity in TF-1 and THP-1 cells, respectively. The strongest molecular concepts emerged around kinases involved in cell-cycle checkpoints and DNA-damage repair. In confirmatory siRNA assays, inhibition of WEE1 resulted in more potent and universal sensitization across myeloid cell lines than siRNA inhibition of PKMYT1, CHEK1, or ATR. Treatment of 8 acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and chronic myeloid leukemia (CML) cell lines with commercial and the first-in-class clinical WEE1 kinase inhibitor MK1775 confirmed sensitization to Ara-C up to 97-fold. Ex vivo, adding MK1775 substantially reduced viability in 13 of 14 AML, CML, and myelodysplastic syndrome patient samples compared with Ara-C alone. Maximum sensitization occurred at lower to moderate concentrations of both drugs. Induction of apoptosis was increased using a combination of Ara-C and MK1775 compared with using either drug alone. WEE1 is expressed in primary AML, ALL, and CML specimens. Data from this first siRNA-kinome sensitizer screen suggests that inhibiting WEE1 in combination with Ara-C is a rational combination for the treatment of myeloid and lymphoid leukemias.
Introduction
Ara-C (cytarabine) is the backbone of acute myeloid leukemia (AML) therapy and has activity as a single agent and in combination with other drugs, including anthracyclines and epipodophyllotoxins.1,2 High-dose Ara-C is essential for consolidation in certain patient groups1 and can salvage and even cure AML. However, Ara-C as a single agent in relapsed and refractory AML is insufficiently active, so there is substantial interest in finding agents that can be combined productively with Ara-C.
Several experimental Ara-C combinations have been tested clinically without major improvements in overall outcomes.3 Apart from using higher doses of anthracyclines in combination with Ara-C in first-line induction treatment of younger patients,4 no regimen has improved survival of AML patients significantly. Therefore, there is considerable room to improve the activity and efficacy of Ara-C, especially in the large group of elderly and relapsed refractory AML patients.5 However, an optimal combination partner to enhance the activity of Ara-C has yet to be determined.
Multiple kinases are aberrantly activated in AML and can contribute to leukemogenesis.6 However, which kinases might need to be inhibited to enhance the activity of Ara-C in leukemias is not known. Current research approaches identifying optimal Ara-C combination partners are limited by throughput. RNA interference (RNAi) screening is a methodology that results in loss-of-function of targeted transcripts (mRNA), which can be used to identify genes that when silenced sensitize to chemotherapeutic agents. Advantages of the RNAi approach are: (1) the high-throughput capacity to interrogate many genes simultaneously; (2) the unbiased nature, allowing target identification despite the “black box” of numerous underlying oncogenic events such as mutations, amplifications, and epigenetic or posttranslational modifications; and (3) the practicality of the screens. Functionally identifying kinases, the inhibition of which by small-interference RNA (siRNA) potentiates the activity of cytotoxics, can accelerate selection and prioritization of top candidate genes to target pharmacologically in combination with Ara-C. Because many novel targeted agents are kinase inhibitors, we hypothesized that a large-scale RNAi screen of the human kinome7-9 would be a rapid, cost-effective, “unbiased,” functional genomic approach to identify candidate targets for combination regimens with Ara-C.
To date, few RNAi screens have been reported in leukemia cells,10,11 and to our knowledge, none have screened for modulation of Ara-C sensitivity in myeloid leukemias. We adapted a siRNA-screening platform in our laboratory7,8,12,13 that uses transfection conditions without electroporation to myeloid suspension cells and performed the first kinome-wide siRNA–Ara-C sensitizer screen in leukemia cells. WEE-1 kinase family genes (WEE1 and PKMYT1) and CHEK1 emerged as top targets potentiating Ara-C activity. In particular, inhibition of WEE1 kinase by siRNA and pharmaceutical WEE1 inhibitors, including the first-in-class WEE1 inhibitor MK1775 in clinical development, potently sensitized myeloid and lymphoid leukemia cells to Ara-C in vitro and ex vivo.
Methods
Cell lines, reagents, and culture conditions
The human cell lines TF-1, THP-1, HEL, MV-4-11, HL-60, K-562, and Jurkat were obtained from the ATCC or DSMZ (www.dsmz.de), and GM03671 was obtained from the Coriell Institute for Medical Research. MDS-L is a myelodysplastic syndrome (MDS) cell line transformed to AML and was kindly provided by Professor Kaoru Tohyama (Department of Laboratory Medicine, Kawasaki Medical School, Kurashiki, Japan).14,15 All tissue-culture reagents were obtained from Invitrogen. Cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 2mM l-glutamine, 100 IU/mL of penicillin, and 100 μg/mL of streptomycin at 37°C under a 5% CO2 atmosphere. Primary patient samples were collected according to Institutional Review Board–approved protocols, separated using Ficoll gradient centrifugation, and cultured in RPMI 1640 medium plus 10% FBS. Ara-C and WEE1 inhibitor I, 4-(2-chlorophenyl)-9-hydroxypyrrolo[3,4-c]carbazole-1,3-(2H,6H)-dione, were obtained from EMD Chemicals. MK1775 was provided by Merck.
High-throughput siRNA screens
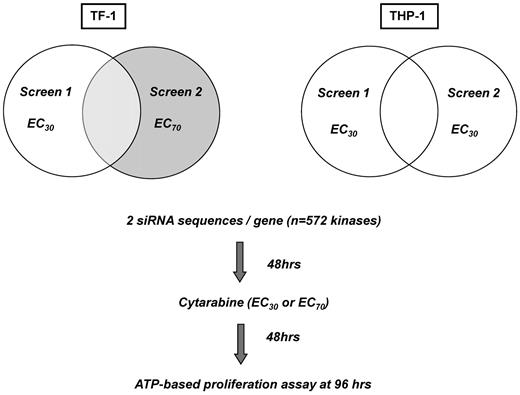
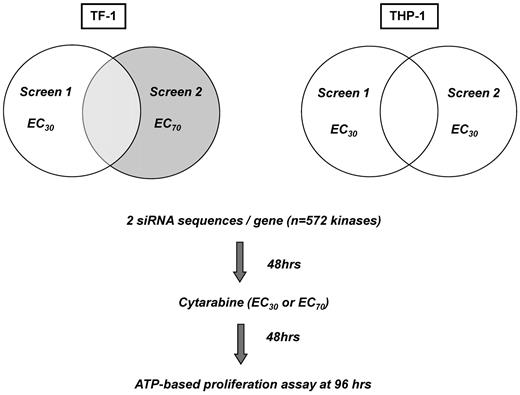
The transfection conditions and siRNA assay platform have been described previously and were adapted to myeloid suspension cells.7,12,13 Briefly, a validated siRNA library targeting 572 kinases (Kinome siRNA Library Version 1.0; QIAGEN) with 2 different siRNA sequences per kinase was printed onto 384-well plates (Fisher Scientific). Cationic lipid-based transfection reagents were diluted in Opti-MEM (Invitrogen) and added to assay plates using a μFill liquid dispenser (Bio-Tek). Subsequently TF-1 and THP-1 cells were added, followed by Ara-C at the corresponding EC30 or EC70 concentrations for each individual cell line at 48 hours, the time point of maximal siRNA gene silencing. After an additional 48 hours, relative cell number was measured using Cell Titer Glo (CTG; Promega) according to the supplier's instructions. Relative luminescence units were measured using an EnVision plate reader (PerkinElmer). Screens in THP-1 cells were performed in duplicate aiming for a target EC30 concentration. For TF-1, 1 screen was performed at the target EC30 dose and 1 at the EC70 dose, to mirror low- versus high-dose Ara-C regimens.
Hit selection criteria and definition of hits
Published criteria for analysis and hit selection were adapted16 in a multistep analysis algorithm. First, the log2 ratio for each individual siRNA per gene was calculated according to the equation: [siRNAx + drug : siRNAx + vehicle], both for individual plates and individual screens. The log2 ratio describes a sensitization effect by which silencing of a kinase with a particular siRNA sequence in combination with Ara-C treatment reduces relative cell number as assessed with CTG. In a second step, a specific siRNA was called a hit within a screen if the log2 ratio of that individual siRNA was greater than 2 SDs below the median of the log2 ratio of all siRNA + Ara-C treatments, both for individual plates and the entire screen. This plate- and screen-based normalization allows intra-plate and intra-screen comparison and hit selection. In a third step, a gene was defined as a candidate (hit) if the same siRNA met the hit selection criteria across screens. A kinase was called a hit only if both siRNA sequences (2 of 2 siRNAs) met the hit selection criteria. A kinase was also a hit if 3 of 4 or all 4 siRNAs for a particular kinase met hit selection criteria (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The RNAi approach screens for a loss-of function in combination with Ara-C and every hit accounts for the siRNA-only effect.
siRNA drug-dose response validation
Selected candidates from initial RNAi screens were validated by siRNA drug-dose–response (siDDR) assays. Following essentially the same protocol as for the siRNA screens and as described previously,7,12 4 different validated siRNA sequences (QIAGEN) against selected candidate genes were assessed. Control wells with nonsilencing siRNA, a lethal positive control (QIAGEN), and buffer/transfection reagent were included on each plate. Serial dilutions of Ara-C across a 10-point concentration range were added at 48 hours and the relative cell number was measured with CTG at 96 hours. Relative cell number was calculated by dividing the average relative luminescence units (RLUs) for the drug-treated wells of individual siRNA/genes by the average of RLU values for the control wells. EC50 values were calculated using Prism Version 5.03 software (GraphPad).
Validation of candidate genes using small-molecule inhibitors
Drug dose response experiments with WEE1 inhibitor and MK1775 were performed with 2 or 3 concentrations in 6-8 cell lines with or without Ara-C. For MK1775, dosing was performed simultaneously and sequentially (Ara-C followed by MK1775 at 24 hours). The relative cell number was determined with CTG at 72 and 96 hours after drug additions. Using Prism Version 5.03 software, Ara-C EC50 was calculated at various concentrations of MK1775, and the -fold change versus Ara-C alone was determined. For the initial primary samples (n = 4), Ara-C at lower (75-150nM range) or higher (300-600nM range) concentrations was added with MK1775 at 200 and 600nM concentrations, and the percentage inhibition of combining Ara-C with MK1775 versus Ara-C alone was calculated. To measure synergy between Ara-C and MK1775, additional primary samples (n = 10) were tested, evaluating 9 concentrations of Ara-C with 6 concentrations of MK1775. Synergy was assessed by calculating the combination index (CI) values with CalcuSyn Version 2.1 software according to the Chou and Talalay model.17 Only doses falling within the linear range of the median-effect plots for both compounds were used, so CI values could not be calculated for all 10 samples.
Quantitative real-time PCR
Transcript levels were measured by quantitative real-time PCR using a 7900HT thermocycler (Applied Biosystems). Cells were treated with siRNA and RNA extracted at 24 hours after transfection. RNA was extracted with TRIzol reagent (Invitrogen) and treated with 10 units/μL of RNase-free DNase I (Roche) to reduce genomic DNA contamination. cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad) according to the supplier's protocol. All primer pairs used were prefabricated mixtures of forward and reverse primers (Quantitect Primer Assay; QIAGEN). Relative quantification was performed with SYBR Green Supermix (Bio-Rad) according to the Pfaffl method,18 using GAPDH as an internal control for normalization. Target and reference samples were set up in triplicate for every experiment. PCR efficiency was determined for every primer pair in each experiment by constructing a standard curve with cDNA containing the target template.
Western blot analysis
TF-1 was transfected with either CHEK1 or WEE1 siRNA (QIAGEN) with or without Ara-C. Lysates were collected using Complete Lysis-M (Roche) supplemented with 10 μL/mL of Phosphatase Inhibitor Cocktail 1 & 2 (Sigma-Aldrich) and quantified by the BCA Protein Assay (Pierce/Thermo Fisher). Forty micrograms of protein was resolved by SDS-PAGE, transferred to PVDF membranes using the iBlot dry transfer system (Invitrogen), and blocked with 5% nonfat dry milk before primary Ab incubation. WEE1 Ab (sc-5285) and CHEK1 Ab (sc-8408) were obtained from Santa Cruz Biotechnology. Phospho-CDC2 (#9111) and antitubulin (#2144) Abs were obtained from Cell Signaling Technology. Anti–rabbit IgG and anti–mouse IgG secondary Abs were obtained from Jackson ImmunoResearch Laboratories.
Cleaved caspase-3 analysis
Cells were processed according to the standard protocol provided by Cell Signaling Technology 48 hours after treatment. Cells were incubated 1 hour with a cleaved caspase-3 (Asp175)–Alexa Fluor 488 Ab conjugate (#9669; Cell Signaling Technology) at a 1:50 dilution. Fluorescence intensity was measured with a CyAn flow cytometer (Beckman Coulter) and data were analyzed with Summit Version 4.3 software (DAKO).
Results
RNAi screens and quality parameters
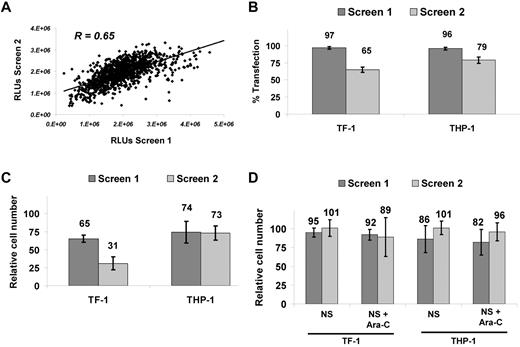
An RNAi screen was developed to identify potentiators of Ara-C activity in AML cells (Figure 1). TF-1 was chosen as an “immature,” stem cell–like, growth-factor–dependent cell line and THP-1 as a complex, MLL-rearranged cell line. We examined the silencing of 572 kinases in these 2 AML cell lines in combination with the DNA-damaging agent Ara-C. Reproducibility was high for the duplicate THP-1 screens for which the same Ara-C concentration (target EC30) was used, with a Pearson correlation coefficient of R = 0.65 (Figure 2A). TF-1 screens were conducted at different Ara-C concentrations, so the correlation was not calculated. Transfection efficiency was determined by reduction in relative cell number after transfection of a lethal siRNA compared with the median relative cell number of all kinase siRNAs. Transfection efficiency was 97% and 65% for TF-1 and 96% and 79% for THP-l in screen 1 and screen 2, respectively (Figure 2B). Reduction in relative cell number because of Ara-C alone was 35% and 69% for TF-1 and 26% and 27% for THP-1 (Figure 2C) in the respective screens, as determined by calculating the relative cell number for Ara-C plus nonsilencing siRNA (NS-siRNA) versus NS-siRNA alone. Nonspecific siRNA toxicity was measured as the reduction in relative cell number by NS-siRNA compared with transfection reagent only. Nonspecific toxicity for TF-1 was 5% and 1% for NS-siRNA and 8% and 11% for NS-siRNA plus Ara-C in screen 1 and screen 2, respectively. For THP-1, the nonspecific toxicity was 14% and 1% for NS-siRNA and 18% and 4% for NS-siRNA plus Ara-C in screen 1 and screen 2, respectively (Figure 2D). The high transfection efficiency, reproducibility, overall low nonspecific toxicity, and EC values of Ara-C at the target doses demonstrate the robustness and accuracy of the siRNA platform.
Performance and robustness of RNAi screens. (A) Reproducibility with Pearson correlation coefficient of R = 0.65 for duplicate RNAi screens conducted in THP-1 at target EC30. (B) Transfection efficiency for TF-1 and THP-1. Reduction in relative cell number was calculated as a percentage of the universal lethal control (n = 16) versus the median value for all 572 siRNA as a readout for transfection efficiency. (C) Ara-C growth-inhibitory effects. Viability calculated as the percentage inhibition of the median of NS-siRNA (n = 16) over NS-siRNA + Ara-C (n = 16). (D) Nonspecific effects on relative cell number calculated as the fraction of NS-siRNA (median, n = 16) versus buffer/control medium for both NS-siRNA–only and NS-siRNA + Ara-C. TF-1 screens were performed at higher (EC70) and lower (EC30) Ara-C concentrations. RLUs indicates relative luminescene units.
Performance and robustness of RNAi screens. (A) Reproducibility with Pearson correlation coefficient of R = 0.65 for duplicate RNAi screens conducted in THP-1 at target EC30. (B) Transfection efficiency for TF-1 and THP-1. Reduction in relative cell number was calculated as a percentage of the universal lethal control (n = 16) versus the median value for all 572 siRNA as a readout for transfection efficiency. (C) Ara-C growth-inhibitory effects. Viability calculated as the percentage inhibition of the median of NS-siRNA (n = 16) over NS-siRNA + Ara-C (n = 16). (D) Nonspecific effects on relative cell number calculated as the fraction of NS-siRNA (median, n = 16) versus buffer/control medium for both NS-siRNA–only and NS-siRNA + Ara-C. TF-1 screens were performed at higher (EC70) and lower (EC30) Ara-C concentrations. RLUs indicates relative luminescene units.
Kinases sensitizing individual cell lines to Ara-C
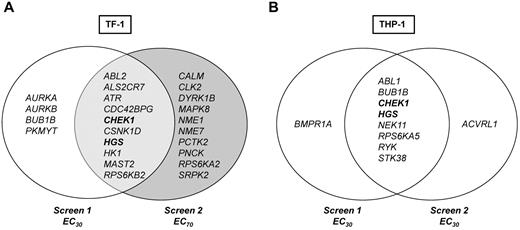
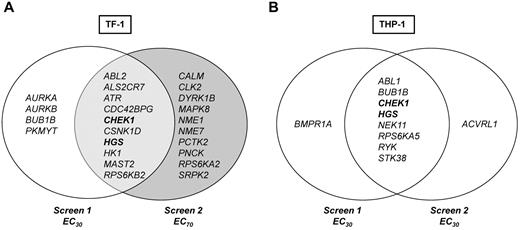
Data were first analyzed to identify kinase hits that overlapped in duplicate RNAi screens for each cell line individually, as outlined in the “Hit selection criteria and definition of hits.” Each RNAi screen included 2 different siRNA sequences per gene, so each gene is represented by 4 siRNA data points per cell line in 2 independent screens. Ten genes, 1.7% of the entire screen, were hits in TF-1. The top hit was ATR (4 of 4 siRNAs from 2 screens), followed by CHEK1, ABL-2, HGS, and MAST2 (each 3 of 4 siRNAs; Figure 3A). For THP-1, 8 genes or 1.4% of the entire screen were hits; for 7 of these genes, 2 of 4 siRNAs (same sequence) and for 1 gene, RYK, 3 of 4 siRNAs significantly reduced viability in combination with Ara-C (Figure 3B).
siRNA screen hits. Venn diagram of hits from primary siRNA-kinome sensitizer screens with Ara-C. Genes/hits depicted in bold are common Ara-C sensitizers identified in both TF-1 and THP-1.
siRNA screen hits. Venn diagram of hits from primary siRNA-kinome sensitizer screens with Ara-C. Genes/hits depicted in bold are common Ara-C sensitizers identified in both TF-1 and THP-1.
Kinases sensitizing at low versus high doses of Ara-C
To determine whether different kinases might contribute to survival at higher versus lower concentrations of Ara-C, TF-1 screens were performed with 2 different Ara-C concentrations, the approximate EC30 and EC70. These screens identified 4 genes, AURKA, AURKB, BUB1B, and PKMYT1 at the lower Ara-C concentration and 10 genes at the higher Ara-C concentration (Figure 3A). Genes that modulated lower-dose Ara-C are involved in cell-cycle regulation and mitotic checkpoint control. Kyoto Encyclopedia of Genes and Genomes pathway analysis showed enrichment for genes involved in nucleoside metabolism and both MAPK and insulin-like growth factor-1/growth hormone signaling pathways at the higher Ara-C concentration (supplemental Figure 2). These data suggest that different kinases and mechanisms may operate to modulate survival at various concentrations of Ara-C.
Kinases “universally” sensitizing myeloid cells to Ara-C
From a therapeutic point of view, it is desirable to identify genes that may be universal sensitizers across molecular and differentiation subtypes (ie, cytogenetics or French-American-British classification) of leukemias. Two genes, CHEK1 and HGS, were identified for which silencing by at least 1 siRNA of the same sequence augmented the antileukemic activity of Ara-C by > 2 SDs in all 4 screens (Figure 3 in bold).
Selection and validation of top candidate kinase hits sensitizing to Ara-C
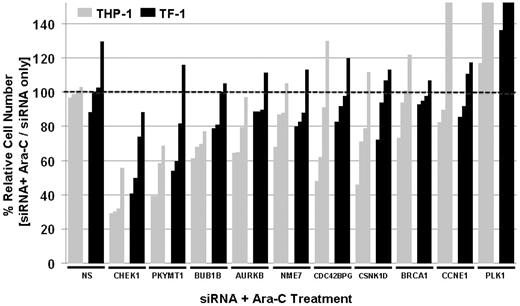
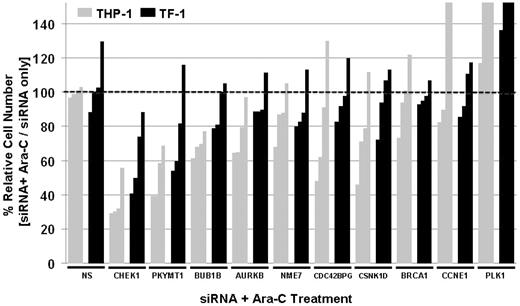
Secondary RNAi screens were performed using an expanded set of siRNAs covering selected hits from primary RNAi screens with a total of 4 different siRNAs (Figure 4). CHEK1 and PKMYT1 were the strongest sensitizers to Ara-C. Other hits from primary screens, such as BUB1B or AURKB, were reproducible but weaker sensitizers to Ara-C compared with CHEK1 and PKYMT1 (Figure 4). To begin assessing the specificity of hits and to determine whether interference with DNA repair or cell-cycle regulation in general had a sensitizing effect, siRNAs against BRCA1, PLK1, and Cyclin E were included in secondary RNAi screens. Silencing of BRCA1 and Cyclin E failed to sensitize to Ara-C, indicating that the observed Ara-C–sensitizing effects of individual genes such as CHEK1 and PKMYT1 are specific and not a general phenomenon of DNA damage or cell-cycle interference. Silencing of PLK1 enhanced survival in the presence of Ara-C (Figure 4) despite PLK1 silencing alone reducing the relative cell number, which highlights the specificity of the screen for identifying sensitizers to Ara-C as opposed to siRNA/gene-only vulnerabilities.
Validation of hits by secondary siRNA screens. Validation of selected top hits from primary siRNA screens with 4 siRNA against each gene. Reduction in relative cell number expressed as Ara-C with siRNA versus Ara-C alone. NS indicates the nonsilencing control. Darkly shaded bars correspond to TF-1 and lightly shaded bars to THP-1.
Validation of hits by secondary siRNA screens. Validation of selected top hits from primary siRNA screens with 4 siRNA against each gene. Reduction in relative cell number expressed as Ara-C with siRNA versus Ara-C alone. NS indicates the nonsilencing control. Darkly shaded bars correspond to TF-1 and lightly shaded bars to THP-1.
We also used a definitive validation assay, which combines siRNA silencing with a drug-dose–response assay (the siDDR assay). For these assays, 2 additional cell lines, HEL and MDS-L, were used together with TF-1 and THP-1. Genes selected for these siDDR validation assays were CHEK1 and HGS as common hits between TF-1 and THP-1 (Figure 3), ATR as the strongest hit with 4 of 4 siRNA positive in initial siRNA screens, and PKMYT1 and WEE1, the latter for clinical interest. WEE1 was not included in the original siRNA library but was added to the validation because of its close relationship to PKMYT1, which was a hit in both primary and secondary screens. The similarity of PKMYT1 and WEE1 in regulating CDC2,19 the intricate involvement of WEE1 in DNA-damage repair and cell-cycle progression,20 and the clinical availability of the first-in-class WEE1 kinase inhibitor MK177521,22 led us to pursue WEE1 as a potential sensitizer.
In siDDR assays, sensitization is demonstrated by a curve shift toward lower Ara-C concentrations in the presence of siRNA compared with nonsilencing siRNA or transfection reagent only. WEE1 siRNA exhibited strong sensitization to Ara-C, and this was observed across all lines. Silencing of CHEK1 strongly sensitized in TF-1 and THP-1, but less so in HEL and not at all in MDS-L. ATR siRNA was a moderate sensitizer across all lines, with the strongest sensitization observed in TF-1 cells, validating ATR as a hit from TF-1 primary screens. PKYMT1 siRNA exhibited sensitization in TF-1 and THP-1, whereas HGS validated as a weak sensitizing kinase (Table 1 and supplemental Figure 3).
siRNA silencing of candidate kinases is specific and results in reduced protein and transcript expression
To ascertain specificity and determine the level of silencing for the top 2 Ara-C–sensitizing gene targets, WEE1 and CHEK1, cells were treated with siRNA alone and in combination with Ara-C, then examined by quantitative RT-PCR and immunoblotting. TF-1, THP-1, and HEL cells exhibited a 92% and 82%, 75% and 76%, and 70% and 64% decrease in their transcript levels after WEE1 and CHEK1 siRNA treatment, respectively (Figure 5A). Immunoblots showed reduction of WEE1 and CHEK1 protein expression at 48 hours after siRNA treatment (Figure 5B), confirming specific silencing of our target genes. Because sensitization was generally observed with all 4 WEE1 siRNAs, only selected WEE1 and CHEK1 siRNAs were used in these assays.
Target silencing by siRNA. (A) WEE1 and CHEK1 transcript levels after treatment with siRNA targeting WEE1 or CHEK1 compared with NS-siRNA. (B) Immunoblots of extracts from cells treated with WEE1 or CHEK1 siRNA with and without 270nM Ara-C. WEE1 (top panel) and CHEK1 (bottom panel). NS indicates the nonsilencing control; and UT, untreated control.
Target silencing by siRNA. (A) WEE1 and CHEK1 transcript levels after treatment with siRNA targeting WEE1 or CHEK1 compared with NS-siRNA. (B) Immunoblots of extracts from cells treated with WEE1 or CHEK1 siRNA with and without 270nM Ara-C. WEE1 (top panel) and CHEK1 (bottom panel). NS indicates the nonsilencing control; and UT, untreated control.
Parallel assays confirmed that reduced transcript and protein levels corresponded to reductions in relative cell number (data not shown). Low nonspecific toxicity was observed with NS-siRNA. Furthermore, WEE1 silencing resulted in a significant reduction of phosphorylated CDC2 levels compared with CHEK1 silencing. These data provide a high probability that the observed biologic effects are due to specific gene silencing.
Validated genes from siRNA secondary screening are expressed in primary leukemia samples
We confirmed the potential biologic disease relevance of these functional genomic targets by assessing their expression in samples from leukemia patients. Examination of the public database (Oncomine, www.Oncomine.org) indicated that all 5 genes from siDDR experiments were expressed in approximately 50% or more of myeloid and lymphoid leukemia samples compared with healthy BM controls (supplemental Figure 4). There was a trend toward higher expression of WEE1, PKMYT1, and CHEK1 in myeloid, as well as B- and T-cell lymphoid leukemias.
WEE1 inhibitors potentiate the antileukemic activity of Ara-C in myeloid cells in vitro
Because WEE1 was the strongest sensitizer in siDDR experiments and is a novel target, we focused our pharmacologic studies primarily on WEE1. A commercial WEE1 inhibitor sensitized a representative panel of cell lines to Ara-C in the range of 2- to 25-fold (supplemental Figure 5A) as calculated by a shift in the EC50 of Ara-C. Because the commercial WEE1 inhibitor targets other kinases, including CHEK1, at higher concentrations at which maximal sensitization occurred,23 we aimed to confirm results with the selective WEE1 kinase inhibitor MK1775.21,22 MK1775 is the first WEE1 kinase inhibitor in clinical trials. In combination dose-response studies, MK1775 sensitized a panel of 8 AML, chronic myeloid leukemia (CML), and acute lymphoblastic leukemia (ALL) cell lines to Ara-C up to 97-fold at 72 hours, as indicated by a shift in EC50 values (Table 2 and supplemental Figure 5B). Sensitization was even more pronounced at 96 hours and was observed at the lower MK1775 concentrations of 50 and 100nM, which are well within the range of plasma concentrations observed in an ongoing phase 1 study.21,22 Interestingly, K562 cells, which often exhibit a plateau in response to cytotoxic agents, were almost completely inhibited by the combination of Ara-C with either the commercial or clinical grade WEE1 inhibitor (supplemental Figure 5A-B).
MK1775 sensitizes primary AML, MDS, and CML specimens to Ara-C ex vivo
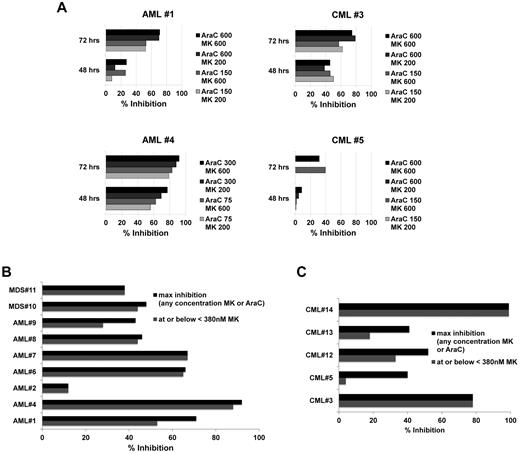
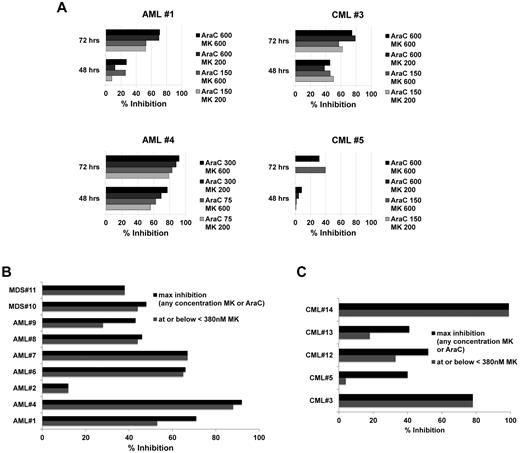
To confirm sensitization ex vivo as proof-of-principle for clinical application, we tested primary leukemia samples from 7 AML, 5 CML, and 2 MDS patients. Initially, 3 AML (AML patients 1, 2, and 4) and 2 CML (CML patients 3 and 5) samples were incubated with Ara-C concentrations of 150, 300, and 600nM with and without MK1775 at 200 or 600nM for 48 and 72 hours. Strong time-dependent reduction of relative cell number, up to 92% at 72 hours, was observed with the combination over Ara-C alone (Figure 6A, AML patient 4 left bottom panel). Interestingly, 200nM MK1775 was similarly potent to 600nM for 3 of the initial samples. Subsequently, additional samples (Figure 6B-C and supplemental Figure 5C) were tested across a dose range of both Ara-C and MK1775 to determine the degree of synergy and the percentage inhibition at each dose combination and to guide selection of clinical dosing schedules based on preclinical information. Data are summarized as the percentage of inhibition combining MK1775 with Ara-C over Ara-C alone for the percentage of inhibition of MK1775 at or below 380nM or the maximal inhibition at any MK1775 concentration (Figure 6B-C). This threshold of 380nM MK1775 was applied as a cutoff for clinically achievable MK1775 plasma concentrations as determined from ongoing phase 1 solid tumor trials22,24 (and personal communication with T.D.), with 380nM being the closest point to these concentrations on the dilution curve. In addition to percentage of inhibition, synergy was assessed by calculating the CI values. Table 3 depicts CI values for MK1775 at 380nM with the corresponding Ara-C concentration that provided the greatest synergy. From these detailed experiments across 14 primary samples, we noted that surprisingly low to moderate Ara-C concentrations often yielded similar to stronger synergy and percentage of inhibition compared with higher Ara-C concentrations (Table 3, Figure 6, and supplemental Figure 5C). For most AML and MDS samples, MK1775 at or below 380nM compared with higher MK1775 concentrations of 1500 or 3000nM. A somewhat different trend was observed for 3 of the 5 CML samples, with a tendency of greater sensitization at higher MK1775 concentrations (Figure 6). Supplemental Figure 5C depicts detailed graphs for representative AML, MDS, and CML samples.
MK1775 is synergistic with Ara-C ex vivo. Percentage inhibition of relative cell number for the combination of Ara-C and MK1775 over Ara-C alone in primary AML, MDS, and CML patient samples. (A) Initial 4 (of 5) primary samples as described in the text. (B) AML and MDS primary samples. (C) CML primary samples. For panels B and C, the maximum percentage inhibition is shown for any concentration of MK1775 or Ara-C (darkly shaded bars) and for the greatest inhibition observed at or below 380nM MK1775 with any concentration of Ara-C (lightly shaded bars; see “MK1775 sensitizes primary AML, MDS, and CML specimens to Ara-C ex vivo” details).
MK1775 is synergistic with Ara-C ex vivo. Percentage inhibition of relative cell number for the combination of Ara-C and MK1775 over Ara-C alone in primary AML, MDS, and CML patient samples. (A) Initial 4 (of 5) primary samples as described in the text. (B) AML and MDS primary samples. (C) CML primary samples. For panels B and C, the maximum percentage inhibition is shown for any concentration of MK1775 or Ara-C (darkly shaded bars) and for the greatest inhibition observed at or below 380nM MK1775 with any concentration of Ara-C (lightly shaded bars; see “MK1775 sensitizes primary AML, MDS, and CML specimens to Ara-C ex vivo” details).
MK1775 and Ara-C in combination induce apoptosis
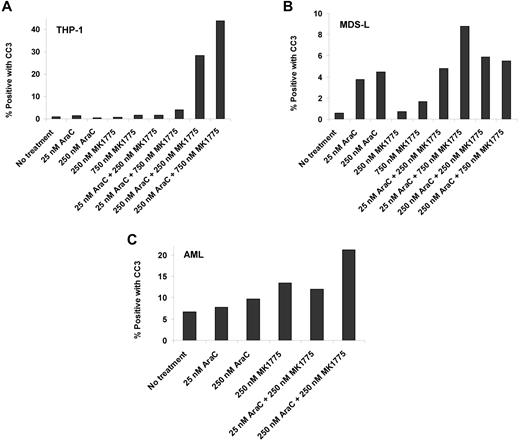
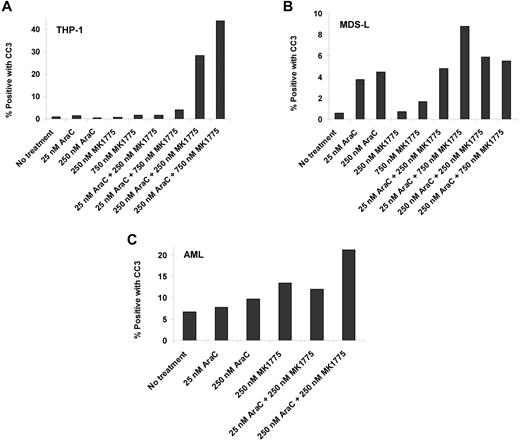
THP-1 and MDS-L cell lines and a primary AML sample were incubated at selected Ara-C and MK1775 concentrations. Cleaved caspase-3 levels increased nearly 40-fold in THP-1 cells, nearly 3-fold in MDS-L cells, and nearly 2-fold in the primary AML sample compared with Ara-C and/or MK1775 alone (Figure 7). These results confirm that reductions in relative cell number are due to induction of apoptosis.
Ara-C and MK1775 induce apoptosis. Percentage of cleaved caspase-3 (CC3)-positive cells after Ara-C, MK1775, or combination treatment in THP-1 cells (A), MDS-L cells (B), and a primary AML sample (C).
Ara-C and MK1775 induce apoptosis. Percentage of cleaved caspase-3 (CC3)-positive cells after Ara-C, MK1775, or combination treatment in THP-1 cells (A), MDS-L cells (B), and a primary AML sample (C).
Discussion
Ara-C is the backbone of many leukemia regimens.1,2 In the present study, we performed a high-throughput RNAi screen silencing 572 kinases to identify kinases for which their inhibition results in the most potent sensitization to Ara-C. This screen has helped to identify WEE1 as a kinase for which inhibition potently and rather universally sensitizes to Ara-C. Furthermore, we determined potential dose ranges for MK1775 and Ara-C in combination, with important implications for clinical trial design.
Previous studies by Mesa et al and other groups25,26 have shown that Ara-C activates a replication checkpoint involving the CHEK1 kinase, and CHEK1 inhibition sensitizes to Ara-C in leukemias.25,26 CHEK1 was among the kinases identified as an Ara-C–sensitizing target in this screen. This confirmed the robustness of the screening platform, data analysis, and hit selection algorithms, as well as subsequent validation experiments. In addition to CHEK1, many of the other hits are also involved in DNA-damage repair and cell-cycle checkpoint regulation, including NEK11, ATR, ABL-1/2, and PKMYT1. NEK11 was shown recently to be a mediator of CHEK1 damage signaling.27 ATR, the kinase immediately upstream of CHEK1 in the replication checkpoint, was positively identified in our screen and was confirmed as a sensitizer in siDDR validation experiments (Table 1). Both ABL-1 and ABL-2, which are known to modulate DNA damage28 and reactive oxygen species responses,29 were hits in TF-1 and THP-1, respectively. These results suggest the involvement of the ABL pathway in DNA-damage repair30 in response to Ara-C, and show that ABL inhibitors may sensitize to Ara-C.
A molecular context for other siRNA hits is less apparent. For example, much less is known about HGS. HGS functions downstream of cytokine and growth factor signaling31 and is involved in vesicular trafficking, autophagosome formation,32 DNA synthesis inhibition, and bone morphogenic protein (BMP) signaling,33 a pathway that is closely coregulated with TGF-β signaling. In fact, the 2 nonoverlapping hits from duplicate screens in THP-1, BMPR1A and ACVRL1 (Figure 3B), also belong to the BMP- and TGF-β–signaling networks, indicating that all hits from replicate THP-1 siRNA screens either overlap directly or share similar molecular concepts.
Through PKMYT1, we arrived at WEE1 kinase, which was the focus of validation experiments involving pharmacologic inhibition. At the time these siRNA screens were performed, we used the most complete commercially available library containing siRNAs against 572 different kinase genes. WEE1 was not part of the siRNA library, but belongs to the same family of proteins as PKMYT1.
WEE1 kinase is an essential G2/M as well as G1/S checkpoint kinase and regulates cellular fate decisions such as cell-cycle progression/delay versus apoptosis induction.34,35 WEE1-inhibitory phosphorylation of CDK1 leads to delayed cell-cycle progression,38 which is thought to provide malignant cells time for DNA-damage repair, thus achieving sufficient genomic integrity to complete cell division.
In the RNAi screens, silencing of CHEK1 and PKMYT1 potentiated Ara-C activity. Because inhibitors of WEE1 are available but have not yet been tested in hematologic malignancies, we focused primarily on this kinase. In validation experiments, inhibition of WEE1 kinase by siRNA (Table 1 and supplemental Figure 3) or pharmacologically with experimental and clinical WEE1 inhibitors (Table 2 and supplemental Figure 5A-B) resulted in the most potent sensitization to Ara-C. Our data suggest that interfering with WEE1 kinase is at least as potent as CHEK1 inhibition in combination with Ara-C. WEE1 siRNA silencing sensitized 4 AML cell lines tested, including HEL and, to a lesser degree, even MDS-L cells to Ara-C, whereas both cell lines were unaffected by CHEK1 siRNA.
MK1775, the first-in-class WEE1 kinase inhibitor, is an oral small molecule in clinical development in combination with gemcitabine and platinum drugs in solid-tumor patients and is generally well tolerated.22,24 Plasma MK1775 concentrations of up to 570nM were readily achieved in an ongoing trial.22 Clinically, there appears to be sufficient MK1775 selectivity for malignant versus normal cells. Excessive or prolonged myelosuppression was not observed in the solid-tumor trial of 146 patients treated with single-agent or cytotoxic combination to date, indicating that normal progenitor cell regeneration is not affected, suggesting a therapeutic window for the Ara-C-MK1775 combination, and minimizing concerns for impaired progenitor recovery.
Adding MK1775 to Ara-C sensitized all myeloid and lymphoid leukemia cell lines and 13 of 14 primary AML, CML, and MDS samples. MK1775 with Ara-C significantly induced apoptosis compared with either drug alone in THP-1 and MDS-L, as well as in a primary AML sample. Combination treatment induced greater levels of cleaved caspase-3 in THP-1 compared with MDS-L (Figure 7). This observation is consistent with the magnitude of sensitization observed in THP-1 compared with MDS-L after treatment with MK1775 and Ara-C (Table 2), demonstrating similar results with parallel assays. DNA damage as determined by γ-H2AX was not significantly increased, whereas cleaved caspase-3 was, suggesting that reduced cell count is mainly because of induction of apoptosis.
Our observations suggest that inhibiting WEE1 kinase could be of broad utility in sensitizing leukemia cells to Ara-C. It is encouraging that dose ranges and trends are similar across in vitro and ex vivo experiments, with lower to moderate Ara-C in combination with moderate MK1775 concentrations being the most efficacious. siDDR experiments also demonstrated that WEE1 inhibition results in greater sensitization at lower Ara-C concentrations (supplemental Figure 2). These preclinical observations will affect the design and dosing schedules for the proposed clinical trial.
In conclusion, we present the first high-throughput siRNA platform in nonadherent cells and show that functional genomics using siRNA drug sensitizer screening provides an attractive experimental concept to rapidly identify molecular targets in leukemias. Hits from RNAi screens can be rapidly validated and results translated into the design of rational combination therapies. WEE1 kinase emerged as a potent sensitizer in myeloid and lymphoid malignant cells, establishing WEE1 as a powerful candidate kinase for inhibition in combination with Ara-C across various leukemia subtypes, including MDS. A clinical trial of MK1775 in combination with Ara-C in advanced leukemias is under development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Professor Kaoru Tohyama (Department of Laboratory Medicine, Kawasaki Medical School, Kurashiki, Japan) for providing the MDS-L cell line.
This work was supported in part by a grant from the Arizona Biomedical Research Commission awarded to R.T. Institutional support was provided by TGen and the Mayo Clinic. R.T. is supported by a Career Development Award of the Conquer Cancer Foundation of the American Society of Clinical Oncology for the data presented herein.
Authorship
Contribution: R.T. designed the project, performed the research, analyzed and interpreted the data, and wrote the manuscript; J.M.B., L.C., R.T.H., and S.A. designed the experiments, performed the research, analyzed the data, and assisted in writing the manuscript; D.C. and I.M.G. performed the research; T.D. helped design research, contributed vital reagents, and interpreted data; H.H.Y. provided essential infrastructure; M.E.B. and E.B. collected and analyzed the data; J.S. and R.A.M. provided essential reagents, samples and clinical information; and D.O.A. designed the experiments, analyzed the data, provided essential supplies, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests directly related to this research. R.T. was the principal investigator for a clinical study with MK1775 and funding for the trial was provided to R.T.'s former institution (Translational Genomics Research Institute/Scottsdale Healthcare, Scottsdale, AZ) by Merck.
Correspondence: Raoul Tibes, MD, PhD, Mayo Clinic, Division of Hematology and Oncology, 13400 E Shea Blvd, Scottsdale, AZ 85259; e-mail tibes.raoul@mayo.edu.
References
Author notes
R.T. and J.M.B. contributed equally to this work.