Comprehensive analysis of the cancer genome has become a standard approach to identifying new disease loci, and ultimately will guide therapeutic decisions. A key technology in this effort, single nucleotide polymorphism arrays, has been applied in hematologic malignancies to detect deletions, amplifications, and loss of heterozygosity (LOH) at high resolution. An inherent challenge of such studies lies in correctly distinguishing somatically acquired, cancer-specific lesions from patient-specific inherited copy number variations or segments of homozygosity. Failure to include appropriate normal DNA reference samples for each patient in retrospective or prospective studies makes it difficult to identify small somatic deletions not evident by standard cytogenetic analysis. In addition, the lack of proper controls can also lead to vastly overestimated frequencies of LOH without accompanying loss of DNA copies, so-called copy-neutral LOH. Here we use examples from patients with myeloid malignancies to demonstrate the superiority of matched tumor and normal DNA samples (paired studies) over multiple unpaired samples with respect to reducing false discovery rates in high-resolution single nucleotide polymorphism array analysis. Comparisons between matched tumor and normal samples will continue to be critical as the field moves from high resolution array analysis to deep sequencing to detect abnormalities in the cancer genome.
Introduction
Global profiling of DNA copy number in cancer cells using microarray platforms holds great appeal, as it offers an unparalleled opportunity to uncover the elusive genetic lesions important for tumor initiation and progression. In contrast to array comparative genomic hybridization, which allows one to record only the DNA copy number at high resolution for the whole genome, single nucleotide polymorphism (SNP) arrays permit the capture of both DNA copy number and SNP-based genotype at a submegabase resolution, facilitating the detection of small areas of genomic loss of heterozygosity (LOH) or uniparental disomy (UPD). This technology began to prove its value early in the current decade, with marked improvements in resolution and performance occurring ever since. Array platforms now interrogate the human genome at a density of 900 000 SNPs with an average intermarker distance of less than 700 bp, and nowhere has the power of genome-wide SNP array analysis been more evident than in the study of hematologic malignancies.
Over the past decade, many pivotal advances in the understanding of the genetics of hematologic diseases have emerged from SNP array analysis. Large-scale analysis of SNP arrays in B-cell acute lymphocytic leukemia, for example, led to the identification of PAX5 as a key target of genetic inactivation in this disease.1 In the same manner, the identification of TET2 as a major tumor suppressor in myelodysplastic syndromes (MDSs) was driven by SNP array analysis.2,3 Thus, SNP arrays afford useful platforms for discovering disease alleles that can shed new light on the pathobiology of leukemias and other hematologic malignancies. Ultimately, these insights should catalyze further advances in diagnosis and risk classification and set the stage for a new era of molecular medicine, in which patients receive personalized treatment based on the unique genetic changes in their malignant cells.
If this promise is to be realized, investigators must meet the challenge of designing studies that unequivocally distinguish acquired, cancer-specific genetic lesions from patient-specific genomic polymorphisms. Otherwise, the false discovery rate associated with SNP array analysis could seriously compromise efforts to translate this important technology to improved patient care. This perspective will consider the potential problem of false discovery in SNP array analysis and will recommend strategies that can be used to generate more reliable datasets for detecting the genetic events underlying malignant transformation.
Essential principles of SNP array analysis
The basic concept of SNP array analysis is the interrogation of genomic loci to obtain the DNA copy number and the genotype. For each SNP, the microarray contains oligonucleotide probes that can hybridize with the fragmented test DNA. In general, a fluorescence signal is obtained for each allele, A or B, at a given SNP. In a straightforward analytical approach, this signal is converted into 2 types of information: a discrete genotype call for the individual SNPs (A, B, or AB as the canonical genotypes) and a copy number value (overall fluorescence intensity) for the specific locus. Because current SNP platforms allow the analysis of more than 900 000 loci simultaneously, genotype and copy number data along the chromosome are readily available.
A typical analysis workflow starts with the normalization of all arrays to a baseline array to adjust the overall brightness of each array and to allow comparability among the arrays. Normal DNA samples are used to convert fluorescence intensities of each SNP to a copy number value (“scaling”). In cases with a high degree of aberrations (complex karyotypes, aneuploid genomes), additional adjustment steps might be required.4 After normalization and data modeling with appropriate software (eg, Dchip),5 the copy number profile can be visualized along the chromosome (Figure 1 far left). A frequently used unit is the log2 ratio, calculated as log2 of the signal of the sample divided by the mean signal of all normal samples at this SNP. Thus, a log2 ratio of 0 reflects no copy number change (white), whereas a lower (blue) or higher (red) ratio indicates loss or gain, respectively. Segmentation algorithms are used to computationally define all regions of copy number loss and gain.6,7
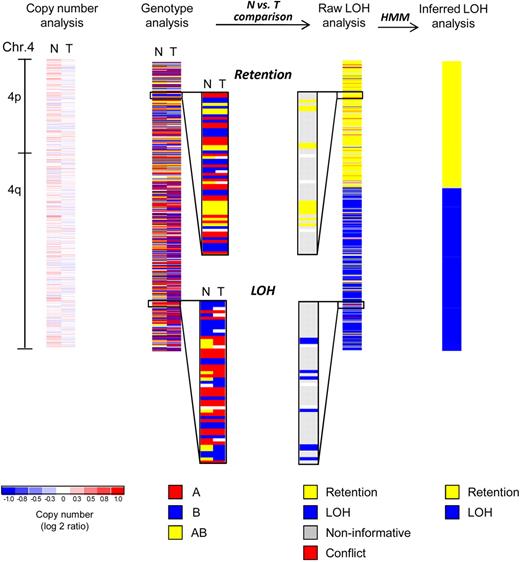
SNP array analysis of chromosome 4 of an MDS patient with a normal karyotype. Copy number analysis of chromosome 4 reveals no deletion or amplification in the tumor sample (T, mononuclear bone marrow). The paired normal DNA sample (N, buccal swab) is also shown. White areas indicate no copy number change (log2 ratio = zero), whereas shades of blue and red designate losses and gains, respectively (see scale at bottom). Minor fluctuations of blue and red are the result of data noise. Genotype analysis detects 3 distinct genotypes (A, red; B, blue; AB, yellow; white, no call). Comparison of normal (N) versus tumor (T) samples reveals that heterozygous calls (AB, yellow) are converted into homozygous calls (A or B, red or blue) in a distal segment of the q-arm indicating loss of heterozygosity (LOH), whereas the single nucleotide polymorphism (SNP) genotypes are retained in the proximal q-arm and the p-arm of the chromosome (retention). Raw LOH analysis is performed by computational comparison of normal-versus-tumor samples, resulting in a single column. Homozygous SNPs are noninformative in such a comparison (gray). Retention is depicted in yellow and LOH in blue. In rare instances, genotype detection errors lead to an apparent heterozygous SNP genotype in the tumor sample paired with a homozygous SNP genotype in the normal sample, and these cases are detected as conflicts (red). Note that yellow and blue have a different meaning in LOH analysis as opposed to genotype analysis (see color codes beneath each type of analysis). Hidden Markov models (HMM) are used to convert the single SNP-based raw LOH data along the chromosome into segments. Finally, inferred LOH analysis represents the last step of the genotype comparison between normal and tumor samples, revealing a copy-neutral LOH (CNLOH) of 4q in this myelodysplastic syndromes (MDS) patient.
SNP array analysis of chromosome 4 of an MDS patient with a normal karyotype. Copy number analysis of chromosome 4 reveals no deletion or amplification in the tumor sample (T, mononuclear bone marrow). The paired normal DNA sample (N, buccal swab) is also shown. White areas indicate no copy number change (log2 ratio = zero), whereas shades of blue and red designate losses and gains, respectively (see scale at bottom). Minor fluctuations of blue and red are the result of data noise. Genotype analysis detects 3 distinct genotypes (A, red; B, blue; AB, yellow; white, no call). Comparison of normal (N) versus tumor (T) samples reveals that heterozygous calls (AB, yellow) are converted into homozygous calls (A or B, red or blue) in a distal segment of the q-arm indicating loss of heterozygosity (LOH), whereas the single nucleotide polymorphism (SNP) genotypes are retained in the proximal q-arm and the p-arm of the chromosome (retention). Raw LOH analysis is performed by computational comparison of normal-versus-tumor samples, resulting in a single column. Homozygous SNPs are noninformative in such a comparison (gray). Retention is depicted in yellow and LOH in blue. In rare instances, genotype detection errors lead to an apparent heterozygous SNP genotype in the tumor sample paired with a homozygous SNP genotype in the normal sample, and these cases are detected as conflicts (red). Note that yellow and blue have a different meaning in LOH analysis as opposed to genotype analysis (see color codes beneath each type of analysis). Hidden Markov models (HMM) are used to convert the single SNP-based raw LOH data along the chromosome into segments. Finally, inferred LOH analysis represents the last step of the genotype comparison between normal and tumor samples, revealing a copy-neutral LOH (CNLOH) of 4q in this myelodysplastic syndromes (MDS) patient.
The genotype analysis (Figure 1) reveals the signal abundance of allele A and allele B for a given SNP and generates a dataset that identifies the genotype as A, B, AB, or “no call” (insufficient signal). Detection of LOH, the somatic conversion of heterozygous germline alleles to homozygosity, is based on the genotype calls in each individual patient at each SNP locus. Thus, for each SNP, comparison of the tumor sample with matched normal DNA will reveal either retention of heterozygosity or LOH (raw LOH analysis). Homozygous SNPs in the matched normal sample are noninformative, as the genotype on both alleles is the same, so that loss or conversion of one allele would not cause any change of the SNP genotypes in the tumor. Occasionally, because of genotype detection errors, one might find a homozygous SNP in the normal DNA sample, but a heterozygous SNP in the tumor sample (“conflict”). Such “noise” in the genotype calls can also lead sporadically to apparently retained heterozygous SNPs within LOH regions. Finally, results for all individual calls along the chromosome are statistically modeled with hidden Markov models, generating an inferred LOH analysis that reveals segments of LOH (Figure 1 far right).
Classically, LOH in cancer cells arises from a chromosomal deletion; however, as shown in the model MDS case (Figure 1), it can also appear in a tumor without loss of DNA (copy-neutral LOH [CNLOH]). During the establishment of the neoplastic clone, a segment of one chromosome is lost and replaced by the same region of its homologous chromosome, resulting in segmental LOH. CNLOH can involve a whole chromosome (also known as UPD) or only a segment of a chromosome (partial UPD). In summary, CNLOH represents an important mechanism by which point mutations or other microlesions can be established in a homozygous state detectable by SNP array analysis.
This abnormality is thought to be positively selected for during clonal evolution because it can result in homozygosity for a mutation in one allele of a tumor suppressor gene or oncogene, together with loss of the wild-type allele. Although the molecular mechanism for the emergence of partial UPD in cancer has not yet been experimentally addressed, a double-strand break repaired by homologous recombination between paternal and maternal chromosome segments is probably involved. As with other acquired mutations and copy number alterations (CNAs), the presence of a segment of CNLOH in a neoplastic clone could have a role in molecular pathogenesis or could be unrelated to pathogenesis, and represent an incidental abnormality that occurred in a clone that was transformed by different events. This passenger-versus-driver issue8 can be partially addressed by analyzing a large patient cohort and using statistical approaches to infer that CNLOH for a particular region is involved in transformation and therefore is a “driver” event because it occurs as a recurrent abnormality.9
A key consideration in the design of a tumor genome SNP array study is the source of the matched normal genomic DNA. Buccal swabs have been used successfully to obtain normal DNA with the advantage that they can be easily obtained in most instances. A buccal swab of a healthy person yields 2 to 3 μg of DNA, which is sufficient for SNP array analysis. However, the swabs may be contaminated with blood, which, for hematologic malignancies, might introduce tumor DNA into the control sample. More importantly, buccal DNA will invariably contain DNA from oral microflora or food remnants. Although this contamination does not affect SNP array analysis, it will pose a major challenge for “next-generation” whole genome resequencing, which will probably replace SNP array analysis of tumor genomes (see “Future prospects”). Long-lived T lymphocytes selected by flow cytometry from the bone marrow sample might also be considered as a source of normal DNA if the cells are free of genetic changes associated with the malignant clone. An optimal source of control DNA is a skin biopsy, which can be taken at the site of the bone marrow aspirate without additional discomfort to the patient (as the skin is anesthetized).10 Indeed, given the rapid progress in the field, it would seem wise to begin including “whole genome sequencing” as well as “SNP array analysis” in the informed consent forms that patients sign, so that samples obtained now can be used in the future for whole genome sequencing.
False discoveries in SNP array analysis
SNP array analysis with paired normal samples is straightforward but can be quite challenging without such matched controls. First, short segments of copy number change in the sample might reflect a disease-associated somatic microaberration or simply the presence of an individual inherited germline copy number variation (CNV).11,12 Regions of germline CNV have a median length of approximately 150 kb but can extend well beyond 1 Mb; larger CNVs could easily be mistaken for somatic CNAs if germline controls were not available.11 Second, individual segments of homozygosity might have arisen from pathogenically significant somatic CNLOH or represent the chance inheritance of 2 copies of the same germline haplotype, reflecting chromosomal segments exhibiting linkage disequilibrium. Thus, several considerations must be taken into account to distinguish acquired somatic aberrations from inherited normal variations.
CNVs can occur as either losses or gains; and although frequent in their appearance, they are not highly recurrent among persons.13 The Database of Genomic Variants (DGV)14 maintained by the Center of Applied Genomics (Toronto, ON) lists more than 8410 CNV loci (Version August 5, 2009). Nonetheless, as shown in our previous MDS SNP array study,15 and as a well-known caveat in resequencing approaches,16,17 CNV and SNP databases are far from complete, as many “private” SNPs or CNVs are detected in persons that do not appear frequently enough to be included in the compiled databases. Another problem with using the DGV to identify CNVs is that the database includes submissions that have not been thoroughly validated. For example, a true somatic aberration might be considered as a CNV because the region coincides with a poorly substantiated CNV identification that found its way into the database. For these reasons, data repositories, such as the DGV, should not be considered as a “gold standard” that can be used to distinguish germline variants from true somatic changes. Thus, to reliably detect somatically acquired chromosomal deletions and gains that are smaller in size than those evident by standard cytogenetic analysis, one must simultaneously analyze nontumor, normal DNA from the same patient.
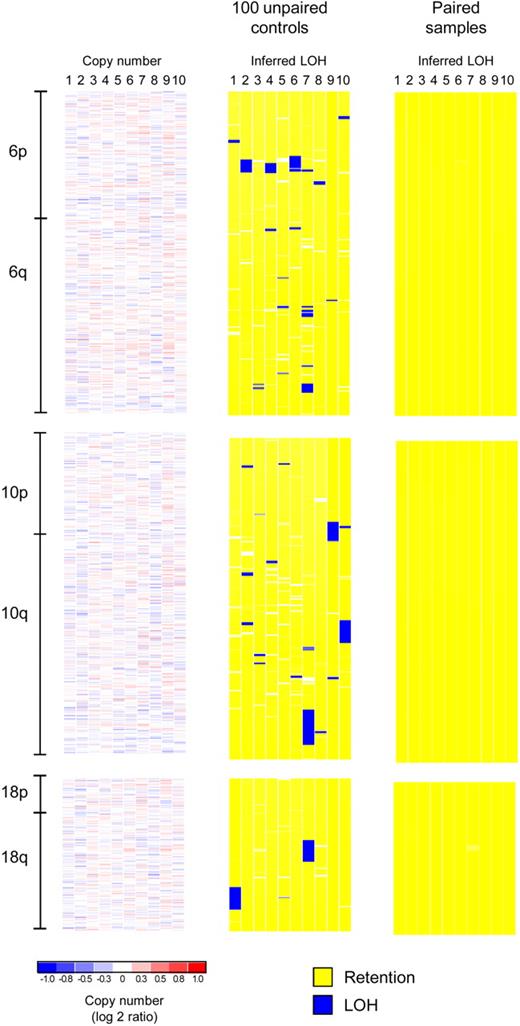
The situation for genotype analysis is even more demanding. The problem with using unpaired normal control samples to detect CNLOH is illustrated in Figure 2. Although computational methods have been proposed to predict the likelihood that a certain segment might represent CNLOH based on the use of unrelated control samples,19 even the use of large numbers of unmatched normal control samples does not ensure the error-free detection of true CNLOH segments. In Figure 2, inferred LOH analysis was performed on SNP arrays of 10 MDS patients (mononuclear bone marrow cell DNA) with either 100 unpaired controls (processed in the same laboratory) or with 10 paired normal DNA samples. Copy number analysis (left panel, showing only tumor samples) revealed an absence of deletions or amplifications on chromosomes 6, 10, and 18 of these patients. Strikingly, analysis of these chromosomes using DNA samples from MDS patients, compared with 100 unpaired controls, detected 9 large and numerous short segments of apparent CNLOH (Figure 2 middle panel). By contrast, analysis of the same MDS DNA samples with the use of paired samples showed a complete lack of LOH.
False discovery of CNLOH in unpaired studies. An SNP array analysis of 10 MDS patients for chromosomes 6, 10, and 18 is shown (Affymetrix StyI 250K arrays). Copy number analysis (only tumor samples are shown) reveals no deletions or amplifications (left panel), whereas inferred LOH analysis based on paired samples (Figure 1) reveals no LOH (right panel). However, an inferred LOH analysis of 10 patients based on 100 unrelated controls, processed in the same laboratory and core facility, reveals 9 large segments (and several short segments) of falsely discovered LOH resulting from the lack of matched normal samples. The segment sizes are: 4.3, 2.3, 5.6, and 3.5 Mb (chromosome 6, patients 2, 4, 6, and 7), 11.3, 10.6, and 12.6 Mb (chromosome 10, patients 7, 9, and 10), and 12.2 and 10.6 Mb (chromosome 18, patients 1 and 7). The organization of the figure corresponds to Supplemental Figure 3 in Radtke et al.18
False discovery of CNLOH in unpaired studies. An SNP array analysis of 10 MDS patients for chromosomes 6, 10, and 18 is shown (Affymetrix StyI 250K arrays). Copy number analysis (only tumor samples are shown) reveals no deletions or amplifications (left panel), whereas inferred LOH analysis based on paired samples (Figure 1) reveals no LOH (right panel). However, an inferred LOH analysis of 10 patients based on 100 unrelated controls, processed in the same laboratory and core facility, reveals 9 large segments (and several short segments) of falsely discovered LOH resulting from the lack of matched normal samples. The segment sizes are: 4.3, 2.3, 5.6, and 3.5 Mb (chromosome 6, patients 2, 4, 6, and 7), 11.3, 10.6, and 12.6 Mb (chromosome 10, patients 7, 9, and 10), and 12.2 and 10.6 Mb (chromosome 18, patients 1 and 7). The organization of the figure corresponds to Supplemental Figure 3 in Radtke et al.18
One could argue that the introduction of a size-exclusion rule for the extent of the CNLOH would reduce the likelihood of false discovery.20 Although stringent exclusion criteria will eliminate the majority of spurious small segments of apparent CNLOH, the false discovery rate could well remain significant. In the example shown in Figure 2, exclusion of segments of up to 2 Mb in size would still leave 9 segments on these chromosomes that would be falsely detected as representing CNLOH. The detection of interstitial CNLOH appears to be rare based on the analysis of paired samples, as one would predict based on the requirement for 2 separate chromosomal breaks to occur during recombination between the 2 homologous chromosomal regions. This criterion can exclude many regions of apparent CNLOH that otherwise would be detected in comparisons with unmatched normal control samples, although at least one large telomeric region of false discovery illustrated in Figure 2 would be difficult to exclude. In addition, any rare interstitial regions of CNLOH occurring in malignant clones would be overlooked without matched normal control DNA samples for comparison. Therefore, both predictive computational methods and the use of size- or physical-location-based exclusion criteria are flawed methods for eliminating the potential for false discovery of CNLOH. Only paired sample analysis can ensure error-free detection of regions of pathobiologic interest.
SNP array analysis in MDS
Among the hematologic malignancies, MDS offers a compelling model for SNP array investigation to discover new clonal molecular changes with implications for pathogenesis. This status is underscored by the apparent lack of cytogenetic abnormalities in 40% to 60% of cases and the general paucity of molecular tests that can be used to distinguish MDS from nonclonal bone marrow diseases. One of the first studies to apply high-density SNP arrays (250K) to MDS focused mainly on CNLOH, identifying this alteration in a remarkably high 46% of 119 patients.21 Unfortunately, this study was based on unpaired DNA samples and used a minimal cut-off size of 2 Mb. A comparative analysis of 13 cases with paired constitutional DNA revealed that 12 of these apparent CNLOH regions were also present in the corresponding normal DNA, suggesting that the majority of the reported instances of CNLOH represented inherited regions of inconsequential apparent homozygosity rather than clonally acquired, disease-associated CNLOH. Similarly, using 50K arrays22,23 and 250K arrays24 in 3 studies of MDS patient samples, Gondek et al found a frequency of acquired CNLOH of 20% to 33%. Moreover, the 250K analysis revealed more than 90 microdeletions in 174 patients with a size less than 1 Mb, well within the range of individual copy number variations. Because these studies used only a limited number of matched controls, the possibility of high false discovery rates must be considered. Indeed, we prospectively analyzed matched pairs of bone marrow and buccal swab DNA for each of 51 MDS patients,15 identifying acquired CNLOH and microdeletions in only 6 and 1 of the patients, respectively. Importantly, recurring CNLOH was found on chromosomes 4q and 7q, indicating that the affected segments might harbor a classic tumor suppressor, which requires inactivation of both alleles. Indeed, more recent studies have confirmed that regions of 4q CNLOH include the TET2 gene locus.2,3 Although the putative tumor suppressor on chromosome 7q has yet to be identified, the presence of CNLOH implies that lesions affecting this region can be reduced to homozygosity. By contrast, CNLOH has not been identified on chromosome 5q, pointing to the presence of a haploinsufficient tumor suppressor gene whose complete inactivation would restrict the proliferation or survival of the malignant clone.
SNP array analysis in AML
Two recent studies of pediatric and adult acute myeloid leukemia (AML) have underscored the value of paired samples in SNP array analysis. Walter et al10 analyzed 86 adult AML cases using ultra-high resolution Affymetrix 6.0 SNP arrays with paired normal DNA in all cases, whereas Radtke et al18 studied 111 pediatric AML cases with high-resolution SNP arrays (combination of 100K and 500K arrays) using paired samples for 65 patients and higher stringency criteria to define abnormalities for the remaining 46 patients. The studies identified only 12 or 18 regions with significantly recurrent CNAs, respectively, and an average of 2.3 CNAs/AML genome.10,18 Thus, given the low frequency of recurring CNAs in AML compared with other cancers, it becomes critically important to discriminate between true acquired CNAs and inherited CNVs. However, even with ultra-high resolution arrays, and the analysis of paired normal DNA, which is needed to identify true CNAs and to eliminate confounding CNVs from the analysis, it can still be challenging to identify small CNAs (> 1 Mb) with certainty, as indicated by a low rate of validation by additional criteria and experimental analysis.10 Both of these studies of AML reported a low frequency of CNLOH, 8% and 13%, in contrast to a frequency of 17% reported in a study without paired analysis.25 Indeed, these lower frequencies are in accord with our study of MDS, in which CNLOH was found in 12% of the cases.15
Future prospects
SNP arrays offer state-of-the-art technology for uncovering somatically altered regions of the genome, which may include important tumor suppressors and oncogenes, and for evaluating their effects on disease progression and response to treatment. Yet, the time is rapidly approaching when this high-resolution analytical tool will be replaced by next-generation whole exome or whole genome sequencing of matched samples of malignant and normal DNA, to detect acquired base-pair changes and regions of altered copy number in the malignant clone.16,17 Until then, there are at least 2 applications of SNP array analysis that could be productively exploited. One is to determine the true pathogenic significance of recurrent regions of CNLOH in the hematologic malignancies. This strategy is warranted because CNLOH is clearly capable of generating homozygosity for mutated tumor suppressor genes or oncogenes involved in malignant transformation; and its presence, if recurrent, indicates that a discrete mutation will probably be found, which is not the case for deletions that cause haploinsufficiency. The second application, which is to define clinically important subsets of patients based on molecular abnormalities linked to pathogenesis, will require a systematic approach based on a careful follow-up of uniformly treated patients. If both tumor and normal tissue samples were routinely collected from patients with hematologic malignancies and subjected to analysis with state-of-the-art SNP arrays, always including matched tumor and normal DNA samples, we envision that the accrued data will constitute a valuable resource for classifying individual cases. In our view, SNP array or deep-sequencing protocols to identify somatic genetic changes linked to transformation, including CNLOH, deletion, and amplification, will prove superior to standard cytogenetic analysis for classification of high-risk MDS and AML cases, thus accelerating the current momentum toward personalized molecular medicine.
Acknowledgments
The authors thank John Gilbert for editorial review and Donna S. Neuberg and David P. Steensma for helpful discussions.
This work was supported in part by the National Institutes of Health (P01 grant CA-108631).
National Institutes of Health
Authorship
Contribution: S.H. designed and performed research, analyzed data, and wrote the paper; C.L. analyzed data and wrote the paper; and A.T.L. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Thomas Look, Department of Pediatric Oncology, Dana-Farber Cancer Institute, Mayer Bldg, Rm 630, 44 Binney St, Boston, MA 02115; e-mail: thomas_look@dfci.harvard.edu.