Abstract
All-trans retinoic acid (ATRA) induces granulocytic differentiation and apoptosis in acute promyelocytic leukemia (APL) cells, although the detailed mechanisms are not fully understood. We investigated ATRA-induced cellular responses mediated by the transcription factor FOXO3A in APL cells. FOXO3A was constitutively phosphorylated and localized in the cytoplasm in both APL-derived NB4 cells and primary APL cells. Upon treating the cells with ATRA, FOXO3A phosphorylation was reduced and FOXO3A translocated into the nucleus. In addition, the expression of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), a target molecule for FOXO3A, was increased at the transcriptional and protein levels. As expected, transfection of a short hairpin RNA (shRNA) oligonucleotide specific for FOXO3A significantly inhibited ATRA-induced granulocytic differentiation and apoptosis in NB4 cells. In NB4-derived ATRA-resistant NB4/RA cells, neither FOXO3A nuclear localization nor subsequent TRAIL induction was observed after ATRA treatment. Furthermore, forced expression of active FOXO3A in the nucleus induced TRAIL production and apoptosis in NB4/RA cells. We conclude that activation of FOXO3A is an essential event for ATRA-induced cellular responses in NB4 cells. FOXO3A is a promising target for therapeutic approaches to overcome ATRA resistance in APL.
Introduction
Acute promyelocytic leukemia (APL) is a subtype of acute myeloid leukemia caused by reciprocal translocations of the long arms of chromosomes 15 and 17, which prevents cellular differentiation into mature neutrophils.1,2 The translocation of the promyelocytic leukemia (PML) gene on chromosome 15 and a retinoic acid receptor α (RARα) gene on chromosome 17 generates a PML-RARα fusion protein that inhibits PML-dependent apoptotic pathways in a dominant negative fashion. This fusion protein also blocks granulocytic differentiation by direct transcriptional inhibition of retinoic acid target genes.3-7
Studies have revealed that ATRA arrests cell growth, granulocytic differentiation, and apoptosis in APL cells via proteasome-dependent degradation of PML-RARα fusion protein and subsequent PML–nuclear body (NB) formation.8,9 PML-NBs are dynamic intranuclear structures that promote specific nuclear events, such as apoptosis and proliferation, in response to various cellular stresses. PML is the essential component of PML-NBs and functions as a tumor suppressor. However, disruption of PML-NBs by the PML-RARα fusion protein inhibits endogenous PML tumor-suppressive functions in APL cells.8,10 Therefore, degradation of PML-RARα fusion protein and reorganization of PML-NBs during ATRA treatment are regarded as critical cellular responses, similar to the cell growth arrest and apoptosis of leukemia cells.8-10
It has been reported that PML, together with protein phosphatase 2A, nuclear AKT, and the transcription factor FOXO3A, orchestrates a tumor suppressor network. PML specifically recruits protein phosphatase 2A into PML-NBs and prevents phosphorylation of AKT inside the nucleus, leading to inactivation of AKT.11 Because FOXO3A is phosphorylated and inactivated by AKT, PML deficiency or loss of PML function may lead to constitutive phosphorylation and activation of AKT, resulting in inactivation of FOXO3A-mediated transcription.
FOXO3A, also named FKHRL1, is a member of the Forkhead family of transcription factors, characterized by a highly conserved forkhead domain with a winged-helix motif and measurable DNA-binding activity. This domain functions as a transcription factor for various genes, including p27/Kip1, p130-Rb2, and cyclin D1/2 (cell-cycle regulation),12-14 tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), Bim, and Fas ligand (apoptosis),15-17 and GADD45a (DNA repair).18 Recently, it was reported that FOXO3A plays an important role in maintenance of the hematopoietic stem cell pool via modulation of the response to physiologic oxidative stress.19,20 The function and localization of FOXO3A depends on its phosphorylation status. FOXO3A is phosphorylated when cells are stimulated with serum or growth factors. FOXO3A is then exported from the nucleus to the cytoplasm, where it interacts with 14-3-3 proteins, resulting in its loss of function as a transcription factor.21,22 By contrast, the dephosphorylated form of FOXO3A translocates into the nucleus and activates target genes when cells are deprived of serum or growth factors.22,23 Thus, the transcriptional activity of FOXO3A is negatively regulated by phosphorylation. FOXO3A is phosphorylated primarily by a serine/threonine kinase, AKT, at threonine 32 (T32), serine 253 (S253), and serine 315 (S315) residues.24 Aberrant activation of AKT due to constitutive activation of the upstream molecules, such as BCR-ABL and FLT3 internal tandem duplication, is often observed in hematologic malignancies.25,26 Consistent with constitutive activation of AKT, FOXO3A phosphorylation has been observed in chronic myeloid leukemia (CML), acute myelogenous leukemia with FLT3 internal tandem duplication, and multiple myeloma.22,26,27 These findings strongly suggest that loss of the transcriptional function of FOXO3A by phosphorylation is involved in the pathophysiology of leukemia and multiple myeloma.
We show here that in a basal state, FOXO3A lies downstream of the PML-RARα fusion protein in its inactivated form, and that ATRA induces apoptosis by converting FOXO3A from its inactive to its active form in the APL-derived NB4 cell line.
Methods
Reagents and antibodies
Fetal calf serum was purchased from Hyclone. RPMI 1640, ATRA, and 4-hydroxytamoxifen (4-OHT) were purchased from Sigma-Aldrich. Nitroblue tetrazolium (NBT) was purchased from Dojindo Laboratories. Antibodies against phospho-FOXO3A (T32) and FOXO3A were purchased from Upstate Biotechnology. Anti-RARα (C20), anti-PML (PG-M3), anti-FOXO1 (H128), and anti–phospho AKT (T308) were purchased from Santa Cruz Biotechnology. Antibody against TRAIL was purchased from BD Biosciences. Antibodies against AKT and β-actin were purchased from Cell Signaling Technology. Recombinant human Apo2L (KillerTRAIL, soluble human recombinant) was purchased from Alexis Biochemicals. pMax vector was purchased from Amaxa. pcDNA containing human FOXO3A-TM-ER cDNA (a fusion gene of FOXO3A-TM, a triple mutant of FOXO3A, in which all three AKT phosphorylation sites have been mutated to alanine, and the estrogen receptor) was provided by Dr Paul Coffer (University Medical Center). pH1Rep7 vector was provided by Dr Masayuki Okada (Kan Research Institute). The human TRAIL promoter gene, beginning 1523 nucleotides upstream of the transcriptional start site, inserted into pGL2-basic luciferase vector (−1523/+55 pGL2m) was provided by Dr B. Mark Evers (University of Texas Medical Branch). pcDNA3 vector containing a dominant negative mutant of AKT cDNA (DN-AKT) was provided by Dr Alfonso Bellacosa (Fox Chase Cancer Research Center).
Cell culture
NB4 and NB4/RA cell lines were provided by Dr Tomoki Naoe (Nagoya University). Informed consent in accordance with the Declaration of Helsinki was obtained from APL patients, and approvals were obtained from the Ethics Committee of the University of Yamanashi. Cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Plasmid construction and transfection
A short hairpin RNA (shRNA) oligonucleotide expression cassette was subcloned into the BamHI-HindIII site of the pH1Rep7 vector. The target sequence for FOXO3A was GAUCCCACCUCAUGGACGACCUGCUUUCAAGAGAAGCAGGUCGUCCAUGAGGUUUUUUUGGAAA. The control sequence for the scramble shRNA was SC1:GAUCCCGCGGCCGAUACUACCCUUAUUCAAGAGAUAAGGGUAGUAUCGGCCGC-UUUUUUGGAAA; SC2:GAUCCCGUCUCCACGCGCAGUACAU-UUCAAGAGAAUGUACUGCGCGUGGAGACUUUUUUGGAAA. FOXO3A-TM-ER cDNA was digested by HindIII and NotI and ligated into the HindIII and NotI sites of the pCEP4 vector (Invitrogen). pH1Rep7 vector encoding shRNA for FOXO3A or scrambled control shRNA were transfected by DMRIE-C according to the manufacturer's instructions (Invitrogen). The pCEP4 vector encoding FOXO3A-TM-ER and pcDNA3 encoding DN-AKT were transfected by electroporation. Electroporations were performed with a Gene PulserXcell unit (Bio-Rad) under conditions of 125 V, 25 ms, and a square-wave pulse. FOXO3A shRNA, scrambled control shRNA, and FOXO3A-TM-ER–expressing clones were selected using 175 μg/mL hygromycin B (Invitrogen). Empty vectors were transfected for control purposes (VCont).
Cell proliferation
Cell growth was assayed using the CellTiter-Glo Luminescent Cell Viability Assay Kit (Promega) according to the manufacturer's instructions. Luminescence was measured using Fluoroskan Ascent FL (Thermo Fisher Scientific). All measurements were performed in triplicate.
Quantitative reverse transcription–PCR
Total RNA was isolated from cells using the RNeasy Mini Kit (QIAGEN) and reverse transcribed into complementary DNA (cDNA) as a template in the real-time quantitative polymerase chain reaction (PCR) analysis. A primer and TaqMan probe set were predesigned by Applied Biosystems (TRAIL: TaqMan Gene Expression Assays Product number Hs00921974_m1). Primers and probes for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used to amplify GAPDH mRNA for normalization (Applied Biosystems). TRAIL and GAPDH mRNAs were detected with an ABI PRISM 7700 Sequence Detection System (Applied Biosystems), and quantification was performed using the relative standard curve method. All measurements were performed in triplicate.
Granulocytic differentiation
Granulocytic differentiation of the cells was determined by a NBT reduction assay as described previously28 and by CD11b expression analysis. To analyze CD11b expression, harvested cells were incubated with phycoerythrin-conjugated anti-CD11b antibody (BD Biosciences), and positive cells were counted by flow cytometry using the Guava EasyCyte Plus System (Guava Technologies). For NBT assays, we counted more than 200 cells across 3 different microscopic fields in a blinded fashion and subsequently calculated the relevant positive cell percentages.
Detection of apoptotic cells
Apoptosis was measured using the Guava Nexin assay kit (Guava Technologies) according to the manufacturer's instructions. Scramble or FOXO3A shRNA-expressing NB4 clone cells were incubated with 1μM ATRA or final volume of 0.1% ethanol. FOXO3A-TM-ER–expressing NB4/RA clone cells were incubated with 0.7μM 4-OHT or final volume of 0.07% ethanol. After incubation, cells were washed with phosphate-buffered saline (PBS) and stained using phycoerythrin-labeled annexin V. Apoptotic cells were analyzed with the Guava EasyCyte Plus System (Guava Technologies). In some experiments, apoptotic cells were detected using 1.0μM Hoechst 33342 (Molecular Probes). Imaging analysis was performed under an Olympus model IX70 microscope. After cotransfection with pMax and FOXO3A-TM vector, cells were incubated with phycoerythrin-labeled annexin V. Proportions of both GFP- and annexin V–positive cells were determined by flow cytometry.
Preparation of cell lysates and Western blotting
Whole protein extraction and Western blotting were performed as described previously.21 The blots were detected with an enhanced chemiluminescence substrate (ECL Western blot detection system; GE Healthcare).
Immunofluorescence microscopy
Cells were incubated on poly-l-lysine–coated slides for 5 minutes, washed gently with PBS, and fixed in 10% formaldehyde/PBS at room temperature (RT). After washing with PBS, the cells were permeabilized with 0.05% Triton X-100 in PBS and then incubated in a blocking buffer (DS Pharma Biomedical) for 60 minutes at RT. The cells were then incubated with the primary antibodies overnight at 4°C. After again washing with PBS, the slides were incubated with the blocking buffer for 60 minutes and then incubated with the secondary antibody, namely Alexa488-conjugated goat anti–rabbit or anti–mouse immunoglobulin G (IgG; Molecular Probes), in blocking buffer for 60 minutes at RT. Nuclei were stained with 10nM TO-PRO-3 iodide (Molecular Probes). Standard epifluorescence was observed under an Olympus IX70 microscope. Confocal images were obtained with a Leica TCS-NT laser confocal microscope (Leica).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed according to the manufacturer's instructions (Upstate Biotechnology). Briefly, NB4 cells were incubated with or without 1.0μM ATRA. Forty-eight hours later, formaldehyde was added to cell culture media to a final concentration of 1% and then the cells were incubated for 10 minutes on a shaking platform at RT. After the addition of glycine at concentrations of up to 0.136M, cells were incubated for 5 minutes and then washed with cold PBS. Cell pellets were obtained, resuspended in sodium dodecyl sulfate lysis buffer, and sonicated with a Bioruptor (Cosmo Bio) for 30 seconds at maximum setting 10 times at 1-minute intervals. Immunoprecipitation was carried out overnight at 4°C with rotation, using antibodies specific for FOXO3A (H-144; Santa Cruz Biotechnology) or rabbit IgG (Santa Cruz Biotechnology). After incubation with protein A, the beads were harvested, washed, and eluted per the manufacturer's instructions. Cross-links were reversed by incubation for 6 hours at 65°C, and DNA was purified from the supernatant by phenol/chloroform extraction and ethanol precipitation. Precipitated DNA was analyzed by conventional PCR. The PCR primer pairs spanned 2 FOXO3A-specific binding sites identified in the 5′-flanking region of the human TRAIL gene: proximal: forward, 5′-AAACAGGCCTTGTGCCTATG-3′; reverse, 5′-GCTCTTGTCTCAAAGTAGTCGTT-3′, distal: forward, 5′-GGCGATAAAGTGAGATTCTGTCA-3′; reverse, 5′-TCCCCACTATCAGATGTCATAGG-3′. Conventional PCR conditions were used: 35 cycles of 94°C for 45 seconds; 60°C for 45 seconds; and 72°C for 45 seconds. The PCR products were analyzed by electrophoresis through a standard 2% agarose gel.
Promoter activity assay
The human TRAIL promoter construct containing the 1.5-kb segment 5′ upstream of the start codon (−1523/+55 pGL2m) was cotransfected into NB4 cells with the pRL-TK plasmid by electroporation. Mutations in the FOXO3A binding sites at the proximal and distal regions of the human TRAIL promoter gene were generated using a KOD-Plus mutagenesis kit (Toyobo) according to the manufacturer's instructions. The proximal FOXO3A binding site at −138 to −121 (TTTCAGTTTCCCTCCTTT) of the TRAIL promoter gene was deleted using the primer set-p (forward 5′-CAACGACTACTTTGAGACAAGAGCTGTCCCTGGGCAGTA-3′, reverse 5′-GAAGCTCCTCTCCTTGCCCTCAGGATCCATGCACCCCT-TATC-3′). The distal FOXO3A-binding site at −986 to −996 was mutated from ATAAATAAATA to CGCCCTCCCGG using the primer set-d (forward 5′-CCGGGAGGGCGTTTTTTGACAGAATCTCACTTTATCGC-CCAGGCTGGAGTGCAGTGGTGCAATGGCACAATCTCGGCCCA-CTACAACCTCCACCTCCCAGATTCAAGCAATTCTCCTGCCTCAG-CCTCCCAAGT-3′, reverse 5′-CATGAAAGAGAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAGAAAG-AAAGAAAGAAGGAAAGAAGGAAAGAATAGAAAAGAAAAGA-AAGAAAGGAAGGAAGAAAAGGAAAGAAAGAAATGC-3′). The human TRAIL promoter gene was mutated at both the proximal and distal FOXO3A binding sites using both primer set-p and set-d. These constructs, tentatively designated as mt1, mt2, and mt3, were transfected into the NB4 cells. Cells were then treated with 1μM ATRA or with a final volume of 0.1% ethanol. After incubation for 48 hours, the cells were extracted with 100 μL of passive lysis buffer (Promega), and the firefly and renilla luciferase activity levels were measured using a Dual-Luciferase Reporter Assay System.
Morphologic study
Cells were cytospun and stained with Wright-Giemsa. Morphology was observed under light microscopy.
Statistical analysis
Data are presented as means plus or minus SE. Student t test was used, and the level of significance was set at a P value of less than .05.
Results
ATRA-induced dephosphorylation of FOXO3A and its nuclear localization and expression of its target molecule TRAIL in NB4 cells
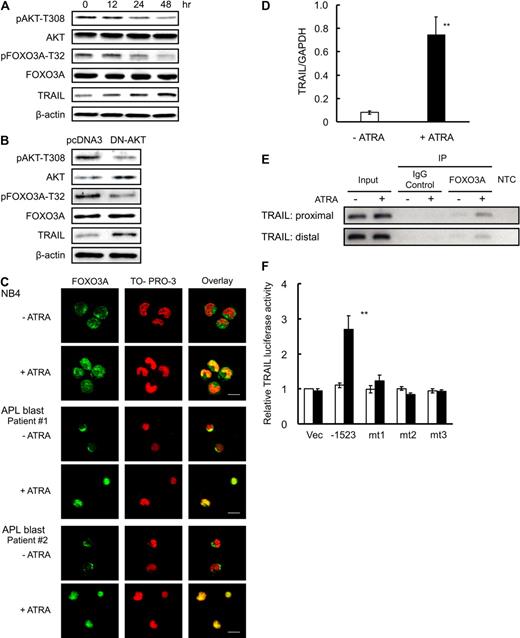
We found that AKT was constitutively phosphorylated at T308 in the basal state of the NB4 cells, and that this phosphorylation was significantly reduced by 24 to 48 hours after ATRA treatment (Figure 1A), indicating that AKT is constitutively activated in the absence of ATRA and its kinase activity is decreased after ATRA treatment in NB4 cells. Because AKT had been reported to directly phosphorylate FOXO3A at T32 (FOXO3A-T32),24 we examined the effect of ATRA on the phosphorylation of FOXO3A-T32. FOXO3A-T32 was constitutively phosphorylated, however, consistent with the kinetics of AKT, the phosphorylation level was significantly reduced at 24 to 48 hours after ATRA treatment in NB4 cells (Figure 1A). In addition, transient expression of a dominant negative mutant of AKT in NB4 cells decreased FOXO3A phosphorylation (Figure 1B). These results indicate that ATRA induces dephosphorylation of FOXO3A at T32 via suppression of AKT kinase activity. Confocal microscopic analysis revealed that FOXO3A was located mainly in the cytoplasm before ATRA treatment and accumulated in the nucleus after 48 hours of ATRA exposure (Figure 1C). Consistent with the results obtained using NB4 cells, our analysis using primary APL blast cells obtained from 2 patients revealed that FOXO3A was constitutively phosphorylated and localized in the cytoplasm before ATRA treatment, and that FOXO3A was dephosphorylated and translocated into the nucleus after ATRA treatment (Figure 1C and supplemental Figure 2A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Thus, ATRA-induced activation of FOXO3A is a common nuclear event in primary APL cells and is not limited to NB4 cells. Morphologic analysis revealed that primary cells from 1 patient (sample 2) became mature granulocyte-like cells after ATRA treatment in vitro (supplemental Figure 2B left and middle panels), and indeed the patient achieved hematologic complete remission after ATRA differentiation therapy (supplemental Figure 2B right panel).
ATRA induces dephosphorylation of FOXO3A and translocation of FOXO3A into the nucleus. (A) NB4 cells were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for the indicated time periods. Total protein was extracted and analyzed by Western blotting using antibodies for the indicated antigens. β-actin was used as an internal control. (B) NB4 cells were transfected with empty vector (pcDNA3) or dominant negative form of AKT (DN-AKT) by electroporation. After incubation for 48 hours, the cells were harvested. Total protein was extracted and submitted to Western blotting with the indicated antibodies. (C) NB4 cells or APL blast cells from patients were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for 48 hours. After fixation and permeabilization, cells were incubated with anti-FOXO3A antibody and Alexa488-conjugated goat anti–rabbit IgG (green). The nuclei were stained with TO-PRO-3 (red). Confocal imaging analysis was performed using a Leica TCS-NT laser confocal microscope. Scale bar represents 10 μm. (D) TRAIL mRNA was detected by quantitative RT-PCR using TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. Results are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with the ethanol control. (E) After cross-link reversal, the recovered DNA was subjected to ChIP analysis with FOXO3A antibody or normal serum IgG (as a control) using conventional PCR. The presence of promoter DNA before immunoprecipitation was confirmed by PCR (input). NTC refers to the no-template control, which lacked input DNA. (F) TRAIL promoter activity, as determined by measurement of luciferase activity, was assayed in NB4 cells transfected with the indicated constructs after a 48-hour incubation with 1.0μM ATRA (■) or a final volume of 0.1% ethanol (□). mt1, mt2, and mt3 represent the human TRAIL promoter gene that had a mutated proximal FOXO3A binding site, a mutated distal FOXO3A binding site, or both mutated sites. Results are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with empty pGL2m vector-transfected control (Vec).
ATRA induces dephosphorylation of FOXO3A and translocation of FOXO3A into the nucleus. (A) NB4 cells were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for the indicated time periods. Total protein was extracted and analyzed by Western blotting using antibodies for the indicated antigens. β-actin was used as an internal control. (B) NB4 cells were transfected with empty vector (pcDNA3) or dominant negative form of AKT (DN-AKT) by electroporation. After incubation for 48 hours, the cells were harvested. Total protein was extracted and submitted to Western blotting with the indicated antibodies. (C) NB4 cells or APL blast cells from patients were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for 48 hours. After fixation and permeabilization, cells were incubated with anti-FOXO3A antibody and Alexa488-conjugated goat anti–rabbit IgG (green). The nuclei were stained with TO-PRO-3 (red). Confocal imaging analysis was performed using a Leica TCS-NT laser confocal microscope. Scale bar represents 10 μm. (D) TRAIL mRNA was detected by quantitative RT-PCR using TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. Results are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with the ethanol control. (E) After cross-link reversal, the recovered DNA was subjected to ChIP analysis with FOXO3A antibody or normal serum IgG (as a control) using conventional PCR. The presence of promoter DNA before immunoprecipitation was confirmed by PCR (input). NTC refers to the no-template control, which lacked input DNA. (F) TRAIL promoter activity, as determined by measurement of luciferase activity, was assayed in NB4 cells transfected with the indicated constructs after a 48-hour incubation with 1.0μM ATRA (■) or a final volume of 0.1% ethanol (□). mt1, mt2, and mt3 represent the human TRAIL promoter gene that had a mutated proximal FOXO3A binding site, a mutated distal FOXO3A binding site, or both mutated sites. Results are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with empty pGL2m vector-transfected control (Vec).
To determine whether FOXO3A is functionally activated as a transcription factor by ATRA in NB4 cells, we examined the expression of TRAIL, reported to be a direct transcriptional target for FOXO3A.17 TRAIL expression was significantly increased at both the mRNA and protein levels after ATRA treatment (Figure 1A,D). To further characterize the ATRA-induced transcriptional activation of TRAIL, we performed ChIP analysis of 2 regions of the transcriptional start site of the TRAIL gene (proximal: −138 to −121; distal: −995 to −986).17,29 As shown in Figure 1E, the FOXO3A binding site was detected in both regions, indicating that ATRA activates the TRAIL gene promoter through FOXO3A binding sites in NB4 cells. Furthermore, we found that the promoter activity of the TRAIL gene, including both FOXO3A binding sites, was significantly increased after ATRA treatment. However, the enhanced promoter activity was completely diminished in the TRAIL gene promoter that had a mutated proximal binding site (mt1), a mutated distal binding site (mt2), or both mutated sites (mt3; Figure 1F). These results suggested that both the proximal and distal FOXO3A binding sites are necessary for ATRA-induced transcriptional activation of the human TRAIL gene promoter. Taken together with the observation that a dominant negative AKT induced up-regulation of TRAIL at the mRNA level (supplemental Figure 1), our data indicate that TRAIL expression is tightly linked to the reduced AKT kinase activity and subsequent activation of FOXO3A observed in ATRA-treated NB4 cells.
FOXO3A lies downstream of PML-RARα fusion protein in NB4 cells
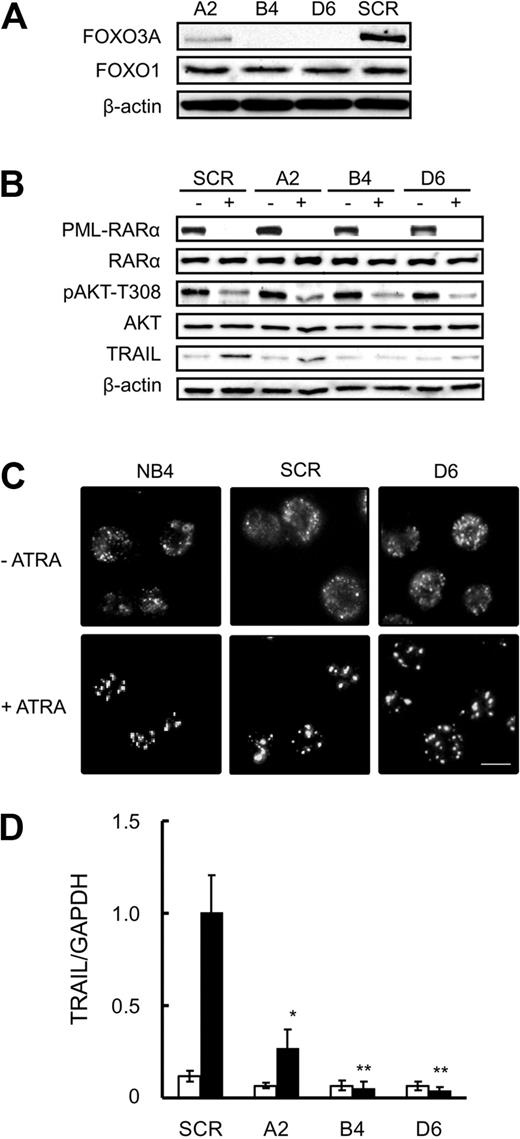
Next, we examined a functional role for FOXO3A in ATRA-induced cellular responses, including apoptosis, using RNA interference-mediated silencing. To this end, we established 3 stable clones expressing shRNA specific to FOXO3A, designated A2, B4, and D6, and a scramble shRNA–expressing clone, designated the SCR. Western blotting analysis with anti-FOXO3A antibody revealed that endogenous FOXO3A protein levels were markedly diminished in B4 and D6 clones and significantly reduced in the A2 clone, compared with the SCR. These FOXO3A knockdown clones and the SCR clone expressed similar levels of FOXO1 protein, another member of the FOXO subfamily (Figure 2A), indicating that the shRNA used in this study specifically down-regulated FOXO3A.
Knockdown of FOXO3A does not affect ATRA-induced PML-RARα degradation or subsequent formation of NBs in NB4 cells. (A) Total protein was extracted from SCR or FOXO3A shRNA clones cells and submitted to Western blotting with antibodies against the indicated antigens. β-Actin was used as an internal control. (B) SCR or FOXO3A shRNA clones cells were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for 48 hours. Total protein was extracted and submitted to Western blotting with antibodies against indicated the antigens. β-Actin was used as the internal control. (C) NB4, SCR, or FOXO3A shRNA clones were incubated with or without 1.0μM ATRA for 48 hours and fixed on poly-l-lysine–treated glass slides. After permeabilization, cells were incubated with anti-PML antibody and Alexa488-conjugated goat anti–mouse IgG. Imaging analysis was performed with an Olympus IX70 microscope. Scale bar represents 10 μm. (D) TRAIL mRNA was detected by quantitative RT-PCR using TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. ■ and □ represent 1.0μM ATRA and 0.1% ethanol, respectively. Results are the mean ± SE of 3 independent experiments. *P < .05 and **P < .01 compared with the SCR control.
Knockdown of FOXO3A does not affect ATRA-induced PML-RARα degradation or subsequent formation of NBs in NB4 cells. (A) Total protein was extracted from SCR or FOXO3A shRNA clones cells and submitted to Western blotting with antibodies against the indicated antigens. β-Actin was used as an internal control. (B) SCR or FOXO3A shRNA clones cells were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for 48 hours. Total protein was extracted and submitted to Western blotting with antibodies against indicated the antigens. β-Actin was used as the internal control. (C) NB4, SCR, or FOXO3A shRNA clones were incubated with or without 1.0μM ATRA for 48 hours and fixed on poly-l-lysine–treated glass slides. After permeabilization, cells were incubated with anti-PML antibody and Alexa488-conjugated goat anti–mouse IgG. Imaging analysis was performed with an Olympus IX70 microscope. Scale bar represents 10 μm. (D) TRAIL mRNA was detected by quantitative RT-PCR using TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. ■ and □ represent 1.0μM ATRA and 0.1% ethanol, respectively. Results are the mean ± SE of 3 independent experiments. *P < .05 and **P < .01 compared with the SCR control.
PML-RARα protein degradation and subsequent PML-NB formation are critical triggers for ATRA-induced cellular responses in APL cells.9,10 As shown in Figure 2B, PML-RARα protein was detected in the SCR and in the 3 FOXO3A knockdown clones, and the related expression level was drastically reduced 48 hours after ATRA treatment. AKT phosphorylation also decreased in the SCR and in all the FOXO3A knockdown clones. As determined by immunofluorescence staining with an anti-PML antibody, PML and PML-RARα fusion proteins exhibited a typical microspeckled nuclear distribution pattern in the absence of ATRA, and NBs were formed in NB4 cells, the SCR, and in all 3 FOXO3A shRNA clones after ATRA treatment (Figure 2C and data not shown). Thus, we conclude that the shRNA-induced down-regulation of FOXO3A did not interfere with ATRA-induced degradation of PML-RARα fusion protein, AKT dephosphorylation, or the reconstitution of NBs with PML protein. Furthermore, TRAIL protein increased in the SCR but not in the B4 and D6 clones after ATRA treatment (Figure 2B). As expected, quantitative reverse-transcription (RT)–PCR analysis revealed that TRAIL expression is regulated at the mRNA level (Figure 2D). ATRA treatment enhanced TRAIL promoter activity in the SCR, but not in the FOXO3A shRNA clones (supplemental Figure 3). These results indicate that FOXO3A lies downstream of the PML-RARα fusion protein and strongly support our hypothesis that FOXO3A plays a pivotal role in ATRA-induced TRAIL expression in NB4 cells.
FOXO3A is required for ATRA-induced granulocytic differentiation and apoptosis
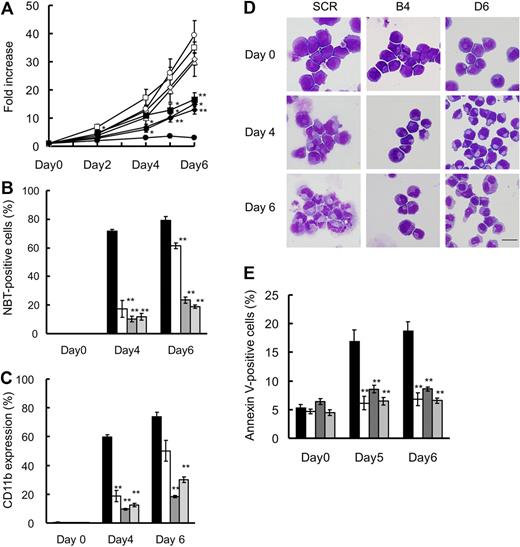
The SCR and 3 FOXO3A shRNA clones proliferated in a comparable manner in the absence of ATRA (Figure 3A open symbols). Cell growth of the SCR clone was completely inhibited by the addition of ATRA, whereas the FOXO3A shRNA clones showed partial suppression on account of ATRA (Figure 3A closed symbols), suggesting that FOXO3A may be involved in the ATRA-induced cell growth inhibition of NB4 cells.
Knockdown of FOXO3A suppresses ATRA-induced TRAIL expression, granulocytic differentiation, and cell death of NB4 cells. (A) SCR or FOXO3A shRNA clones were incubated with 1.0μM ATRA (closed) or 0.1% ethanol (open) for the indicated time periods. Cell growth was assessed by the CellTiter-Glo Luminescent Cell Viability Assay Kit. Results are presented as the mean ± SE of 3 independent experiments relative to the results obtained at day 0. SCR (circles), A2 (squares), B4 (triangles), D6 (diamonds). *P < .05: A2 (days 2, 4, and 5), B4 (day 6), D6 (days 4 and 5); **P < .01: A2 (day 6), B4 (day 5), D6 (day 6), compared with SCR with ATRA. Granulocytic differentiation was determined by an NBT reduction assay (B) and CD11b expression (C) by FACS analysis. (D) Morphologic changes were examined by Wright-Giemsa staining and use of an Olympus BX51. Scale bar represents 10 μm. (E) Annexin V–positive cells were detected by FACS analysis. Results of panels B, C, and E are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with SCR; SCR ( ), A2 (░), B4 (
), A2 (░), B4 ( ), D6 (
), D6 ( ).
).
Knockdown of FOXO3A suppresses ATRA-induced TRAIL expression, granulocytic differentiation, and cell death of NB4 cells. (A) SCR or FOXO3A shRNA clones were incubated with 1.0μM ATRA (closed) or 0.1% ethanol (open) for the indicated time periods. Cell growth was assessed by the CellTiter-Glo Luminescent Cell Viability Assay Kit. Results are presented as the mean ± SE of 3 independent experiments relative to the results obtained at day 0. SCR (circles), A2 (squares), B4 (triangles), D6 (diamonds). *P < .05: A2 (days 2, 4, and 5), B4 (day 6), D6 (days 4 and 5); **P < .01: A2 (day 6), B4 (day 5), D6 (day 6), compared with SCR with ATRA. Granulocytic differentiation was determined by an NBT reduction assay (B) and CD11b expression (C) by FACS analysis. (D) Morphologic changes were examined by Wright-Giemsa staining and use of an Olympus BX51. Scale bar represents 10 μm. (E) Annexin V–positive cells were detected by FACS analysis. Results of panels B, C, and E are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with SCR; SCR ( ), A2 (░), B4 (
), A2 (░), B4 ( ), D6 (
), D6 ( ).
).
Because cell growth arrest is often associated with cell differentiation, we examined whether ATRA-induced cell growth arrest may be due to blockage of granulocytic differentiation in FOXO3A shRNA clones. Using the NBT reduction assay, we determined that more than 70% of the SCR cells were positive for NBT staining after 4 days (Figure 3B). By contrast, fewer than 20% of the cells were positive among ATRA-treated B4 and D6 clone cells during the observation periods. For the A2 clone, in which FOXO3A knockdown was incomplete, approximately 60% of ATRA-treated cells were positive on the sixth day after exposure to ATRA (Figure 3B). Similar results were obtained from our CD11b expression analysis (Figure 3C). In addition, morphologic analysis revealed that B4 and D6 clone cells remained blastlike even after ATRA treatment (Figure 3D). These observations suggest that ATRA-induced granulocytic differentiation is dependent on the presence of FOXO3A in NB4 cells.
Next, we examined the involvement of FOXO3A in ATRA-induced apoptosis. As shown in Figure 3E, the ratio of annexin V–positive cells was increased in a time-dependent manner, and approximately 20% of the SCR cells were positive on the sixth day after ATRA treatment. By contrast, apoptosis was significantly reduced during the observation periods up to day 6 in all FOXO3A knockdown clones (Figure 3E). Consistent with these results, TRAIL expression was suppressed in the B4 and D6 clones (Figure 2B), suggesting that FOXO3A is a prerequisite for ATRA-induced cell death in NB4 cells.
Active FOXO3A overcomes ATRA resistance in NB4/RA cells
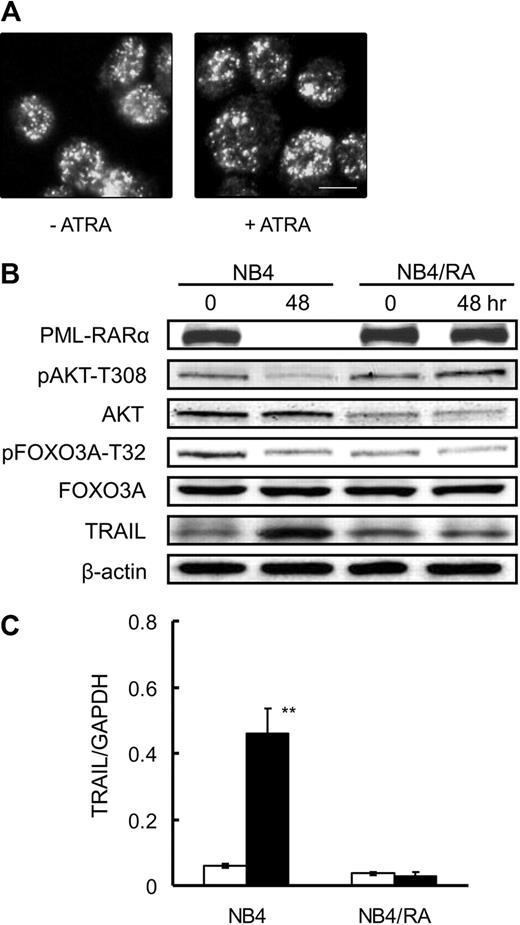
Consistent with previous reports,30 degradation of PML-RARα and formation of PML-NBs were observed in NB4 cells treated with ATRA (Figure 2B); however, neither finding was observed in ATRA-resistant NB4/RA cells (Figure 4A). In addition, the phosphorylation of FOXO3A was unchanged, and TRAIL was not up-regulated at the protein or mRNA level (Figure 4B-C), indicating that FOXO3A is functionally inactive as a transcription factor in NB4/RA cells, even in the presence of ATRA. Considering that there is a loss of ATRA binding capacity as a consequence of a C-to-T substitution (P900L) in the AF2 domain of the RARα gene in NB4/RA cells,30 binding of ATRA to PML-RARα and subsequent degradation of the fusion protein are likely to be critical events for the activation of FOXO3A.
ATRA does not affect PML-RARα degradation or the dephosphorylation of FOXO3A in ATRA-resistant NB4/RA cells. NB4/RA cells were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for 48 hours. (A) Cells were fixed on poly-l-lysine–treated glass slides. After permeabilization, cells were incubated with anti-PML antibody and with Alexa488-conjugated goat anti–mouse IgG. Imaging analysis was performed with an Olympus IX70 microscope. Scale bar represents 10μm. (B) Total protein was extracted and submitted to Western blotting with antibodies for each indicated antigen. (C) TRAIL mRNA was detected by quantitative RT-PCR using TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. Results are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with ethanol control.
ATRA does not affect PML-RARα degradation or the dephosphorylation of FOXO3A in ATRA-resistant NB4/RA cells. NB4/RA cells were incubated with 1.0μM ATRA or a final volume of 0.1% ethanol for 48 hours. (A) Cells were fixed on poly-l-lysine–treated glass slides. After permeabilization, cells were incubated with anti-PML antibody and with Alexa488-conjugated goat anti–mouse IgG. Imaging analysis was performed with an Olympus IX70 microscope. Scale bar represents 10μm. (B) Total protein was extracted and submitted to Western blotting with antibodies for each indicated antigen. (C) TRAIL mRNA was detected by quantitative RT-PCR using TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. Results are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with ethanol control.
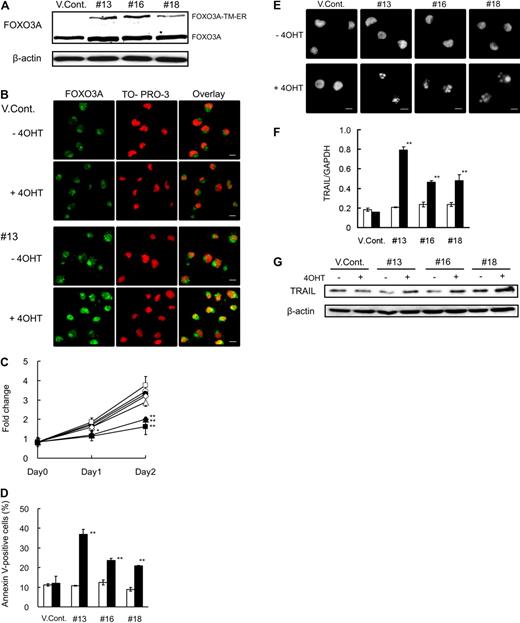
Transient transfection of FOXO3A-TM induced TRAIL expression and apoptosis in both NB4 and NB4/RA cells (supplemental Figure 4A-C). Based on these findings, we speculated that FOXO3A activation is a prerequisite for ATRA-induced cell death and that forced expression of active FOXO3A in the nucleus may be able to overcome ATRA resistance in NB4/RA cells. To evaluate this hypothesis, we generated 3 stable NB4/RA transfectants (nos. 13, 16, and 18) that expressed FOXO3A-TM-ER (Figure 5A). ER fusion proteins become active when cells are exposed to 4-OHT, the ligand for the modified ER.31 The subcellular localization of FOXO3A-TM as determined by confocal microscopy revealed that FOXO3A remained in the cytoplasm of the vector control (VCont) cells treated with or without 4-OHT, whereas FOXO3A-TM-ER clones were selectively expressed in the nucleus after the addition of 4-OHT (Figure 5B). In addition, 4-OHT exerted no inhibitory effect on cell growth of the VCont cells, whereas 4-OHT inhibited the cell growth of clones (Figure 5C). We confirmed that annexin V–positive cell numbers were significantly increased after treatment with 4-OHT in clones (Figure 5D). Furthermore, morphologic analysis after Hoechst 33342 staining revealed apoptosis-specific nuclear fragmentation in 4-OHT–treated clones (Figure 5E). We conclude that the forced expression of active FOXO3A in the nucleus induced apoptosis in NB4/RA cells. In addition, we found that TRAIL expression was significantly increased at the mRNA and protein levels after exposure to 4-OHT in all clones, but not in VCont (Figure 5F-G). These results suggest that expression of active FOXO3A may overcome ATRA resistance, resulting in the induction of TRAIL expression and apoptosis in APL cells.
Forced expression of active FOXO3A induces apoptosis of NB4/RA cells. (A) Total protein was harvested from vector control (VCont) and FOXO3A-TM-ER clone cells and submitted to Western blotting with antibodies for FOXO3A and β-actin. (B) Cells were incubated with 0.7μM 4-OHT or final volume of 0.07% ethanol for 48 hours and fixed on poly-l-lysine–treated glass slides. After permeabilization, cells were incubated with anti-FOXO3A antibody and Alexa488-conjugated goat anti–rabbit IgG (green). Nuclei were stained with TO-PRO-3 (red). Confocal imaging analysis was performed using a Leica TCS-NT laser confocal microscope. Scale bar represents 10 μm. (C) Cells were incubated with 0.7μM 4-OHT (closed) or a final concentration of 0.07% ethanol (open) at the indicated times. Cell growth was assessed using the CellTiter-Glo Luminescent Cell Viability Assay Kit. Results are presented as the mean ± SE of 3 independent experiments relative to the results obtained at day 0; VCont (circles), no. 13 (squares), no. 16 (triangles), and no. 18 (diamonds). *P < .05: no. 13 (day 1); **P < .01: nos. 13, 16, and 18 at day 2 compared with VCont treated with 4-OHT. (D) Annexin V–positive cells were detected by FACS analysis. (E) Nuclei were stained using Hoechst 33342. Imaging analysis was performed with an Olympus IX70 microscope. Scale bar represents 10 μm. (F) TRAIL mRNA was detected by quantitative RT-PCR using predesigned TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. (G) Total protein was extracted at 48 hours and blotted. The blots were probed with antibody for TRAIL and were reprobed with anti–β-actin antibody to confirm equal protein loading. Results of panels D and F are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with the VCont treated with 4-OHT.
Forced expression of active FOXO3A induces apoptosis of NB4/RA cells. (A) Total protein was harvested from vector control (VCont) and FOXO3A-TM-ER clone cells and submitted to Western blotting with antibodies for FOXO3A and β-actin. (B) Cells were incubated with 0.7μM 4-OHT or final volume of 0.07% ethanol for 48 hours and fixed on poly-l-lysine–treated glass slides. After permeabilization, cells were incubated with anti-FOXO3A antibody and Alexa488-conjugated goat anti–rabbit IgG (green). Nuclei were stained with TO-PRO-3 (red). Confocal imaging analysis was performed using a Leica TCS-NT laser confocal microscope. Scale bar represents 10 μm. (C) Cells were incubated with 0.7μM 4-OHT (closed) or a final concentration of 0.07% ethanol (open) at the indicated times. Cell growth was assessed using the CellTiter-Glo Luminescent Cell Viability Assay Kit. Results are presented as the mean ± SE of 3 independent experiments relative to the results obtained at day 0; VCont (circles), no. 13 (squares), no. 16 (triangles), and no. 18 (diamonds). *P < .05: no. 13 (day 1); **P < .01: nos. 13, 16, and 18 at day 2 compared with VCont treated with 4-OHT. (D) Annexin V–positive cells were detected by FACS analysis. (E) Nuclei were stained using Hoechst 33342. Imaging analysis was performed with an Olympus IX70 microscope. Scale bar represents 10 μm. (F) TRAIL mRNA was detected by quantitative RT-PCR using predesigned TaqMan probes for TRAIL and GAPDH. Quantification was performed using the relative standard curve method. (G) Total protein was extracted at 48 hours and blotted. The blots were probed with antibody for TRAIL and were reprobed with anti–β-actin antibody to confirm equal protein loading. Results of panels D and F are presented as the mean ± SE of 3 independent experiments. **P < .01 compared with the VCont treated with 4-OHT.
Discussion
In this study, we examined the involvement of FOXO3A in ATRA-induced cellular events, including granulocytic differentiation and apoptosis, using the APL-derived NB4 cell line. Constitutive phosphorylation of FOXO3A in a basal state, ATRA-induced dephosphorylation, and nuclear localization of FOXO3A were all observed in NB4 cells. Our results indicate that FOXO3A is in an inactive state as a transcription factor and is converted to an active form by ATRA in NB4 cells. The transcriptional activation of FOXO3A on account of ATRA was confirmed by up-regulation of the TRAIL gene, a target gene for FOXO3A. Furthermore, knockdown of endogenous FOXO3A by shRNA blocked the ATRA-induced granulocytic differentiation and apoptosis in NB4 cells. Taken together with the observation that FOXO3A translocates into the nucleus after ATRA treatment of primary APL cells, our data strongly suggest that FOXO3A may be a key molecule in a series of cellular responses induced by ATRA differentiation therapy of APL.
Previously, Altucci et al demonstrated that ATRA causes cell death of NB4 and APL blast cells by enhancing TRAIL expression in a paracrine mode of action.32 However, the precise mechanism by which ATRA induced TRAIL expression was unclear. In this study, we identified FOXO3A as a key transcriptional factor involved in TRAIL induction and subsequent apoptosis in ATRA-treated NB4 cells. Importantly, several recent studies have suggested that chemotherapeutic agents, such as glucocorticoid, arsenic trioxide, cisplatin, and gefitinib, induce apoptosis via activation of FOXO3A in various types of cancer cells.33-36 Previous research demonstrated that FOXO3A is activated by imatinib mesylate, a specific inhibitor of BCR-ABL tyrosine kinase, in several CML-derived cell lines and that activation of FOXO3A overcomes resistance to imatinib mesylate.22,37 These studies suggest that FOXO3A is a key mediator of apoptosis in many cancer cells treated with anticancer therapies.
PML-RAR mutations in the ligand-binding domain of the RAR region often lead to ATRA resistance in APL cells, including NB4/RA, NB4-R2, and UF-1.30,38,39 Our observation that neither FOXO3A dephosphorylation nor TRAIL expression was induced by ATRA in NB4/RA cells suggests that the binding of ATRA to the RARα portion of PML-RARα protein and subsequent degradation of PML-RARα are critical events for FOXO3A activation and for subsequent cellular events, including granulocytic differentiation and apoptosis. The addition of recombinant human TRAIL to induce apoptosis in both NB4 and ATRA-resistant NB4-R2 cells in a time- and dose-dependent fashion has been described previously.32 We obtained similar results using NB4 and NB4/RA cells (data not shown). These results suggest that the apoptosis-inducing signaling pathway involving TRAIL remains intact in NB4 and ATRA-resistant NB4/RA cells. Importantly, the FOXO3A knockdown clones, such as the parental NB4 cells, still had high sensitivity to exogenous TRAIL (supplemental Figure 5). This finding suggests that loss of TRAIL induction is involved in the acquisition of ATRA resistance in the FOXO3A knockdown clones. Taken together with our observation that active FOXO3A induced endogenous TRAIL expression and subsequent apoptosis in NB4/RA cells, ATRA resistance in APL cells may be overcome by forced activation of FOXO3A and subsequent induction of TRAIL, in accordance with the findings on imatinib resistance in CML cells.37
We note that the capacity to differentiate into mature granulocytes after ATRA treatment is dependent on the levels of FOXO3A expression in NB4 cells (Figures 2A, 3B-C). Complete knockdown clones of endogenous FOXO3A showed a marked decrease in NBT reduction activity and in the expression of CD11b. These findings indicate that FOXO3A is essential for ATRA-induced granulocytic differentiation of NB4 cells, although the downstream FOXO3A molecule(s) have not been fully identified to date. We also note that forced expression of active FOXO3A (FOXO3A-TM) in the nucleus induced apoptosis without granulocytic differentiation in NB4/RA cells, apparently contradicting the notion that granulocytic differentiation is required for apoptotic processes to occur. There are several possible explanations for these results. One explanation is that ATRA may trigger separate biologic events that affect differentiation and apoptosis in APL cells. Another possibility is that deacetylation of FOXO3A may be required for differentiation. The FOXO3A-TM used in this study is a triple mutant of FOXO3A in which all 3 AKT phosphorylation sites are mutated to alanine, which is predominantly expressed in the nucleus. Considering that FOXO3A-TM does not interact with the nicotinamide adenine dinucleotide–dependent mammalian SIR2 homolog SIRT1 in the absence of stress signals,40 the FOXO3A-SIRT1 interaction may be a prerequisite for the induction of those FOXO3A target genes that are involved in differentiation. It is also possible that rapid apoptosis caused within 48 hours of exposure to 4-OHT does not permit differentiation into mature granulocytic cells.
In conclusion, we have demonstrated that FOXO3A plays a pivotal role in ATRA-induced granulocytic differentiation and apoptosis in NB4 cells. Forced expression of active FOXO3A in the nucleus rapidly induces apoptosis in the ATRA-resistant NB4-derived NB/RA cell line. We conclude that forced activation of FOXO3A may be a promising therapeutic strategy for overcoming ATRA resistance in APL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mirai Iwaya, Hidemi Yamazoe, and Mariko Ozawa for their technical assistance.
This work was supported by Grants-in-Aid for Cancer Research and Scientific Research from the Ministry of Education, Science and Culture of Japan.
Authorship
Contribution: Y.S. and K.S. performed the experiments, interpreted results, and contributed to writing the paper; K.K. and K.O. interpreted results; and N.K. was the group lead, coordinated the study, and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Norio Komatsu, Department of Hematology, Juntendo University School of Medicine, 2-1-1 Hongo, Bunkyo-ku, Tokyo, 113-8421 Japan; e-mail: komatsun@juntendo.ac.jp.