Abstract
Runx1 is required for the emergence of hematopoietic stem cells (HSCs) from hemogenic endothelium during embryogenesis. However, its role in the generation and maintenance of HSCs during adult hematopoiesis remains uncertain. Here, we present analysis of a zebrafish mutant line carrying a truncation mutation, W84X, in runx1. The runx1W84X/W84X embryos showed blockage in the initiation of definitive hematopoiesis, but some embryos were able to recover from a larval “bloodless” phase and develop to fertile adults with multilineage hematopoiesis. Using cd41–green fluorescent protein transgenic zebrafish and lineage tracing, we demonstrated that the runx1W84X/W84X embryos developed cd41+ HSCs in the aorta-gonad-mesonephros region, which later migrated to the kidney, the site of adult hematopoiesis. Overall, our data suggest that in zebrafish adult HSCs can be formed without an intact runx1.
Introduction
Runx1 plays a critical role in the emergence of hematopoietic stem cells (HSCs) in the aorta-gonad-mesonephros (AGM) at the beginning of definitive hematopoiesis.1 The function of Runx1 in adult HSCs, however, has not been fully resolved. Some studies suggest that Runx1 is required for the maintenance of HSCs, whereas others indicate that it negatively regulates the number of HSCs.2,3 Overall, though, it has been difficult to explore the role of Runx1 in initiation of adult hematopoiesis because of limitations of conditional knockout mouse models. We recently reported on the generation of a zebrafish line, W84X, with a premature truncation in the runt domain of the runx1 protein.4 This truncation removes most of the residues important in runx1 activity, such as CBFβ and DNA binding, and nuclear localization signal.5,6 Consistent with the Runx1 knockout mice,7 the mutant embryos had normal primitive hematopoiesis but blockage of definitive hematopoiesis.4 In this study, we analyzed the role of runx1 in adult hematopoiesis using the W84X mutant line.
Methods
Zebrafish were maintained under an approved National Institutes of Health animal use protocol. All zebrafish handling and breedings were performed as described.8 Peripheral blood smears were prepared from tail clips of killed fish. Whole kidney marrow preparation and flow cytometric analysis were performed as described.9 Killed adult fish were fixed in 10% formalin for histology. Lineage tracing by uncaging was performed as described.10 Protocols for genotyping, microscopy and imaging, morpholino design, microinjections, reverse-transcription polymerase chain reaction (RT-PCR), and time lapse are described in the supplemental Methods (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Results and discussion
All runx1W84X/W84X embryos lack definitive hematopoiesis, as demonstrated by lack of expression of HSC and lineage-specific markers between 36 hours after fertilization and 5 days after fertilization4 (supplemental Figure 1A-G). Despite lack of runx1-expressing cells by whole mount in situ hybridization, we detected the mutant runx1 transcript by RT-PCR (supplemental Figure 1H), suggesting that nonhematopoietic cells expressing the mutant runx1 were not significantly affected in terms of their survival at this time point. Because of defective definitive hematopoiesis, all runx1W84X/W84X larvae gradually lost circulating blood cells and became bloodless between 8 and 12 days after fertilization (supplemental Video 1). Most runx1W84X/W84X larvae died during the bloodless phase (11-20 days after fertilization); however, approximately 20% of them gradually regained circulating blood cells between 15 and 21 days after fertilization and survived to adulthood. This was a surprising finding because none of the Runx1−/− mouse embryos could survive past embryonic day 13.5.7 The progeny of runx1W84X/W84X fish incrosses went through a similar bloodless phase and recovered with a comparable survival rate, thus ruling out the possibility that the surviving runx1W84X/W84X adults acquired genetic modifiers that compensated for runx1 deficiency. We speculate that the runx1W84X/W84X larvae died during the bloodless phase because their increased oxygen consumption was not met by diffusion.11 The surviving runx1W84X/W84X adults were fertile and appeared normal grossly. Because runx1 also plays a role in development of some nonhematopoietic tissues, particularly the nervous system,12,13 our mutant line provides a viable resource to probe the role of runx1 in those tissues.
Because recovery of hematopoiesis in runx1W84X/W84X adults was an unexpected finding, we next examined their kidneys and blood to determine whether cells of all hematopoietic lineages are present in their recovered blood. We observed hematopoietic cells in kidney sections of adult runx1W84X/W84X fish, similar to the wild-type (WT) adults (Figure 1A). Fluorescence-activated cell sorter analysis revealed the presence of all hematopoietic cell lineages in the kidney of the runx1W84X/W84X fish (Figure 1B). However, quantitative analysis showed significant reduction in total numbers of whole kidney marrow cells (3 adults in each group, of 100 000 events collected: WT = 71 426 cells, mutant = 62 682 cells, P = .017), especially myeloid and progenitor lineages (Figure 1B), suggesting lineage-specific differentiation defects similar to the Runx1 conditional knockout mice.14,15 When crossed to an lck-green fluorescent protein (GFP) transgenic fish line, which marks developing T cells,16 lck-GFP+ cells were present in similar numbers in the thymi of the runx1W84X/W84X and the WT fish (data not shown). In the peripheral blood of the runx1W84X/W84X fish, mature erythrocytes were readily identified, whereas lymphocytes, monocytes, and neutrophils appeared to be reduced in numbers (Figure 1C). Mature thrombocytes were almost undetectable in the peripheral blood; instead, we observed progenitor-like cells, which could be immature thrombocytes (green arrow in Figure 1C). Defective thrombocyte differentiation has also been described in Runx1 conditional knockout mice,14,15 and patients with heterozygous mutations in RUNX1 develop familial platelet disorder with thrombocytopenia.6 Overall, our data suggest that zebrafish with the truncated runx1 are capable of establishing multilineage adult hematopoiesis, albeit with defects in specific lineages.
Hematopoiesis with multilineage potential in the kidney and blood of adult runx1W84X/W84X zebrafish. (A) Histologic sections of kidneys from WT and runx1W84X/W84X zebrafish. Red arrows indicate hematopoietic cells; and yellow arrows, kidney tubules. The pictures are representative of kidneys from 10 each of the WT and runx1W84X/W84X fish (original magnification ×400). (B) Flow cytometric analysis of kidney cells. FSC-A indicates forward scatter; and SSC-A, side scatter. Cell populations labeled with red circles are erythroid, blue for lymphoid, green for progenitors, and brown for myeloid. The numbers next to the circles indicate percentages for each cell population. The panels are representative of kidneys from 3 each of WT and runx1W84X/W84X fish. (C) Peripheral blood cells in the WT and runx1W84X/W84X zebrafish. Red arrows indicate erythrocytes; brown arrows, monocytes; blue arrows, lymphocytes; purple arrow, thrombocytes; green arrow, progenitor cell. Original magnifications ×100.
Hematopoiesis with multilineage potential in the kidney and blood of adult runx1W84X/W84X zebrafish. (A) Histologic sections of kidneys from WT and runx1W84X/W84X zebrafish. Red arrows indicate hematopoietic cells; and yellow arrows, kidney tubules. The pictures are representative of kidneys from 10 each of the WT and runx1W84X/W84X fish (original magnification ×400). (B) Flow cytometric analysis of kidney cells. FSC-A indicates forward scatter; and SSC-A, side scatter. Cell populations labeled with red circles are erythroid, blue for lymphoid, green for progenitors, and brown for myeloid. The numbers next to the circles indicate percentages for each cell population. The panels are representative of kidneys from 3 each of WT and runx1W84X/W84X fish. (C) Peripheral blood cells in the WT and runx1W84X/W84X zebrafish. Red arrows indicate erythrocytes; brown arrows, monocytes; blue arrows, lymphocytes; purple arrow, thrombocytes; green arrow, progenitor cell. Original magnifications ×100.
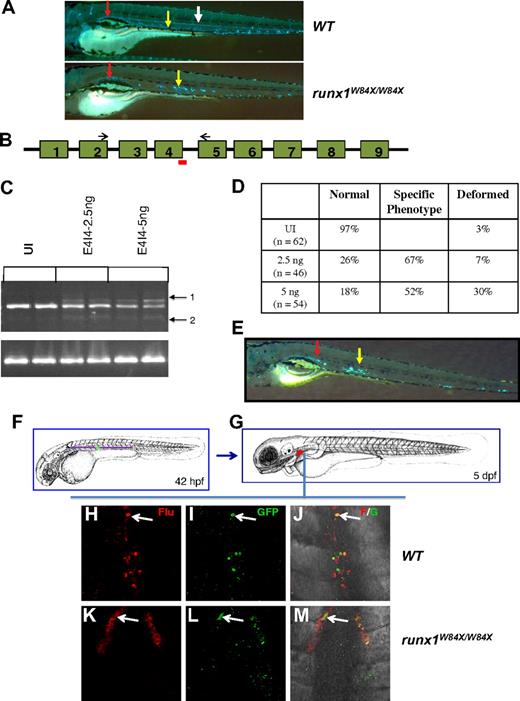
The presence of multilineage hematopoiesis in the runx1W84X/W84X zebrafish suggested that HSCs were produced in the absence of full-length runx1. We crossed the runx1W84X/W84X fish to cd41-GFP transgenic fish17 in which GFP marks HSCs,18-20 in addition to thrombocytes. Interestingly, stationary cd41-GFPlow HSCs were present in the AGM and caudal hematopoietic tissue of all runx1W84X/W84X embryos starting at 3 days after fertilization, even though they lacked cd41-GFP+ circulating cells resulting from defective larval hematopoiesis (Figure 2A). To further demonstrate that the observed phenotype is the result of the W84X mutation, we designed a splice-blocking morpholino (E4I4-MO) to the exon 4 splice acceptor site, which is predicted to skip exon 4, thus generating a protein only 11 amino acids longer than W84X (Figure 2B). By RT-PCR and sequencing, we confirmed that E4I4-MO caused either skipping of exon 4 or cryptic splicing in intron 4, and both events caused premature truncation in the runt domain (Figure 2C; supplemental Figure 2). The morphant embryos displayed similar stationary cd41-GFPlow cells (Figure 2D-E), confirming that the observed phenotype is the result of the runx1 truncation mutation.
cd41-GFPlow cells in the AGM and kidney regions of runx1W84X/W84X and E4I4-MO–injected embryos. (A) cd41-GFP+ cells in the WT and runx1W84X/W84X larvae at 6 days after fertilization. The red and yellow arrows indicate stationary cd41-GFP+ cells in the pronephric duct and the AGM, respectively; and white arrow, cd41-GFP+ cells in circulation in the WT larva. (B-E) E4I4-MO data showing that the phenotype of the morphants is similar to that of the runx1W84X/W84X larvae at 6 days after fertilization. (B) Schematic of the genomic organization of the runx1 gene: rectangles represent exons 1 to 9; lines connecting the exons (not to scale), introns; red line, E4I4-MO; and black arrows, RT-PCR primers. (C) RT-PCR with runx1 primers (top panel) and β-actin primers (bottom panel) from 2 embryos of each of the following groups: uninjected (UI), E4I4-2.5 ng injected, and E4I4-5 ng injected. (D) Table showing the number of injected embryos with different phenotypes at 2.5-ng and 5-ng doses of E4I4-MO. (E) cd41-GFP+ cells in the AGM (yellow arrow) and pronephric duct (red arrow) in the 2.5-ng injected morphant embryo at 6 days after fertilization (A,E: original magnifications ×50). (F-G) Lineage tracing showing colocalization of uncaged fluorescence and cd41-GFP in the same cells in the pronephros. (F) Schematic view of embryos 42 hours after fertilization, indicating the uncaged position of cd41-GFP+ cells in the ventral to dorsal aorta region (VDA), marked by a green cross. (G) Schematic view of embryos 5 days after fertilization, showing the pronephros region (marked red), which were imaged in panels H to M. (H-J) Double staining of flu (H), cd41:GFP (I), and merged view (J) of pronephros in WT embryos. (K-M) Double staining of flu (K), cd41:GFP (L), and merged view (M) of pronephros in runx1W84X/W84X embryos. Arrows indicate flu, cd41:GFP costained cells. Flu indicates antiflu antibody detected using TSA-cy3 as substrate; and GFP, goat anti-GFP antibody as primary and anti–goat Alexa 488 antibody a secondary. The numbers of uncaged embryos exhibiting kidney signals were 3 of 12 and 8 of 12 for runx1W84X/W84X and WT embryos, respectively; on average, there were 2 signals in each runx1W84X/W84X embryo and 5 signals in each WT embryo. The GFP expression intensity in the runx1W84X/W84X embryos seemed to be lower than that in the WT embryos, which may reflect a distinct cd41-GFP+ population that were formed or maintained in the presence of a truncated runx1.
cd41-GFPlow cells in the AGM and kidney regions of runx1W84X/W84X and E4I4-MO–injected embryos. (A) cd41-GFP+ cells in the WT and runx1W84X/W84X larvae at 6 days after fertilization. The red and yellow arrows indicate stationary cd41-GFP+ cells in the pronephric duct and the AGM, respectively; and white arrow, cd41-GFP+ cells in circulation in the WT larva. (B-E) E4I4-MO data showing that the phenotype of the morphants is similar to that of the runx1W84X/W84X larvae at 6 days after fertilization. (B) Schematic of the genomic organization of the runx1 gene: rectangles represent exons 1 to 9; lines connecting the exons (not to scale), introns; red line, E4I4-MO; and black arrows, RT-PCR primers. (C) RT-PCR with runx1 primers (top panel) and β-actin primers (bottom panel) from 2 embryos of each of the following groups: uninjected (UI), E4I4-2.5 ng injected, and E4I4-5 ng injected. (D) Table showing the number of injected embryos with different phenotypes at 2.5-ng and 5-ng doses of E4I4-MO. (E) cd41-GFP+ cells in the AGM (yellow arrow) and pronephric duct (red arrow) in the 2.5-ng injected morphant embryo at 6 days after fertilization (A,E: original magnifications ×50). (F-G) Lineage tracing showing colocalization of uncaged fluorescence and cd41-GFP in the same cells in the pronephros. (F) Schematic view of embryos 42 hours after fertilization, indicating the uncaged position of cd41-GFP+ cells in the ventral to dorsal aorta region (VDA), marked by a green cross. (G) Schematic view of embryos 5 days after fertilization, showing the pronephros region (marked red), which were imaged in panels H to M. (H-J) Double staining of flu (H), cd41:GFP (I), and merged view (J) of pronephros in WT embryos. (K-M) Double staining of flu (K), cd41:GFP (L), and merged view (M) of pronephros in runx1W84X/W84X embryos. Arrows indicate flu, cd41:GFP costained cells. Flu indicates antiflu antibody detected using TSA-cy3 as substrate; and GFP, goat anti-GFP antibody as primary and anti–goat Alexa 488 antibody a secondary. The numbers of uncaged embryos exhibiting kidney signals were 3 of 12 and 8 of 12 for runx1W84X/W84X and WT embryos, respectively; on average, there were 2 signals in each runx1W84X/W84X embryo and 5 signals in each WT embryo. The GFP expression intensity in the runx1W84X/W84X embryos seemed to be lower than that in the WT embryos, which may reflect a distinct cd41-GFP+ population that were formed or maintained in the presence of a truncated runx1.
Lineage tracing by laser-activated cell labeling10 further established that cd41-GFPlow HSCs migrated from AGM to the kidney in the runx1W84X/W84X embryos, albeit at reduced efficiency compared with WT embryos (Figure 2F-M). Finally, time-lapsed imaging disclosed that the cd41-GFPlow HSCs migrated from AGM to the kidney in both WT and runx1W84X/W84X embryos (supplemental Figure 3; supplemental Video 2). These data suggest that both larval and adult hematopoiesis in the zebrafish start from cd41+ precursors, which can be formed in the absence of full-length runx1. Further steps of hematopoiesis during the larval stage are blocked without runx1. However, at least some adult HSCs can be formed from the cd41+ precursors in the absence of the full-length runx1.
Over the past few years, several studies have eluded to a distinction between fetal and adult stages of definitive hematopoiesis.21,22 A schematic comparing the timeline and anatomic locations of fetal and adult hematopoiesis between zebrafish and mouse is shown in supplemental Figure 4. In mice, these 2 stages have been termed as transient definitive and long-term definitive.23,24 In zebrafish, a transient wave of erythromyeloid progenitors is proposed as the first wave of definitive hematopoiesis before HSCs capable of giving rise to adult hematopoiesis are produced.19 An overwhelming body of evidence indicates that mouse HSCs cannot be generated in the absence of Runx1. However, our data provide genetic evidence that HSCs and adult definitive hematopoiesis can be formed in the absence of functional runx1 in the zebrafish. The mechanism for this observation is not clear. One possible explanation is compensation by other runx family members (runx2a, runx2b, and runx3) during adult hematopoiesis but not during the larval stage. However, roles of these genes in HSC initiation have never been demonstrated. The runx1W84X/W84X fish could also carry additional mutations that compensate runx1 loss as it was derived from N-ethyl-N-nitrosourea mutagenesis. Although we cannot exclude the possibility of cosegregation of a closely linked mutation, we are confident that mutations in other parts of the genome have been separated because we have outcrossed the mutant fish for more than 5 generations with no significant phenotypic changes. We hypothesize that this runx1-“independent” HSC generation is through a secondary or salvage pathway, which is not used when normal runx1 is present. Consistent with this hypothesis, Runx1 knockout embryonic stem cells were capable of contributing to adult hematopoiesis in chimeric mice at very low but detectable levels.25
In conclusion, the runx1W84X/W84X zebrafish provides an animal model to dissect the differences between larval and adult HSCs and their differentiation, allowing us to uncover the role of runx1 in later stages of hematopoiesis, which was not possible in the mouse models because of their embryonic lethality.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Robert Handin for providing the Tg:cd41-GFP fish, Leonard Zon for providing the rag1 and runx1 plasmids, Graham Lieschke for providing the ikaros plasmid, Martha Kirby for assistance with fluorescence-activated cell sorter, and Julia Fekecs for help with illustrations.
This work was supported in part by the Intramural Research Programs of National Human Genome Research Institute and National Institute of Child Health and Human Development, NIH.
National Institutes of Health
Authorship
Contribution: R.S., M.A.E., and P.P.L. designed the research; R.S., M.A.E., C.L.B., R.H., J.C., and K.B. performed most of the experiments; H.J. and Z.W performed lineage tracing; M.C.M. and B.M.W. performed time-lapse imaging; and R.S. and P.P.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: P. Paul Liu, Genetics and Molecular Biology Branch, National Human Genome Research Institute, National Institutes of Health, 49 Convent Dr, Rm 3A26, Bethesda, MD 20892; e-mail: pliu@mail.nih.gov.