Abstract
c-Myb is a transcription factor with functions in many hematopoietic lineages. c-Myb–deficient mice display reduced numbers of B cells; however, it is unknown what role c-Myb plays in B lymphopoiesis because no critical target genes have been identified in the B-cell lineage. We demonstrate that conditional deletion of c-Myb in B-cell progenitors completely abolishes B-cell development. c-Myb is required for lymphoid progenitors to respond to the cytokines interleukin-7 and thymic stromal lymphopoietin; in the absence of sufficient c-Myb activity, mice display a B lymphopenia that closely resembles that observed in interleukin-7 receptor α–deficient animals. Analysis of the multipotent progenitor compartment indicates that c-Myb is also required for up-regulation of multiple lymphoid-associated genes, including Il7r, and for the subsequent development of the common lymphoid progenitor population. These data show that c-Myb plays a critical role in the regulatory pathways governing lymphoid specification and early B-cell differentiation.
Introduction
The development of B cells from hematopoietic stem cells (HSCs) proceeds through several well-defined intermediate stages. HSCs are enriched within the Lineage−Sca-1hic-kithi (LSK) population in the bone marrow.1 The LSK population also includes progenitors with strong lymphoid and myeloid potential but limited erythro-megakaryocyte potential, termed lymphoid-primed multipotent progenitors (LMPPs).2 LMPPs can be identified by their high level of Flt3 and display expression of several lymphoid transcripts.3 Downstream of the LMPPs, the common lymphoid progenitor (CLP) up-regulates expression of interleukin-7 receptor α (IL-7Rα) and depends on signaling through this receptor for further development to B-cell progenitor stages.4-6 Together, signals through Flt3 and IL-7Rα are required for all B-cell development.7
Transcription factors act in concert with these cytokine signals to regulate B-cell development.8 Although factors such as Ebf1, E2a, and Pax5 control B-cell commitment and pro-B cell differentiation,9-12 less is understood about the earlier stages of lymphoid specification from HSCs. Pu.1 and Ikaros have been implicated in the development of LMPPs; however, this could reflect roles for these factors in Flt3 expression.13,14 The process of lymphoid priming appears to depend on Ikaros and E2a,15,16 but additional transcription factors are probably involved.
The transcription factor c-Myb is highly expressed in hematopoietic progenitor cells17 ; however, its role in lymphopoiesis has remained elusive. Mice bearing a germline deletion of c-Myb die embryonically because of anemia.18 More recently, viable mutant alleles of c-Myb have been developed, enabling the role of c-Myb in adult hematopoiesis to be examined. Mice with impaired c-Myb activity or expression display reduced numbers of B cells.19-23 Conditional deletion of c-Myb in pro-B cells ablates further B-cell development, indicating that c-Myb is stringently required in the B-cell lineage.24 The precise role of c-Myb during B-cell development remains undefined, however, and no critical target genes have been described. In addition, it is unclear whether c-Myb also plays a role within multipotent progenitors to influence B-cell development.
Here, we have used a range of alleles to dissect the role of c-Myb during B-cell development. Conditional deletion of c-Myb at the earliest B-cell progenitor stage resulted in a complete lack of B cells, confirming that c-Myb is directly required in the B-cell lineage at the point of lineage commitment. Mice expressing a severely hypomorphic allele of c-Myb throughout hematopoiesis display an even earlier defect in lymphoid development, including impaired lymphoid priming in multipotent progenitor cells. Using these hypomorphic mice, we found that c-Myb is required for normal Il7r expression and IL-7–dependent pro-B cell proliferation. Collectively, these data suggest that c-Myb plays a central role in B-cell development by integrating external signals with the transcription factor network in lymphoid progenitors.
Methods
Experimental animals
Antibodies and flow cytometry
Single-cell suspensions were stained with fluorophore- or biotin-conjugated antibodies and analyzed on an LSRII (BD Biosciences) with dead cells excluded by propidium iodide staining. Antibodies CD11cAPC (HL3), Flt3PE (A2F10.1), CD34FITC (RAM34), NK1.1PECy7 (PK136), CD43PE (S7), B220PECy7 (RA3-6B2), CD25PE (7D4), and streptavidinPE were from BD PharMingen; B220Alexa750 (RA3-6B2), CD19PECy7 (1D3), Sca-1PECY7 (E13161.7), and IL-7RαBio (B12-1) were from eBioscience; CD27PerCPCy5.5 (LG.3A10) was from Biolegends; anti–ratAlexa680 and anti–ratAlexa700 were from Invitrogen Molecular Probes; CD3 (KT3), CD19 (1D3), B220 (RA3-6B2), Mac-1 (M1-70), Gr-1 (1A8 and RB68C5), CD2 (RM2.1), CD8 (53-6.7), Ter119 (Ter119), c-kit (ACK2), IgM (331.1), Thy1 (30H12), and CD4 (GK1.5) were purified and conjugated to fluorophores in our laboratory.
Phosphorylated STAT5 intracellular flow cytometry
Bone marrow cell suspensions were washed 3 times with phosphate-buffered saline (PBS) and resuspended in RPMI. Cells were incubated for 15 minutes at 37°C and then stimulated with 2% of the supernatant of an IL-7–producing cell line for 15 minutes at 37°C; control samples were rested for a further 15 minutes at 37°C. Cells were fixed and permeabilized with BD Phosflow Lyse/Fix buffer III according to instructions. Cells were then stained with B220, CD43, and Stat5(pY694)Alexa488 (BD PharMingen) and analyzed on an LSRII.
Cell sorting
To isolate pro-B cells (defined as B220+CD19+c-kit+) from the fetal liver of embryos at embryonic day 15.5 (e15.5) to e16.5 or the bone marrow of 3-week-old mice, cells were stained with antibodies to B220, CD19, and c-kit. Hematopoietic progenitor cells were isolated as described.27 LSK cells were defined as Lineage−Sca-1hic-kithiIL-7Rα−; CLPs were defined as lineage−Sca-1+c-kitintIL-7Rα+Thy1−. Cells were sorted by flow cytometry on a FACSDiva (BD Biosciences) or a MoFlo (Dako).
OP9 and OP9-DL1 cultures
OP9 and OP9-DL1 cells were cultured as described.28 To determine B-cell precursor frequencies, OP9 stromal cells were plated in 96-well culture plates. Pro-B cells, LSK cells, or CLPs were seeded in limiting dilution onto the OP9 cells in the presence of either 2% IL-7 supernatant or 10 ng/mL thymic stromal lymphopoietin (TSLP; R&D Systems). For LSK and CLP assays, the media was supplemented with 5 ng/mL Flt3L. After 7 to 10 days, wells were scored for the presence of a B-cell colony; cells were harvested for flow cytometric analysis, and B-cell precursor frequency was calculated according to Poisson statistics. To determine T-cell precursor frequency, the same procedure was followed except that cells were plated on OP9 cells overexpressing DL1 ligand (OP9-DL1).29
Microarrays
LSK cells were isolated by flow cytometry, and RNA was extracted with the RNeasy Micro kit (QIAGEN). RNA was labeled and hybridized to Illumina MouseWG-6 V1.1 Expression BeadChips according to Illumina standard protocols. Data were analyzed with the R Project for Statistical Computing, and arrays were subjected to norm exp transformation30 and quantile normalization. Linear modeling with an empirical Bayes approach, including a batch factor, was applied to the data.31 Data were corrected for multiple testing with the Benjamini-Hochberg correction. Pro-B microarrays were performed as for LSK cells, except for the use of Illumina WG Beadarrays V2. The probes were filtered by Illumina presence calls to include only probes expressed on at least one sample. Full data for the pro-B microarrays are available online at ArrayExpress under accession no. E-TABM-690 (http://www.ebi.ac.uk/microarray-as/ae/); data for the LSK arrays will be reported separately.
Quantitative reverse transcription–polymerase chain reaction expression analysis
cDNA was synthesized from total RNA with an oligo-dT primer with the use of Superscript III Reverse Transcriptase (Invitrogen). For semiquantitative reverse transcription–polymerase chain reaction (RT-PCR), cDNA was normalized with the use of Hprt1 expression, and 3-fold serial dilutions of the cDNA were prepared. Primers used for semiquantitative RT-PCR are described in supplemental Table 4 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Quantitative PCR reactions to quantify expression of Fcrl1, Il10ra, Dntt, Il7r, Lef1, Robo4, Slamf7, Tcf12, and Hprt were performed with the use of predesigned Taqman gene expression assays (Mm00462105_m1, Mm00434151_m1, Mm00493500_m1, Mm00434295_m1, Mm00550265_m1, Mm00452963_m1, Mm00513807_m1, Mm00441699_m1, Mm01545399_m1; Applied Biosystems). All quantitative PCR reactions were performed on an ABI 7900 HT real-time PCR platform (Applied Biosystems). Ct values were derived with the use of SDS2.2 software (Applied Biosystems), and relative gene expression was calculated with the 2−ΔΔCt method.32
Retrovirus production
Retroviral supernatants were prepared by transient transfection of 293T cells with plasmids that encode viral proteins (pMD1-gag-pol and pCAG-Eco) and a specific expression vector (pMIG) encoding either c-Myb, Ebf1, IL-7Rα, STAT5(1*6), or Bcl2 followed by an IRES-GFP cassette. Empty pMIG vector expressing GFP alone acted as a control. pMIG-Ebf1, pMXIG-STAT5A(1*6), and pMIG-hBcl2 have been previously described.33-35 Viral supernatants were harvested after 48 to 72 hours.
Retroviral infection of progenitors
Fetal liver cells from e14.5 embryos were depleted of Ter119+ cells and cultured overnight in IMDM with 10% fetal calf serum, 1mM l-glutamine, 50μM β-mercaptoethanol, 100 ng/mL stem cell factor, 50 ng/mL thrombopoietin, 5 ng/mL Flt3L, and 1% IL-6 supernatant. Retroviral supernatants were applied to culture dishes pretreated with RetroNectin and centrifuged at 4000 rcf for 1 hour at 4°C. Fetal liver cells were cocultured with the virus in the presence of polybrene (4 μg/mL) for 24 hours. Cells were then washed and transferred to OP9 cells. After 4 to 9 days, cells were harvested and analyzed for the formation of CD19+ B cells by flow cytometry; cell counts were performed by adding a known number of Calibrite beads (BD Biosciences) to the samples before analysis. CLPs were sorted as described in “Cell sorting” and transduced with the use of the same procedure.
Statistical analysis
The 2-sided t test was used for all statistical analyses unless otherwise specified. P values were adjusted by the Bonferroni correction when multiple tests were done with the same sample.
Results
c-Myb is essential for B-cell development
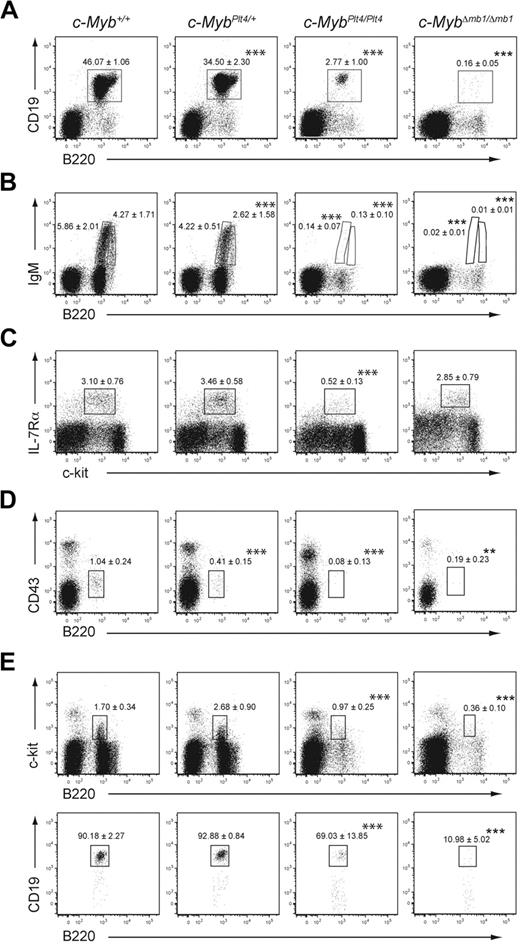
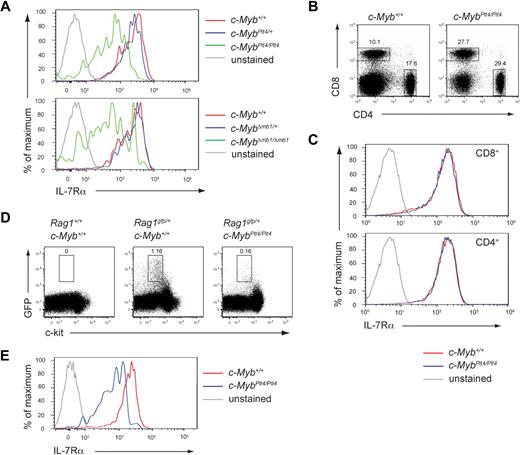
To examine the role of c-Myb early in the B-cell lineage, we conditionally inactivated c-Myb at the pre-pro-B cell stage of development. Mice with loxP sites flanking exons 3 to 6 of c-Myb (c-Mybfl)21 were crossed with mice that express Cre recombinase under control of the Cd79a promoter (mb1Cre mice). The mb1Cre mice express Cre from an early B-lymphoid progenitor stage and mediate efficient deletion of loxP-flanked DNA at an earlier stage of development than observed in Cd19Cre mice.9,24,25 Flow cytometric characterization showed that CD19+ B cells, including immature and mature subsets, were essentially absent in the bone marrow of c-MybΔmb1/Δmb1 mice (Figure 1A-B), in agreement with a recent study.24 c-MybΔmb1/+ mice also showed a reduction in the percentage of B cells in the bone marrow (supplemental Figure 1A-B), confirming a previous report that the dosage of c-Myb is important for B-cell development.23
c-Myb is essential for B-cell development. (A-B) Flow cytometry of bone marrow of 3- to 4-week-old c-Myb+/+, c-MybPlt4/+, c-MybPlt4/Plt4, and c-MybΔmb1/Δmb1 mice. Numbers adjacent to the boxes indicate proportion of (A) total B cells (B220+CD19+) and (B) immature (B220loIgM+) and mature (B220hiIgM+) B cells among all live cells (mean ± SD of 4-10 mice per genotype). (C) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 6- to 9-week-old c-Myb+/+, c-MybPlt4/+, c-MybPlt4/Plt4, and c-MybΔmb1/Δmb1 mice. CLPs were identified as Sca-1+Thy1− c-kitint IL-7Rα+; the gating strategy is available in supplemental Figures 1 and 3. Numbers in plots indicate the proportion of cells in each gated area (mean ± SD of 5-6 mice per genotype). (D-E) Flow cytometry of bone marrow of 3- to 4-week-old c-Myb+/+, c-MybPlt4/+, c-MybPlt4/Plt4, and c-MybΔmb1/Δmb1 mice. (D) Pre-pro-B cells were identified as CD19− CD11c− NK1.1− B220+CD43lo; the gating strategy is available in supplemental Figures 1 and 3. Numbers in plots indicate the proportion of cells in each gated area (mean ± SD of 4-10 mice per genotype). (E) The numbers above the boxes in the top plots indicate the proportion of pro-B cells (B220+c-kit+) among all live cells. The numbers above the boxes in the bottom plots indicate the proportion of committed B cells (CD19+) among B220+c-kit+ cells. Data are the mean ± SD of 4 to 10 mice per genotype. *P < .05; **P < .01; ***P < .001 in test group vs c-Myb+/+ cells.
c-Myb is essential for B-cell development. (A-B) Flow cytometry of bone marrow of 3- to 4-week-old c-Myb+/+, c-MybPlt4/+, c-MybPlt4/Plt4, and c-MybΔmb1/Δmb1 mice. Numbers adjacent to the boxes indicate proportion of (A) total B cells (B220+CD19+) and (B) immature (B220loIgM+) and mature (B220hiIgM+) B cells among all live cells (mean ± SD of 4-10 mice per genotype). (C) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 6- to 9-week-old c-Myb+/+, c-MybPlt4/+, c-MybPlt4/Plt4, and c-MybΔmb1/Δmb1 mice. CLPs were identified as Sca-1+Thy1− c-kitint IL-7Rα+; the gating strategy is available in supplemental Figures 1 and 3. Numbers in plots indicate the proportion of cells in each gated area (mean ± SD of 5-6 mice per genotype). (D-E) Flow cytometry of bone marrow of 3- to 4-week-old c-Myb+/+, c-MybPlt4/+, c-MybPlt4/Plt4, and c-MybΔmb1/Δmb1 mice. (D) Pre-pro-B cells were identified as CD19− CD11c− NK1.1− B220+CD43lo; the gating strategy is available in supplemental Figures 1 and 3. Numbers in plots indicate the proportion of cells in each gated area (mean ± SD of 4-10 mice per genotype). (E) The numbers above the boxes in the top plots indicate the proportion of pro-B cells (B220+c-kit+) among all live cells. The numbers above the boxes in the bottom plots indicate the proportion of committed B cells (CD19+) among B220+c-kit+ cells. Data are the mean ± SD of 4 to 10 mice per genotype. *P < .05; **P < .01; ***P < .001 in test group vs c-Myb+/+ cells.
To identify the point at which B-cell development is affected by the loss of c-Myb, we examined various lymphocyte and B-cell progenitor fractions within the bone marrow. The CLP fraction (defined as Sca-1+Thy1−c-kitintIL-7Rα+) appeared normal in c-MybΔmb1/Δmb1 mice (Figure 1C; supplemental Figure 1C), consistent with the absence of Cre expression at this stage of development. In contrast, the pre-pro-B cell fraction (defined as B220+CD19−NK1.1−CD11c−CD43lo) was absent in c-MybΔmb1/Δmb1 mice and markedly reduced in MybΔmb1/+ mice (Figure 1D; supplemental Figure 1D). Similarly, pro-B cells (defined as B220+CD19+c-kit+) were virtually absent in c-MybΔmb1/Δmb1 mice (Figure 1E). These results show that B-cell development in c-MybΔmb1/Δmb1 mice is completely blocked at the transition from the CLP to the pre-pro-B cell stage. The point of the defect observed in c-Myb–deficient mice closely parallels the induction of Cre in mb1Cre mice,9 indicating that deletion of c-Myb in early B-cell progenitors is incompatible with further development.
B-cell development is sensitive to the level of c-Myb activity
Although the profound defect observed in c-MybΔmb1/Δmb1 mice illustrates a requirement for c-Myb after the CLP stage, the lack of B-cell progenitors in these mice made it difficult to further characterize the nature of the defect. Instead, we decided to analyze mice with a hypomorphic allele of c-Myb (c-MybPlt4). c-MybPlt4/Plt4 mice are viable and carry a point mutation in c-Myb that leads to the substitution of valine for aspartic acid at residue 384 in the leucine zipper domain of the protein.20 This mutation does not affect the c-Myb expression level, nuclear localization, or DNA binding capacity but leads to a reduced capacity to transactivate a model promoter containing 5 c-Myb binding sites (supplemental Figure 2). This allele offered the additional advantage that it is expressed throughout hematopoiesis and hence could be used to examine the role of c-Myb in developmental stages before Cd79a (mb1) expression.
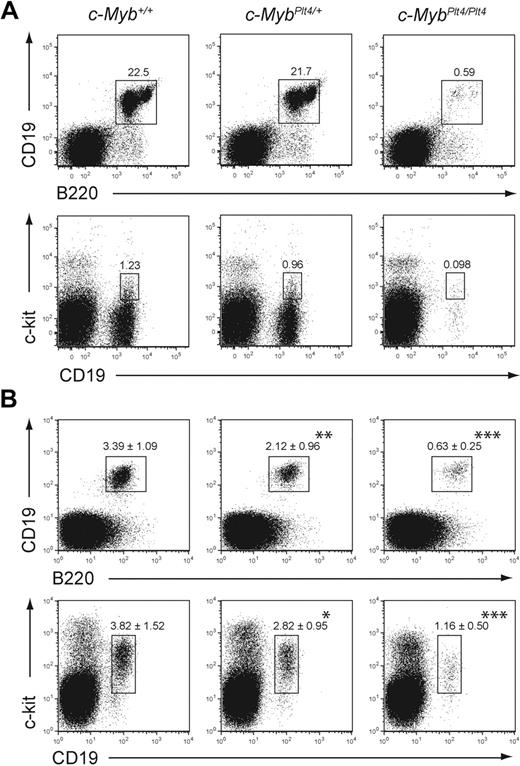
Flow cytometric analysis of B-cell development in c-MybPlt4/Plt4 mice showed an approximate 20-fold reduction in the percentage of B cells within their bone marrow (Figure 1A). Immature and mature recirculating B-cell subsets were drastically reduced in c-MybPlt4/Plt4 bone marrow (Figure 1B), and a more subtle decrease was observed in c-MybPlt4/+ mice (Figure 1A-B). Closer examination of the B-cell progenitor fractions showed that both CLPs and pre-pro-B cells were severely reduced in c-MybPlt4/Plt4 mice (Figure 1C-D; supplemental Figure 3). A reduction was also observed in the proportion of pro-B cells in MybPlt4/Plt4 mice, although some pro-B cells could be observed in young mice (Figure 1E). The phenotype of c-MybPlt4/Plt4 mice appeared to become more severe with age, with pro-B cells clearly present in the fetal liver of e16.5 embryos but essentially absent in adult (> 8 weeks old) mice (Figure 2A-B). A similar phenotype, albeit less severe, was observed in mice bearing a mutant allele of p300 that disrupts the interaction between c-Myb and p300 (supplemental Figure 4).
The B-cell defect in c-MybPlt4/Plt4 mice is more severe in older mice. (A) Flow cytometry of adult (8-10 weeks old) c-Myb+/+, c-MybPlt4/+, and c-MybPlt4/Plt4 mice. Top indicates total B cells; bottom, pro-B cells. Populations were identified as described in Figure 1. Plots are representative of 3 mice per genotype. (B) Flow cytometry of fetal liver cells from e16.5 embryos. Numbers in the plots represent the proportion of cells within the gated population expressed as the mean ± SD of 7 to 21 embryos per genotype. (Top) Numbers above the boxes indicate percentages of total B cells (B220+CD19+) among all live cells. (Bottom) Numbers above the boxes indicate percentages of pro-B cells (CD19+c-kit+) among all live cells. *P < .05; **P < .01; ***P < .001 in test group vs c-Myb+/+ cells.
The B-cell defect in c-MybPlt4/Plt4 mice is more severe in older mice. (A) Flow cytometry of adult (8-10 weeks old) c-Myb+/+, c-MybPlt4/+, and c-MybPlt4/Plt4 mice. Top indicates total B cells; bottom, pro-B cells. Populations were identified as described in Figure 1. Plots are representative of 3 mice per genotype. (B) Flow cytometry of fetal liver cells from e16.5 embryos. Numbers in the plots represent the proportion of cells within the gated population expressed as the mean ± SD of 7 to 21 embryos per genotype. (Top) Numbers above the boxes indicate percentages of total B cells (B220+CD19+) among all live cells. (Bottom) Numbers above the boxes indicate percentages of pro-B cells (CD19+c-kit+) among all live cells. *P < .05; **P < .01; ***P < .001 in test group vs c-Myb+/+ cells.
c-Myb is required for lymphoid progenitors to respond to IL-7
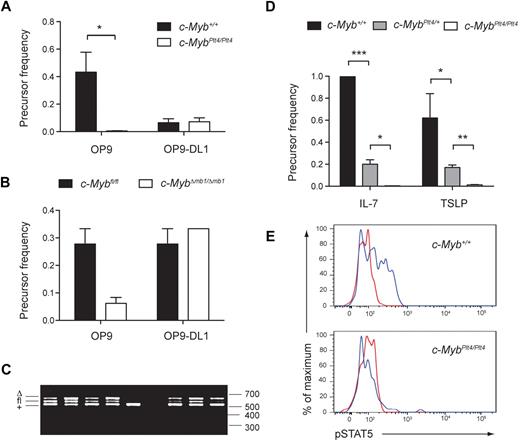
To test whether c-MybPlt4/Plt4 hematopoietic progenitors had an intrinsic defect in B-cell potential, we analyzed whether they could generate B-cell colonies in a well-defined culture system. We isolated progenitor cells from bone marrow and performed a limiting dilution analysis on OP9 stromal cells in the presence of IL-7 and Flt3L. In parallel, we performed a limiting dilution analysis on OP9-DL1 stromal cells in the presence of IL-7 and Flt3L to examine T-cell potential. LSK cells isolated from c-MybPlt4/Plt4 bone marrow displayed a normal T-cell precursor frequency but a dramatically reduced B-cell precursor frequency (Figure 3A). A similar phenotype was observed for CLPs isolated from c-MybΔmb1/Δmb1 bone marrow (Figure 3B). All of the B-cell colonies generated from c-MybΔmb1/Δmb1 CLPs contained some cells retaining the floxed allele (Figure 3C), indicating that complete deletion of c-Myb is not compatible with the generation of a B-cell colony. Thus, c-Myb is required for hematopoietic progenitors to form B-cell colonies in vitro.
c-Myb is required for lymphoid progenitors to respond to IL-7 and TSLP. (A) Limiting dilution analysis of LSK cells isolated from the bone marrow of 6- to 8-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Cultures were performed on OP9 or OP9-DL1 stromal cells in the presence of IL-7 and Flt3L. *P < .025. Data represent the mean ± SEM of 5 samples per genotype. (B) Limiting dilution analysis of CLPs isolated from the bone marrow of 6- to 9-week-old c-Myb+/+ and c-MybΔmb1/Δmb1 mice. Cultures were performed on OP9 or OP9-DL1 stromal cells in the presence of IL-7 and Flt3L. The difference in B-cell precursor frequency was not significant (P > .025), probably because of incomplete deletion of the floxed allele; see panel C. Data represent the mean ± SEM of 2 to 3 samples per genotype. (C) PCR genotyping of B-cell colonies generated from c-MybΔmb1/Δmb1 CLPs. CLPs were isolated from c-Mybfl/fl or c-MybΔ/fl mice carrying the mb1Cre allele. After 8 days of culture on OP9 stromal cells, individual B-cell colonies were harvested, and DNA was extracted for genotyping at the c-Myb locus. The deleted (Δ), floxed (fl), and wild type (+, derived from the OP9 cells) alleles are indicated. Data are representative of 26 colonies analyzed. Sizes in base pairs are indicated on the right. (D) Limiting dilution analysis of pro-B cells (B220+CD19+c-kit+) isolated from the fetal liver of c-Myb+/+, c-MybPlt4/+, and c-MybPlt4/Plt4 e15.5 to e16.5 embryos. Cultures were performed on OP9 stromal cells in the presence of either IL-7 or TSLP. *P < .025; **P < .005; ***P < .001. Data represent the mean ± SEM of 2 to 8 embryos per genotype. Only 2 small colonies were observed from c-MybPlt4/Plt4 pro-B cells stimulated with IL-7; hence, the number in the graph represents an overestimate of precursor frequency. The precursor frequency in panels A, B and D was defined as the inverse of the number of cells per well at which more than 37% of the wells lacked a colony. (E) Intracellular flow cytometry for phosphorylated STAT5 (pSTAT5) after IL-7 stimulation of c-Myb+/+ and c-MybPlt4/Plt4 bone marrow. Plots are gated on B220+CD43+ pro-B cells and are representative of 4 mice per genotype. The red line indicates cells without IL-7; the blue line indicates cells stimulated with IL-7.
c-Myb is required for lymphoid progenitors to respond to IL-7 and TSLP. (A) Limiting dilution analysis of LSK cells isolated from the bone marrow of 6- to 8-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Cultures were performed on OP9 or OP9-DL1 stromal cells in the presence of IL-7 and Flt3L. *P < .025. Data represent the mean ± SEM of 5 samples per genotype. (B) Limiting dilution analysis of CLPs isolated from the bone marrow of 6- to 9-week-old c-Myb+/+ and c-MybΔmb1/Δmb1 mice. Cultures were performed on OP9 or OP9-DL1 stromal cells in the presence of IL-7 and Flt3L. The difference in B-cell precursor frequency was not significant (P > .025), probably because of incomplete deletion of the floxed allele; see panel C. Data represent the mean ± SEM of 2 to 3 samples per genotype. (C) PCR genotyping of B-cell colonies generated from c-MybΔmb1/Δmb1 CLPs. CLPs were isolated from c-Mybfl/fl or c-MybΔ/fl mice carrying the mb1Cre allele. After 8 days of culture on OP9 stromal cells, individual B-cell colonies were harvested, and DNA was extracted for genotyping at the c-Myb locus. The deleted (Δ), floxed (fl), and wild type (+, derived from the OP9 cells) alleles are indicated. Data are representative of 26 colonies analyzed. Sizes in base pairs are indicated on the right. (D) Limiting dilution analysis of pro-B cells (B220+CD19+c-kit+) isolated from the fetal liver of c-Myb+/+, c-MybPlt4/+, and c-MybPlt4/Plt4 e15.5 to e16.5 embryos. Cultures were performed on OP9 stromal cells in the presence of either IL-7 or TSLP. *P < .025; **P < .005; ***P < .001. Data represent the mean ± SEM of 2 to 8 embryos per genotype. Only 2 small colonies were observed from c-MybPlt4/Plt4 pro-B cells stimulated with IL-7; hence, the number in the graph represents an overestimate of precursor frequency. The precursor frequency in panels A, B and D was defined as the inverse of the number of cells per well at which more than 37% of the wells lacked a colony. (E) Intracellular flow cytometry for phosphorylated STAT5 (pSTAT5) after IL-7 stimulation of c-Myb+/+ and c-MybPlt4/Plt4 bone marrow. Plots are gated on B220+CD43+ pro-B cells and are representative of 4 mice per genotype. The red line indicates cells without IL-7; the blue line indicates cells stimulated with IL-7.
To more closely examine B-cell progenitors, we isolated pro-B cells and performed a limiting dilution analysis on OP9 stromal cells in the presence of IL-7 (Figure 3D). We used e16.5 embryos for these experiments, because, in contrast to adult mice, pro-B cells were clearly present in c-MybPlt4/Plt4 fetal livers (Figure 2). Our analysis showed that essentially all c-Myb+/+ pro-B cells were able to proliferate in response to IL-7 and to generate B-cell colonies, whereas c-MybPlt4/Plt4 pro-B cells were unable to generate colonies under the same conditions, and an intermediate effect was observed for c-MybPlt4/+ pro-B cells. Pro-B cells isolated from bone marrow of young c-MybPlt4/Plt4 or c-MybΔmb1/Δmb1 mice also showed markedly reduced B-cell precursor frequency (< 0.005 for c-MybPlt4/Plt4 and c-MybΔmb1/Δmb1 compared with 0.47 for c-Myb+/+). IL-7 stimulation led to phosphorylation of STAT5 in c-Myb+/+ pro-B cells but not in c-MybPlt4/Plt4 pro-B cells (Figure 3E), indicating a profound failure of signaling immediately downstream of the IL-7Rα.
The cytokine TSLP has been shown to support the development of pro-B cells from fetal liver but not adult bone marrow.7 Like IL-7, TSLP signals through the IL-7Rα chain. We performed a limiting dilution analysis to test whether pro-B cells isolated from c-MybPlt4/Plt4 fetal liver could respond to TSLP. Although the precursor frequency for c-Myb+/+ pro-B cells grown in TSLP was 0.62, this frequency was reduced to 0.17 for c-MybPlt4/+ pro-B cells and 0.012 for c-MybPlt4/Plt4 pro-B cells (Figure 3D). Thus, c-Myb is required for B-cell progenitors to respond to both IL-7 and TSLP.
c-Myb is required for normal expression of IL-7Rα
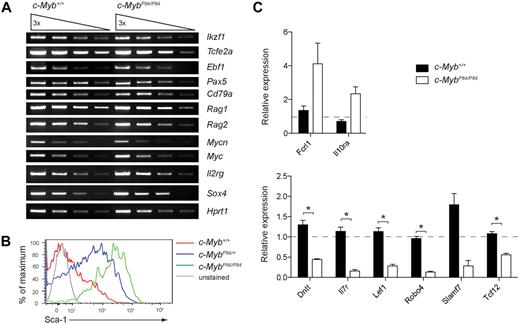
A key advantage of c-MybPlt4/Plt4 mice is that, unlike c-MybΔmb1/Δmb1 mice, they retain a small pool of pro-B cells that can be used to examine the molecular mechanism by which c-Myb controls early B-cell development. Importantly, c-MybPlt4/Plt4 pro-B cells displayed relatively normal expression of several genes known to be critical for early B-cell differentiation and hence were bona fide pro-B cells (Figure 4A). In addition, c-MybPlt4/Plt4 pro-B cells had undergone Igh rearrangements appropriate for their developmental stage (supplemental Figure 5). To identify genes dependent on c-Myb activity in the B-cell lineage, we performed a microarray comparing gene expression between c-Myb+/+ and c-MybPlt4/+ pro-B cells (supplemental Table 1). We used c-MybPlt4/+ pro-B cells because they displayed a haploinsufficient phenotype (Figures 1–3) but were present in sufficient numbers for microarray analysis. Expression changes were confirmed by real-time quantitative PCR and flow cytometry in c-MybPlt4/Plt4 pro-B cells (Figure 4B-C). Several genes that function during lymphoid development were significantly down-regulated in c-MybPlt4/Plt4 pro-B cells, including Dntt, Il7r, Lef1, and Tcf12. The majority of differentially expressed genes, however, had no known role in lymphoid development.
Analysis of genes differentially expressed in c-MybPlt4/Plt4 pro-B cells. (A) Semiquantitative RT-PCR of gene expression in pro-B cells (B220+CD19+c-kit+) isolated from the bone marrow of 3- to 4-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Wedges indicate serial dilution of cDNA. Data are representative of 2 experiments. (B) Flow cytometry of Sca-1 (Ly6a) surface expression on pro-B cells from 3-week-old mice of the indicated genotypes. Plots are representative of 3 mice per genotype. (C) Quantitative real-time PCR of gene expression in pro-B cells (B220+CD19+c-kit+) isolated from the bone marrow of 3- to 4-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Expression is normalized to Hprt1 and is presented as relative to that in one of the wild-type samples, set as 1. *P < .05 in test group vs c-Myb+/+ cells. Data represent the mean ± SEM of 3 to 6 samples of each genotype.
Analysis of genes differentially expressed in c-MybPlt4/Plt4 pro-B cells. (A) Semiquantitative RT-PCR of gene expression in pro-B cells (B220+CD19+c-kit+) isolated from the bone marrow of 3- to 4-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Wedges indicate serial dilution of cDNA. Data are representative of 2 experiments. (B) Flow cytometry of Sca-1 (Ly6a) surface expression on pro-B cells from 3-week-old mice of the indicated genotypes. Plots are representative of 3 mice per genotype. (C) Quantitative real-time PCR of gene expression in pro-B cells (B220+CD19+c-kit+) isolated from the bone marrow of 3- to 4-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Expression is normalized to Hprt1 and is presented as relative to that in one of the wild-type samples, set as 1. *P < .05 in test group vs c-Myb+/+ cells. Data represent the mean ± SEM of 3 to 6 samples of each genotype.
We hypothesized that reduced Il7r expression in c-MybPlt4/Plt4 pro-B cells could be a contributor to the phenotype of these mice. The inability of c-Myb mutant progenitors to respond to IL-7 in vitro strongly implicated a defect in IL-7 signaling. Moreover, the B-cell phenotype of c-MybPlt4/Plt4 mice in vivo closely phenocopies that observed in mice with defective IL-7 signaling. Mice lacking either IL-7 or the IL-7Rα chain display a profound reduction in B-cell numbers.36,37 The main developmental block in these mice occurs at the transition from the CLP to the pre-pro-B cell stage.5,6 The few B cells that develop in the absence of IL-7 signaling are produced during fetal and neonatal life, because B-cell development ceases in the adult.7,38 Given these parallels, we decided to further investigate the role of reduced Il7r expression in the phenotype of c-MybPlt4/Plt4 mice. To assess whether the reduction in Il7r expression in c-MybPlt4/Plt4 pro-B cells had an effect at the protein level, we analyzed IL-7Rα expression by flow cytometry. Pro-B cells from c-MybPlt4/Plt4 and c-MybΔmb1/Δmb1 mice displayed a markedly reduced level of IL-7Rα (Figure 5A), despite these cells expressing normal levels of Pu.1, the transcription factor previously implicated in IL-7Rα expression in B cells (supplemental Figure 6). In contrast, c-MybPlt4/Plt4 mice displayed normal expression of IL-7Rα on peripheral T cells (Figure 5B-C).
c-Myb is required for normal expression of IL-7Rα on lymphoid progenitors. (A) Flow cytometry of IL-7Rα surface expression on pro-B cells from 3- to 4-week-old mice. The upper panel is gated on B220+CD19+c-kit+ cells; the lower panel is gated on B220+c-kit+ cells. Plots are representative of 3 to 9 mice per genotype. (B) Flow cytometry of splenocytes from c-Myb+/+ and c-MybPlt4/Plt4 mice. Numbers adjacent to the boxes indicate percentages of CD4+ and CD8+ T cells among all live cells. Dot plots are representative of 3 mice per genotype. Note that the relative increase in CD8+ and CD4+ cells is due to the loss of B cells in c-MybPlt4/Plt4 mice. (C) Flow cytometry of IL-7Rα surface expression on splenic T cells from c-Myb+/+ and c-MybPlt4/Plt4 mice, gated for CD8+ or CD4+ cells as indicated. Plots are representative of 3 mice per genotype. (D) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 5- to 9-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice on a Rag1gfp/+ background. Cells at the CLP stage of development were identified as Sca-1+c-kitint GFP+; numbers in plots indicate the percentage of cells in each gated area. (E) Flow cytometry of IL-7Rα surface expression on Sca-1+c-kitint GFP+ cells. Plots are representative of 4 to 5 mice per genotype.
c-Myb is required for normal expression of IL-7Rα on lymphoid progenitors. (A) Flow cytometry of IL-7Rα surface expression on pro-B cells from 3- to 4-week-old mice. The upper panel is gated on B220+CD19+c-kit+ cells; the lower panel is gated on B220+c-kit+ cells. Plots are representative of 3 to 9 mice per genotype. (B) Flow cytometry of splenocytes from c-Myb+/+ and c-MybPlt4/Plt4 mice. Numbers adjacent to the boxes indicate percentages of CD4+ and CD8+ T cells among all live cells. Dot plots are representative of 3 mice per genotype. Note that the relative increase in CD8+ and CD4+ cells is due to the loss of B cells in c-MybPlt4/Plt4 mice. (C) Flow cytometry of IL-7Rα surface expression on splenic T cells from c-Myb+/+ and c-MybPlt4/Plt4 mice, gated for CD8+ or CD4+ cells as indicated. Plots are representative of 3 mice per genotype. (D) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 5- to 9-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice on a Rag1gfp/+ background. Cells at the CLP stage of development were identified as Sca-1+c-kitint GFP+; numbers in plots indicate the percentage of cells in each gated area. (E) Flow cytometry of IL-7Rα surface expression on Sca-1+c-kitint GFP+ cells. Plots are representative of 4 to 5 mice per genotype.
Because c-Myb is required for normal expression of IL-7Rα in the B-cell lineage, it was unclear whether the CLP defect in c-MybPlt4/Plt4 mice was due to a failure in development or a lack of IL-7Rα expression, the marker that defines this population. To examine CLPs independently of IL-7Rα expression, we crossed c-MybPlt4/Plt4 mice to mice expressing gfp under the control of the Rag1 promoter.39 On the Rag1gfp background, Sca-1+c-kitintGFP+ cells form a population that includes conventionally defined CLPs (Sca-1+c-kitintIL-7Rα+). Like CLPs, Sca-1+c-kitintGFP+ cells were reduced in c-MybPlt4/Plt4 mice (Figure 5D). In addition, the Sca-1+c-kitintGFP+ cells in c-MybPlt4/Plt4 mice displayed reduced expression of IL-7Rα compared with their c-Myb+/+ counterparts (Figure 5E). These data indicate that c-Myb both promotes the development of CLPs and is required for normal expression of IL-7Rα on lymphoid progenitors.
c-Myb is required for lymphoid priming
Our finding that c-MybPlt4/Plt4 mice have fewer CLPs suggested that c-Myb must play a role earlier in B-cell development than shown by analysis of c-MybΔmb1/Δmb1 mice. Recent evidence suggests that lymphoid-specific gene transcription is primed very early in hematopoiesis, before the CLP stage. Lymphoid transcripts including Dntt, Notch1, and Satb1 can be detected in HSCs, whereas Il7r, Rag1/2 transcripts are up-regulated in LMPPs.3,16 Given that c-Myb is expressed in hematopoietic progenitor cells, we examined whether c-Myb is required for priming of lymphoid genes, including Il7r, in multipotent progenitors.
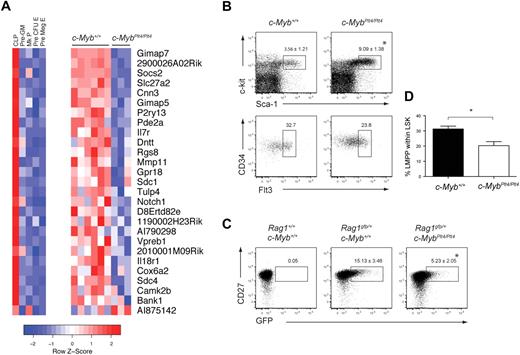
To identify global changes in gene expression, we isolated LSK cells from c-Myb+/+ and c-MybPlt4/Plt4 mice and performed a microarray analysis. Within the microarray data, we focused our attention on genes previously characterized as lymphoid specific, based on their expression in CLPs rather than myeloid, megakaryocyte, or erythroid progenitors.40 Of 108 lymphoid-specific genes in the dataset, 27 were differentially expressed between c-Myb+/+ and c-MybPlt4/Plt4 LSK cells (Figure 6A). Strikingly, of these 27 differentially expressed genes, 26 were down-regulated in the c-MybPlt4/Plt4 LSK cells, including Il7r, Notch1, and Dntt. This down-regulation of lymphoid transcripts was not due to a simple lack of LMPPs. Although c-MybPlt4/Plt4 mice had an increased proportion of LSK cells, the frequency of LMPPs, as well as early lymphoid progenitors defined by Rag1gfp expression,39 within the c-MybPlt4/Plt4 LSK fraction was only partially reduced (Figure 6B-D). Importantly, c-MybPlt4/Plt4 LSK cells displayed normal expression of 81 lymphoid-specific genes, including Rag1/2, Igh6, Satb1, Ccr9, and Ebf1 (supplemental Table 2). c-MybPlt4/Plt4 LSK cells also expressed normal levels of other genes implicated in lymphoid priming and/or the development of LMPPs, including Flt3, Ikzf1 (Ikaros), Sfpi1 (Pu.1), and Tcfe2a (E2a; supplemental Table 3). Overall, these data indicate that c-Myb plays an important role in priming expression of a subset of lymphoid genes within the LSK compartment, including Il7r.
c-MybPlt4/Plt4 LSKs display reduced expression of lymphoid genes. (A) Heat map depicting expression of 27 lymphoid-specific genes that display altered expression in c-MybPlt4/Plt4 LSK samples (P < .1). (Left) Expression pattern of these genes in progenitor populations. Pre-GM indicates pre–granulocyte-macrophage progenitor; Mk progenitor, megakaryocyte progenitor; pre–CFU-E, erythroid progenitor pre–colony forming unit; pre-MegE, pre–megakaryocyte-erythroid progenitor. (Right) Expression pattern in c-Myb+/+ and c-MybPlt4/Plt4 LSK samples. Red squares denote up-regulated genes, blue squares denote down-regulated genes, with scale denoting z score, ie, standard deviation from mean expression. (B) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 6- to 9-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Upper plots display the proportion of Sca-1+c-kithi cells among lineage− cells (mean ± SD of 5-6 mice per genotype). Lower plots display the percentage of CD34+Flt3+ cells within the LSK fraction. Plots are representative of 5 to 6 mice per genotype. (C) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 5- to 7-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice on a Rag1gfp/+ background. Early lymphoid progenitors were identified as CD27+ GFP+ cells within the LSK fraction, identified as in panel B. Numbers in plots indicate the proportion of cells in the gated area. Data are the mean ± SD of 6 to 8 mice per genotype. (D) Graph depicting the percentage of LMPPs within the LSK fraction of c-Myb+/+ and c-MybPlt4/Plt4 bone marrow. *P < .05 in test group vs c-Myb+/+ cells. Data represent the mean ± SEM of 5 to 6 mice per genotype.
c-MybPlt4/Plt4 LSKs display reduced expression of lymphoid genes. (A) Heat map depicting expression of 27 lymphoid-specific genes that display altered expression in c-MybPlt4/Plt4 LSK samples (P < .1). (Left) Expression pattern of these genes in progenitor populations. Pre-GM indicates pre–granulocyte-macrophage progenitor; Mk progenitor, megakaryocyte progenitor; pre–CFU-E, erythroid progenitor pre–colony forming unit; pre-MegE, pre–megakaryocyte-erythroid progenitor. (Right) Expression pattern in c-Myb+/+ and c-MybPlt4/Plt4 LSK samples. Red squares denote up-regulated genes, blue squares denote down-regulated genes, with scale denoting z score, ie, standard deviation from mean expression. (B) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 6- to 9-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice. Upper plots display the proportion of Sca-1+c-kithi cells among lineage− cells (mean ± SD of 5-6 mice per genotype). Lower plots display the percentage of CD34+Flt3+ cells within the LSK fraction. Plots are representative of 5 to 6 mice per genotype. (C) Flow cytometry of density-fractionated and lineage-depleted bone marrow of 5- to 7-week-old c-Myb+/+ and c-MybPlt4/Plt4 mice on a Rag1gfp/+ background. Early lymphoid progenitors were identified as CD27+ GFP+ cells within the LSK fraction, identified as in panel B. Numbers in plots indicate the proportion of cells in the gated area. Data are the mean ± SD of 6 to 8 mice per genotype. (D) Graph depicting the percentage of LMPPs within the LSK fraction of c-Myb+/+ and c-MybPlt4/Plt4 bone marrow. *P < .05 in test group vs c-Myb+/+ cells. Data represent the mean ± SEM of 5 to 6 mice per genotype.
Overexpression of Il7r is not sufficient to rescue B-cell development from c-MybPlt4/Plt4 fetal liver cells
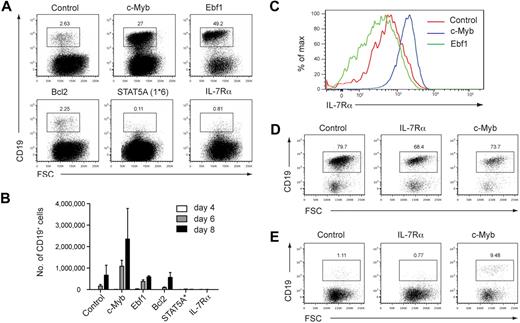
To further examine the basis of the B-cell defect observed in c-MybPlt4/Plt4 mice, we performed complementation experiments. c-MybPlt4/Plt4 fetal liver progenitors were transduced with a control retrovirus encoding GFP, or one encoding a protein of interest and an IRES-GFP to identify infected cells. After infection, cells were plated on OP9 stromal cells in the presence of IL-7 and Flt3L and assessed for efficiency of GFP+ CD19+ B-cell generation after 6 to 9 days of culture. Cells transduced with GFP alone generated a small percentage of B cells that proliferated poorly over the culture period (Figure 7A-B). Cells transduced with c-Myb generated a larger percentage of B cells that showed greater proliferation, achieving comparable numbers of CD19+ cells to c-Myb+/+ progenitors (Figure 7A-B; data not shown). The B cells generated by c-Myb overexpression showed higher levels of IL-7Rα expression than B cells expressing GFP alone, again suggesting that c-Myb is required for normal Il7r expression (Figure 7C).
IL-7Rα cannot rescue B-cell development from c-Myb deficient fetal liver cells. (A) Generation of CD19+ B cells from retrovirally infected c-MybPlt4/Plt4 fetal liver cells. Fetal liver cells from e14.5 embryos were depleted of Ter119+ cells and transduced with control retrovirus or retrovirus encoding c-Myb, Ebf1, Bcl2, constitutively active STAT5A(1*6), or IL-7Rα; all retroviruses included an IRES GFP to identify infected cells. Retrovirally infected cells were grown on OP9 stromal cells for 8 days in the presence of IL-7 and Flt3L. Numbers above the boxes indicate percentages of B cells (CD19+) among GFP+ cells. Plots are representative of 6 independent experiments. (B) Quantitation of CD19+ B cells generated from c-MybPlt4/Plt4 fetal liver cells transduced with retrovirus as in panel A. The number of GFP+ CD19+ cells was counted after 4, 6, and 8 days of culture. Data represent the mean ± SEM of 3 independent experiments. (C) Flow cytometry of IL-7Rα surface expression on B cells generated from c-MybPlt4/Plt4 fetal liver cells transduced with control, c-Myb, or Ebf1 retrovirus. Plots are gated on GFP+ CD19+ cells. Data are representative of 6 independent experiments. (D-E) Generation of CD19+ B cells from retrovirally infected (D) c-Myb+/+ or (E) c-MybΔmb1/Δmb1 CLPs. Cells were transduced and cultured as in panel A. Plots are representative of 2 independent experiments.
IL-7Rα cannot rescue B-cell development from c-Myb deficient fetal liver cells. (A) Generation of CD19+ B cells from retrovirally infected c-MybPlt4/Plt4 fetal liver cells. Fetal liver cells from e14.5 embryos were depleted of Ter119+ cells and transduced with control retrovirus or retrovirus encoding c-Myb, Ebf1, Bcl2, constitutively active STAT5A(1*6), or IL-7Rα; all retroviruses included an IRES GFP to identify infected cells. Retrovirally infected cells were grown on OP9 stromal cells for 8 days in the presence of IL-7 and Flt3L. Numbers above the boxes indicate percentages of B cells (CD19+) among GFP+ cells. Plots are representative of 6 independent experiments. (B) Quantitation of CD19+ B cells generated from c-MybPlt4/Plt4 fetal liver cells transduced with retrovirus as in panel A. The number of GFP+ CD19+ cells was counted after 4, 6, and 8 days of culture. Data represent the mean ± SEM of 3 independent experiments. (C) Flow cytometry of IL-7Rα surface expression on B cells generated from c-MybPlt4/Plt4 fetal liver cells transduced with control, c-Myb, or Ebf1 retrovirus. Plots are gated on GFP+ CD19+ cells. Data are representative of 6 independent experiments. (D-E) Generation of CD19+ B cells from retrovirally infected (D) c-Myb+/+ or (E) c-MybΔmb1/Δmb1 CLPs. Cells were transduced and cultured as in panel A. Plots are representative of 2 independent experiments.
Surprisingly, c-MybPlt4/Plt4 cells transduced with IL-7Rα generated even fewer CD19+ cells than control cells transduced with GFP (Figure 7A-B) and instead generated large numbers of myeloid lineage cells (data not shown). To exclude the possibility that overexpression of IL-7Rα had led to phosphorylation of inappropriate STAT molecules, we transduced cells with a constitutively active form of STAT5 (STAT5A(1*6)). This, however, had the same effect as overexpression of IL-7Rα (Figure 7A-B). Overexpression of IL-7Rα and STAT5A(1*6) also inhibited B-cell development from c-Myb+/+ progenitors (data not shown), supporting previous studies demonstrating that ectopic expression of IL-7Rα in multipotent progenitors is inhibitory to B-cell development.41,42
To overcome this limitation we expressed IL-7Rα in CLPs, because these cells are IL-7Rα+ and express a range of lymphoid transcription factors. Overexpression of IL-7Rα in c-Myb+/+ CLPs permitted B-cell development, suggesting that increased IL-7Rα signaling is not detrimental in an appropriate cellular context (Figure 7D). B-cell development from c-MybΔmb1/Δmb1 CLPs was partially rescued by overexpression of c-Myb but not by IL-7Rα, showing that IL-7Rα is not sufficient to replace the role of c-Myb in B-cell development (Figure 7E).
B-cell development from Il7−/− and Il7r−/− progenitors can be promoted by overexpression of Ebf1.5,6 We reasoned that if c-MybPlt4/Plt4 mice are similar to Il7r−/− mice then overexpression of Ebf1 should lead to enhanced B-cell formation. Indeed, overexpression of Ebf1 in c-MybPlt4/Plt4 fetal liver progenitors led to a marked increase in CD19+ cells (Figure 7A); however, these B cells failed to expand to the same extent as cells overexpressing c-Myb (Figure 7B). A possible explanation for this finding was that, unlike overexpression of c-Myb, overexpression of Ebf1 did not rescue IL-7Rα expression (Figure 7C). These results indicate that, as for Il7−/− and Il7r−/− mice, Ebf1 can promote B-cell development from c-MybPlt4/Plt4 progenitors. Also similar to Il7r−/− mice, the B-lymphopenia in c-MybPlt4/Plt4 mice could not be rescued by overexpression of Bcl2, indicating it is not due to a survival defect (Figure 7A-B).
Discussion
Despite evidence that c-Myb is required for B-cell development, the precise role of c-Myb in this process has remained enigmatic. We used a range of alleles of c-Myb to dissect the requirement for this gene during early lymphopoiesis. We found that c-Myb plays critical roles at multiple stages of lymphopoiesis, driving both the development of lymphoid progenitors and B-cell differentiation from the CLP.
Using c-MybPlt4/Plt4 mice, which express a hypomorphic allele of c-Myb throughout hematopoiesis, we showed that c-Myb is required at the very onset of lymphopoiesis. Although phenotypic LMPPs and the transcription of some lymphoid genes, including Rag1/2 and Ebf1, were found in the LSK compartment of c-MybPlt4/Plt4 mice, a large set of lymphoid transcripts, including Il7r, failed to be properly up-regulated. These data suggest that c-Myb is required to activate expression of several lymphoid genes within multipotent progenitor cells. Subsequent stages of B-cell development, including the CLP, pre-pro-B and pro-B cell stages, were all dramatically reduced, yet not absent, in c-MybPlt4/Plt4 mice, perhaps because these mice retain a small amount of c-Myb activity. In contrast, T-cell development was relatively unaffected in c-MybPlt4/Plt4 mice, suggesting that, as has been observed in Ikzf1−/− mice,43 T-cell development in c-MybPlt4/Plt4 mice may occur independently of the CLP. The defect in lymphoid priming in c-MybPlt4/Plt4 mice was not due to perturbations in expression of other transcriptional regulators of early lymphoid development, including Ikzf1, Sfpi1, and Tcfe2a. c-Myb is therefore a novel component of the regulatory network controlling lymphoid specification.
However important c-Myb is within the multipotent progenitor compartment, c-Myb also plays an essential role in B-cell progenitors. In agreement with another recent study,24 we found that deletion of c-Myb specifically in the B-cell lineage using mb1Cre led to an absolute block in B-cell development after the CLP stage. The complete absence of B cells in c-MybΔmb1/Δmb1 mice places c-Myb in the same category as other key regulators of B-cell development, including Ebf1 and Pax5. We took advantage of the residual pool of B-cell progenitors in c-MybPlt4/Plt4 mice to gain insights into the function of c-Myb during B-cell development. A key role of c-Myb is to enable B-cell progenitors to respond to IL-7, thereby providing a vital link between the intrinsic and extrinsic signals governing B-cell development. c-MybPlt4/Plt4 pro-B cells failed to respond to IL-7 or TSLP, either by phosphorylating STAT5 or by generating B-cell colonies. Although c-MybPlt4/Plt4 pro-B cells could not respond to IL-7, they did display VH-DJH rearrangements of the Igh locus, in agreement with a recent report that Il7r and Stat5a/b are not required for this process.44
The B-lymphopenia in c-MybPlt4/Plt4 mice shares striking similarities with that observed in mice with defective IL-7 signaling. All of these mice display a profound reduction in B-cell numbers, with the most significant developmental block occurring at the transition from the CLP to the pre-pro-B cell stage.5,6 The B lymphopenia becomes more severe with age, with most residual lymphopoiesis occurring during fetal life.7,38 The defect in B-cell development is magnified as the cells mature, such that hematopoietic progenitors retain a small capacity to generate mature B cells that is lost by the pre-pro-B or pro-B cell stage.45 As for Il7r−/− progenitors, B-cell development from c-MybPlt4/Plt4 progenitors could be promoted by overexpression of Ebf1.5,6 Given that Ebf1 can promote the differentiation of LSK cells toward the B-cell lineage,46 it is possible that this effect of Ebf1 is predominantly due to effects on early progenitors rather than on pro-B cell differentiation itself. Overall, the striking parallels between c-MybPlt4/Plt4 mice and mice with defects in IL-7 signaling suggest that failure of IL-7 signaling makes an important contribution to the phenotype of c-Myb–deficient mice.
A possible reason why c-MybPlt4/Plt4 pro-B cells display defective IL-7 signaling is that they express decreased IL-7Rα on their cell surface. IL-7Rα is a component of both the IL-7 and TSLP receptors; hence, reduced expression of IL-7Rα could underlie the defective response of c-MybPlt4/Plt4 cells to both of these cytokines. Regulation of the Il7r promoter in B cells is incompletely understood. Although there is evidence that Pu.1 binds to the Il7r promoter and can activate expression in B cells,47 B cells expressing Il7r can be generated from Sfpi1−/− cells, either by culturing for long periods48 or by overexpressing Ebf1.10 Moreover, Sfpi1 can be deleted from committed B cells without any effect on Il7r expression.49 These findings suggest that regulation of Il7r in B cells is complex and that other factors in addition to Pu.1 must be able to activate and/or maintain Il7r expression. c-Myb may contribute to regulation of the Il7r promoter in the B-cell lineage. Il7r expression was reduced in both LSK cells and pro-B cells from c-MybPlt4/Plt4 mice, despite these cells expressing normal levels of Pu.1. Complementation of MybPlt4/Plt4 pro-B cells with c-Myb restored IL-7Rα expression and IL-7 responsiveness. Although these data suggest that c-Myb regulates Il7r expression, at present it is unclear whether this is due to a direct role for c-Myb at the Il7r promoter. Reporter gene assays suggest that c-Myb can activate the promoter in vitro (data not shown), but at present we lack functional anti–c-Myb antibodies that would allow us to test by chromatin immunoprecipitation whether this is due to a direct interaction between c-Myb and regulatory elements of the Il7r promoter in B cells.
Overexpression of IL-7Rα did not rescue B-cell development from c-MybPlt4/Plt4 or c-MybΔmb1/Δmb1 progenitors, showing that, in the absence of c-Myb function, activation of Il7r is not sufficient for B lymphopoiesis. Moreover, the large number of lymphoid transcripts down-regulated in c-MybPlt4/Plt4 LSK cells indicates that c-Myb has important roles before the stage when IL-7 signaling becomes critical. This may indicate that the regulation of Il7r expression is not the sole function of c-Myb. It is possible that the residual IL-7R expression on c-Myb–deficient B-cell progenitors is sufficient for productive signaling and that the defective IL-7 responses result from further defects in other signaling components, such as the JAK proteins, and/or their activation downstream of the receptor. In support of this possibility, c-MybPlt4/+ and c-MybΔmb1/+ heterozygous mice displayed reduced B-cell development despite relatively normal IL-7Rα surface expression. The pro-B cell microarray showed several additional genes dysregulated in the absence of sufficient c-Myb activity. These included several genes with known roles in B-cell development (eg, Lef1, Tcf12) as well as genes whose function in B-cell development is yet to be characterized (eg, Robo4, Il10ra, Fcrl1). Future studies will be directed toward understanding how these genes are involved in B-cell development and the extent to which they contribute to the phenotype of c-MybPlt4/Plt4 mice.
Collectively, these findings show c-Myb to be a critical player in lymphoid specification and B-cell development. c-Myb controls the up-regulation of a subset of lymphoid genes within multipotent progenitors and is required for normal development of CLPs as well as in committed B-cell progenitors. Recognition that c-Myb is essential for the B-cell lineage makes it imperative to incorporate c-Myb into existing models of the factors regulating early lymphoid development and B-cell differentiation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Corbin, J. Brady, K. Elder, K. Trueman, C. Evans, and S. Ross for technical assistance; M. Reth (Max Planck Institute of Immunology) for the mb1Cre mice; N. Sakaguchi (Kumamoto University) for the Rag1gfp mice; J.C. Zuniga-Pflucker (University of Toronto) for the OP9-DL1 cell line; R. DeKoter (University of Cincinnati), T. Kitamura (University of Tokyo), and E. Mullner (Max F. Perutz Laboratories) for plasmid constructs.
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia and the National Heart, Lung, and Blood Institute (grant R01 HL080019). S.L.N. is supported by the Pfizer Australia Research Fellowship, D.J.H. and J.M.M. by NHMRC Fellowships, C.A.d.G. by an Australian Postgraduate Award, B.T.K. by a QEII Fellowship from the Australian Research Council, S.H.M.P. by the Leukemia Foundation of Australia, and K.T.G. by a Stella Mary Langford Scholarship from the University of Melbourne.
National Institutes of Health
Authorship
Contribution: K.T.G. performed most of the experiments and cowrote the manuscript; C.A.d.G., J.M.M., M.R.C., and S.H.M.P. performed experiments; J.F. provided essential reagents; B.T.K. and D.J.H. provided input into the experimental design and manuscript preparation; and S.L.N. supervised the experimental design and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for K.T.G. is Division of Molecular Genetics, The Netherlands Cancer Institute, The Netherlands.
Correspondence: Stephen L. Nutt, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria 3052, Australia; e-mail: nutt@wehi.edu.au.