Abstract
Tumor–bone marrow microenvironment interactions in multiple myeloma (MM) are documented to play crucial roles in plasma-cell growth/survival. In vitro coculture of MM cells with osteoclasts supported cell survival and significantly down-regulated JUN expression. JUN expression in myeloma cells from late-stage and high-risk MM was significantly lower than in plasma cells from healthy donors, monoclonal gammopathy of undetermined significance, smoldering MM, and low-risk MM; patients with low-JUN–expressing MM cells had earlier disease-related deaths. JUN overexpression in MM cells induced cell death and growth inhibition and up-regulated expression of early growth response protein 1 (EGR-1), whose low expression also carried unfavorable clinical implications. EGR-1 knockdown in MM cells abrogated JUN overexpression-induced MM cell death and growth inhibition, indicating that EGR-1 acts directly downstream of JUN. JUN modulates myeloma cell apoptosis through interacting with EGR-1, which down-regulates Survivin and triggers caspase signaling. Importantly, high JUN or EGR-1 expression was associated with improved outcome in Total Therapy 3, in which bortezomib is given throughout therapy, versus Total Therapy 2, in which bortezomib is given only at relapse. Consistently, JUN or EGR-1 knockdown in cultured MM cells enhanced their resistance to bortezomib, demonstrating the crucial role of low JUN/EGR-1 expression in MM resistance to bortezomib.
Introduction
Multiple myeloma (MM) is a clonal B-cell malignancy characterized by bone marrow plasmacytosis, expansion of monoclonal immunoglobulin, bone lesions, renal failure, and immunodeficiency.1 The bone-marrow microenvironment plays a key role in the growth and survival of myeloma cells,2 and the interactions of myeloma cells with the microenvironment are believed to be critical in the pathophysiology of MM.2,3
Activator protein 1 (AP-1) transcription factor, a heterodimer consisting of proteins of the Jun (JUN, JunB, and JunD) and Fos (c-Fos, FosB, Fra-1, and Fra-2) families, has been called a double-edged sword in tumorigenesis because it has been implicated in induction of apoptosis as well as in promotion of cell survival and proliferation.4 AP-1 regulates transcriptional activation of different target genes, depending on the different physiologic and pathophysiologic stimuli, and thus executes distinct biologic functions. AP-1 is considered a key mediator in the pathogenesis of cancer.5-7 It can alter target gene expression, including activation of CCND1 and inhibition of TP53 and CDKN1A, to promote cell-cycle progression from G1 to S phases to induce cell proliferation in response to tumor promoters,8 such as 12-O-tetradecanoylphorbol-13-acetate9,10 and ultraviolet radiation.11 However, AP-1 also can induce apoptosis by activating the expression of cell-death genes,12-14 although the mechanisms of AP-1–induced apoptosis in MM are still unclear.
We previously demonstrated that osteoclasts can support myeloma plasma cell survival.15 We also have analyzed the gene expression profiles of myeloma plasma cells after 2 days of coculture with osteoclasts. In 14 of 19 myeloma plasma cell samples cocultured with osteoclasts, we identified simultaneously decreased signals from AP-1 transcription factors JUN, JunD, Fos, and FosB. In recent studies16,17 authors have shown that activation of the JUN NH2-terminal kinase (JNK) pathway modulates the proliferation of myeloma plasma cells and participates in the apoptosis of these cells induced by treatment with therapeutic agents, including proteasome inhibitor bortezomib and arsenic trioxide. JUN′s activities are regulated by phosphorylation through the JNK pathway and thus mediate JNK-associated myeloma plasma cell proliferation and apoptosis regulation.
The gene encoding early growth response protein 1 (EGR-1) is a member of the immediate-early gene family and is rapidly induced by various mitogens, growth factors, tissue injury, ischemia, and apoptotic signals in distinct cell types.18-20 The EGR-1 protein plays a pivotal role in the regulation of cell growth, differentiation, and apoptosis.21-23 The proapoptotic activity of EGR-1 may depend on the cell type and the nature of the stimulus. EGR-1 induces apoptosis via regulation of expression of tumor suppressor genes TP5324 and PTEN25 and transcription factor JUN.26,27 Levkovitz et al26,27 reported that the EGR-1/JUN complex induced neurite outgrowth and promoted cerebellar granule cell apoptosis. However, Hoffmann et al28 identified JUN as an essential effector of EGR-1 transcriptional regulation in inflammatory processes. Although the authors of recent studies29-32 have implicated EGR-1 as a tumor suppressor in many tumor cell types, little attention has been paid to its role in MM. In the present report, we describe our findings delineating the functional role of JUN and EGR-1 in myeloma cell growth and survival, as well as in the development of resistance to bortezomib.
Methods
Study subjects
Purified CD138+ plasma cells were obtained from patients with MM enrolled in the National Institutes of Health–sponsored clinical trials UARK 98-026 (Total Therapy 2 [TT2]; n = 339) and UARK 03-033 (Total Therapy 3 [TT3]; n = 205).33-36 Both protocols use induction regimens followed by melphalan-based tandem autotransplantation, consolidation chemotherapy, and maintenance treatment. Proteasome inhibitor bortezomib was incorporated in TT3. The Institutional Review Board of the University of Arkansas for Medical Sciences approved the research studies, and all subjects provided written informed consent in accordance with the Declaration of Helsinki. The plasma cells were isolated by automated CD138 immunomagnetic bead selection (Miltenyi Biotec). CD38/CD45 flow cytometry determined that plasma cell purity was routinely 85% or greater.
Human myeloma cell lines OCI-My5,OPM2, and U266 were cultured in RPMI1640 containing 10% heat-inactivated fetal calf serum, 2mM l-glutamine (Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C in humidified 95% air and 5% carbon dioxide.
Coculture of osteoclasts with myeloma plasma cells
Multinucleate osteoclast cultures were prepared as previously described.37 Peripheral-blood mononuclear cells obtained from patients with MM were cultured at 2.5 × 106 cells/mL in osteoclast medium α-minimal essential medium supplemented with 10% fetal bovine serum, 50 ng/mL receptor activator of nuclear factor-κB ligand, and 25 ng/mL macrophage colony-stimulating factor for 10 to 14 days, by which time large numbers of multinucleate osteoclasts with bone-resorbing activity had formed.
Osteoclasts were washed 3 times with phosphate-buffered saline (PBS) to remove nonadherent cells. Purified myeloma plasma cells (0.5 × 106 cells/mL) from patients with MM were added to the osteoclasts in osteoclast medium in 6-well plates (5 mL/well). Myeloma cells were collected for analysis after 2 days of culturing.
Gene expression profiling of myeloma cells with mesenchymal stem cells in coculture
Mesenchymal stem cells (MSCs) were obtained from Dr D. Prockop (Tulane University) and seeded in 24-well plates at 40 000 cells per well in complete culture medium 1 at least 24 hours before the addition of MM cells, at which time the media were removed and 106 CD138-sorted (> 95% viability) myeloma cells in complete culture media were added to each well. The plates were kept in a humidified atmosphere at 37°C and 5% CO2. After 18 hours' incubation, the medium was removed; total RNA was extracted by the use of the RNeasy kit (QIAGEN) and DNA was digested by the use of DNase (QIAGEN). For the control group (t = 0 hours group), the medium was removed from MSCs, and 106 CD138-sorted (> 95% viability) myeloma cells were added per well in PBS for a total volume of 20 μL or less. Immediately afterward, the MSC plus myeloma cell mixture was lysed, and total RNA was extracted by the use of QIAGEN's RNeasy kit with DNAse digestion. Primary MM cells from 18 different patients and MSCs from 7 healthy donors were used in the experiment. Gene expression profiling (GEP) was performed on the 18 RNA specimens with Affymetrix U133Plus2 chips. GeneChip Operating Software (http://www.affymetrix.com/products/software/specific/gcos.affx) normalized output data were further examined by the use of Acuity 4 bioinformatics software for analysis of microarrays. A paired t test was performed to determine significance between the groups. Ratios of signal means and standard deviations for t = 0 and t = 18 hours were calculated and plotted.
Real-time reverse-transcriptase PCR
Total RNA was extracted with RNeasy kit (QIAGEN) and reverse-transcribed with M-MLV reverse transcriptase III (Invitrogen) to form cDNA. To amplify JUN and EGR-1 transcripts, the cDNA was subjected to a SYBR green–based method for real-time polymerase chain reaction (PCR) relative quantification. Real-time PCR was performed on an ABI PRISM 7900 analytical thermal cycler (Applied Biosystems) according to the manufacturer's recommendations. The real-time PCR primers were as follows: for JUN, forward, 5′GGCTGGTGTTTCGGGAGTGT3′ and reverse, 5′CGCCGCCTTCTGGTCTTTAC3′; for EGR-1, forward, 5′CTTCAACCCTCAGGC GGACA3′ and reverse, 5′GGAAAAGCGGCCAGTATAGGT3′. JUN and EGR-1 expression levels were calculated relative to the level of the glyceraldehyde-3-phosphate dehydrogenase housekeeping gene. Each sample was analyzed in duplicate, and the results were expressed as means plus or minus SEM.
Evaluation of DNA binding activity of JUN by ELISA
The DNA binding activity of JUN was detected by enzyme-linked immunosorbent assay (ELISA) with the Trans-AM AP-1 transcription factor assay kit (Active Motif North America) according to the instructions of the manufacturer. In brief, nuclear extracts were prepared and incubated in 96-well plates coated with immobilized oligonucleotide (5′-CGCTTGATGAGTCAGCCGGAA-3′) containing a JUN binding site. JUN binding to the target oligonucleotide was detected by the use of phospho-JUN antibody (Active Motif North America) and quantified at 450 nm with a reference wavelength of 655 nm. Each sample was analyzed in duplicate, and the results were expressed as the mean plus or minus SEM.
Transfection of myeloma cell lines
JUN and Survivin cDNA sequences derived by PCR amplification were cloned into pWPI lentiviral vectors (a generous gift from Didier Trono, Ecole Polytechnique Fédérale de Lausanne School of Life Sciences). Synthetic double-stranded oligonucleotide sequences specific for genes encoding EGR-1 (5′ GATCCCCGTTACTACCTCTTATCCATTTCAAGAGAATGGATAAGAGGTAGTAACTTTTTA 3′) and JUN (5′GATCCCCAACGACCTTCTATGACGATGCTTCAAGAGAGCATCGTCATAGAAGGTCGTTTTTTTA 3′) and a nonsense scrambled oligonucleotide (5′GATCCCCGACACGCGACTTGTACCACTTCAAGAGAGTGGTACAAGTCGCGTGTCTTTTTA 3′) were obtained from OligoEngine. shRNA double-stranded oligonucleotides were cloned into lentiviral pLVTH vectors (kindly provided by Didier Trono). Recombinant lentivirus was produced by transient transfection of 293T cells according to a standard protocol. Crude virus was concentrated by ultracentrifugation at 90 000g for 100 minutes. Viral titers were determined by measuring the amount of HIV-1 p24 antigen by ELISA (NEN Life Science Products). A 99% transduction efficiency of myeloma cells was achieved with a concentration of lentiviral p24 particles of 3 μg/106 cells. All transfection experiments were performed in duplicate.
Western blotting
To test JUN protein levels in myeloma cell lines and for JUN overexpression studies, the Western Breeze (Invitrogen) chemiluminescent immunodetection protocol was used for Western blotting according to the manufacturer's instructions. Antibodies (Cell Signaling Technology) included anti-JUN; anti–EGR-1; anti-Survivin; anti-XIAP; anti-cIAP1; anti–phosphorylated-Elk-1; anti–Bcl-2; anti-Bax; anti–caspase-3, -7, -8, and -9; and anti–β-actin.
Cell-proliferation assay
OCI-My5, OPM2, and U266 cell lines infected with the JUN coding DNA sequence (CDS) and JUN and with EGR-1 shRNA, together with their controls, were seeded at a density of 0.2 × 106 cells/mL. Cell number and viability were determined by trypan blue exclusion at various time intervals.
Cell apoptosis analysis
Cells (106) from each sample were fixed in 75% ethanol at −20°C overnight. On the following day, the cells were washed with cold PBS, treated with 100 μg of RNase A (QIAGEN), and stained with 50 μg of propidium iodide (Roche Applied Science). For flow cytometry, a 3-color FACScan was used, together with CellQuest software (Becton Dickinson). For each sample, 10 000 events were gated. The data were analyzed with Modfit LT software (Verity Software House). Apoptotic cell fractions were determined as means plus or minus SEM of the percentage of cells with sub–G0-phase DNA content in 2 independent experiments.
Caspase inhibition assay
To test whether JUN overexpression induced apoptosis through caspase activation, OCI-My5 and OPM2 cells infected with JUN-CDS-pWPI constructs were treated with pancaspase inhibitor Z-VAD-fmk (100μM; R&D Systems) after 16 hours of lentiviral infection. All experiments were performed in duplicate.
ChIP
Chromatin immunoprecipitation (ChIP) experiments were performed with the use of the ChIP-IT kit (Active Motif). In brief, 107 OCI-My5 cells or JUN-overexpressing OCI-My5 cells were fixed in 1% formaldehyde at room temperature for 10 minutes; the fixation reaction was stopped by adding a 1:10 volume of 10× glycine to the tube at room temperature for 5 minutes. The cell pellets were resuspended and incubated for 30 minutes in ice-cold lysis buffer with phenylmethylsulfonyl fluoride and proteinase inhibitor cocktail. The nuclear pellets were resuspended in shearing buffer, and chromatin was sheared to an average size of 200 to 1500 bp by sonication at 25% power for 12 pulses of 20 seconds each, with a 30-second rest on ice between each pulse. Chromatin (10 μL) was saved for input DNA control. Sheared chromatin (6.3 μg) was incubated in ChIP buffer 1 with 25 μL of protein G magnetic beads and 3 μg of anti-JUN antibody (Active Motif) or rabbit IgG antibody as a negative control on a rolling shaker at 4°C for 4 hours. The immunoprecipitated chromatin was purified from the chromatin-antibody mixture by several washing steps, and the chromatin-immunoprecipitated DNA was eluted in 50 μL of elution buffer AM2 (Active Motif). Crosslinks were reversed by adding 50 μL of reverse-crosslink buffer. After removing proteins by digestion with proteinase K, the DNA was purified and examined by real-time PCR. The EGR-1 primer sequences were as follows: forward, 5′ATTCAGAGCTAGAGCAGGAGGAG C3′; and reverse, 5′GGTGGCGAGGGAGAACGATT3′.
GEP and data analyses
GEP with the Affymetrix U133 Plus 2.0 microarray (Affymetrix) was performed on CD138-purified plasma cells as previously described.38,39 GEP data on the 544 patients with MM used in this study have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE2658. Significant association with outcome was determined by log-rank test for survival. Hazard ratios were calculated by the Cox proportional model. A multivariate survival analysis was applied to select independent features that were most significantly associated with outcome. All statistical analyses were performed with the use of the statistical software R (Version 2.6.2), available from The R Project for Statistical Computing website (http://www.r-project.org), and with R packages developed by the BioConductor project (http://www.bioconductor.org); all the software is freely available.
Results
Osteoclasts and MSCs down-regulate EGR-1 expression
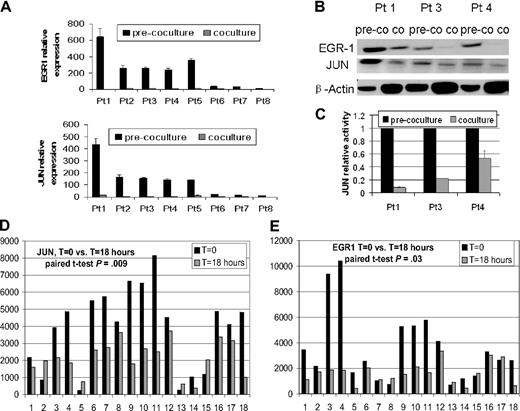
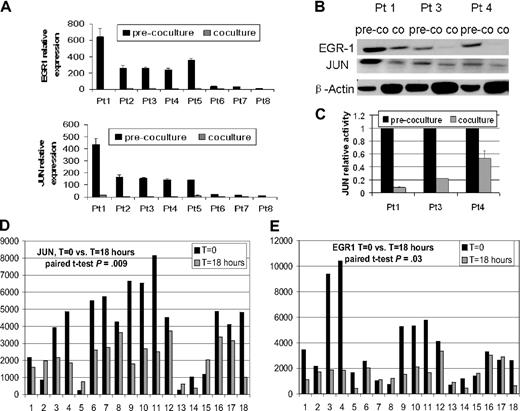
Real-time PCR analysis revealed a 7.5- to 32-fold decrease in EGR-1 signal in primary myeloma plasma cells from 8 patients after coculture with osteoclasts compared with controls in culture without osteoclasts (Figure 1A). Western blot analysis of samples from 3 patients also demonstrated down-regulation of EGR-1 protein (Figure 1B). In addition, EGR-1 down-regulation was highly correlated with expression and binding activity of AP-1 family member JUN (Figure 1B-C). The down-regulation of JUN in 77.8% (14 of 18) patients and EGR-1 in 72.2% (13 of 18) patients was identified by GEP after coculture of primary myeloma plasma cells with MSCs (t = 18 hours) compared with controls (t = 0 hours; P < .05; Figure 1D-E). The remarkable decrease in EGR-1 expression in plasma cells after coculture with osteoclasts and MSCs indicated that survival of MM cells through interaction with cells in the bone marrow microenvironment is associated with decreased EGR-1 expression.
Osteoclasts and MSCs down-regulated JUN and EGR-1 in myeloma plasma cells. (A-C) Primary myeloma plasma cells from 8 patients were cultured in the presence (coculture) or absence (pre-coculture) of osteoclasts for 2 days. (A) JUN and EGR-1 gene expression levels in these cells were evaluated by real-time PCR, expressed as relative values by the comparative Ct method. (B) Protein levels of JUN and EGR-1 were determined by Western blot analysis in 3 patient samples, shown in panel A, that had sufficient quantities of protein extracts available. Protein levels were compared in cultured cells in the presence (co) or absence (pre-co) of osteoclasts. β-Actin was used as a loading control. (C) JUN binding activity was evaluated in nuclear extracts of the same 3 samples of myeloma plasma cells shown in panel B by the use of an ELISA-based assay (expressed as relative value compared with each control); each sample is expressed as the mean ± SEM of 2 duplicates. JUN (D) and EGR-1 (E) gene expression from GEP data of myeloma cells cultured with (t = 18 hours) or without (t = 0 hours) MSCs.
Osteoclasts and MSCs down-regulated JUN and EGR-1 in myeloma plasma cells. (A-C) Primary myeloma plasma cells from 8 patients were cultured in the presence (coculture) or absence (pre-coculture) of osteoclasts for 2 days. (A) JUN and EGR-1 gene expression levels in these cells were evaluated by real-time PCR, expressed as relative values by the comparative Ct method. (B) Protein levels of JUN and EGR-1 were determined by Western blot analysis in 3 patient samples, shown in panel A, that had sufficient quantities of protein extracts available. Protein levels were compared in cultured cells in the presence (co) or absence (pre-co) of osteoclasts. β-Actin was used as a loading control. (C) JUN binding activity was evaluated in nuclear extracts of the same 3 samples of myeloma plasma cells shown in panel B by the use of an ELISA-based assay (expressed as relative value compared with each control); each sample is expressed as the mean ± SEM of 2 duplicates. JUN (D) and EGR-1 (E) gene expression from GEP data of myeloma cells cultured with (t = 18 hours) or without (t = 0 hours) MSCs.
High risk and poor prognosis in MM are highly correlated with low expression of both JUN and EGR-1
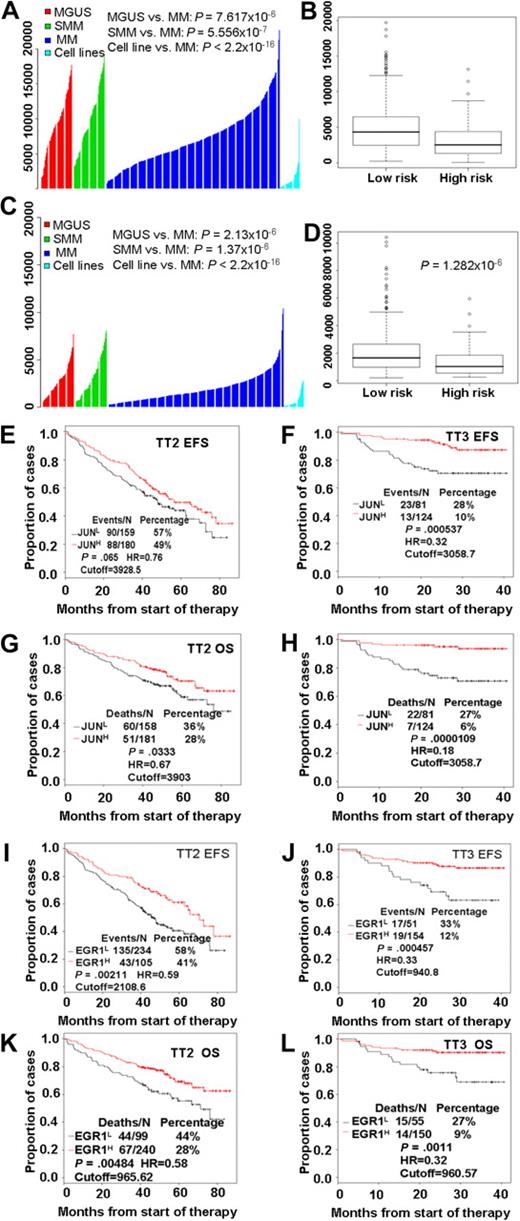
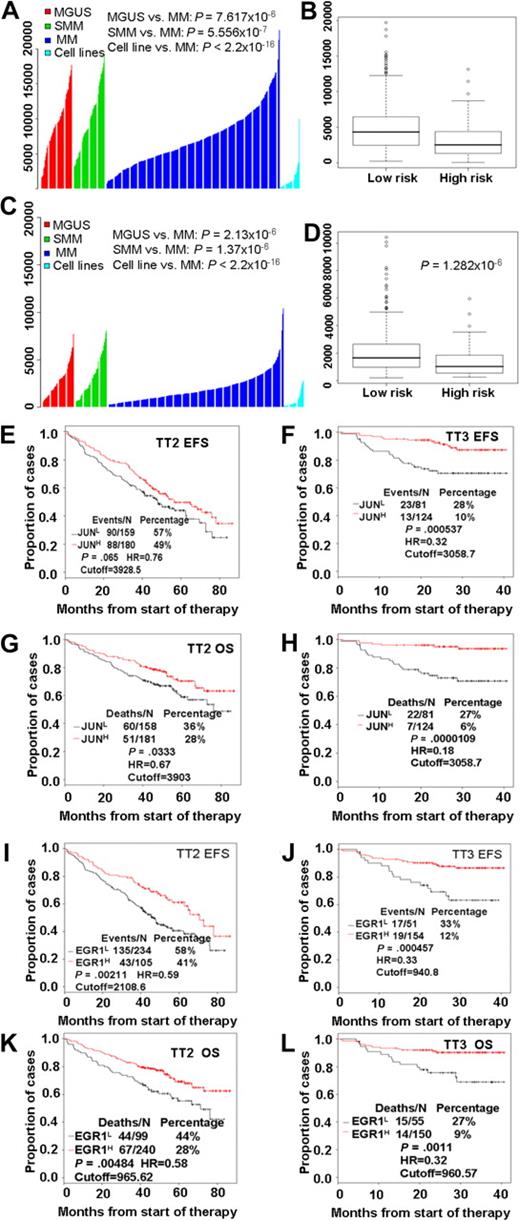
Our examination of the clinical relevance of JUN expression revealed that highly purified plasma cells from patients with monoclonal gammopathy of undetermined significance (MGUS; n = 98) or smoldering MM (n = 104) had significantly greater (P < .001; t test) JUN expression than was found in patients with MM (n = 544) or in 63 myeloma cell lines derived from end-stage disease (Figure 2A). Furthermore, a t test analysis of JUN expression levels in high- and low-risk disease groups showed that low levels were highly associated with high-risk MM (P < .001; Figure 2B). EGR-1 expression exhibited expression patterns similar to those of JUN; low EGR-1 expression was highly correlated with disease progression (Figure 2C) and high risk (P < .001; Figure 2D).
High risk and poor prognosis in MM is highly correlated with low expression of both JUN and EGR-1. JUN (A) and EGR-1 (C) gene expression levels (y-axis) in plasma cells from 98 subjects with MGUS, 104 with smoldering MM (SMM), and 544 newly diagnosed MM in the TT2 trial, as well as in 63 myeloma cell lines. The height of each bar indicates a sample's level of JUN (A) or EGR-1 (C) gene expression, as measured by Affymetrix microarray signal. Low JUN (B) and EGR-1 (D) expression levels were highly correlated with high-risk MM. JUN and EGR-1 expression levels, as measured by Affymetrix microarray signal, in 75 high-risk MM patients are significantly lower than in 469 low-risk MM patients. By the use of the optimal cutoffs, EFS (E, F, I, and J) and OS (G, H, K, and L), analyses were performed on JUN (E-H) or EGR-1 (I-L) mRNA expression levels, measured by Affymetrix microarray signal, in the TT2 and TT3 datasets.
High risk and poor prognosis in MM is highly correlated with low expression of both JUN and EGR-1. JUN (A) and EGR-1 (C) gene expression levels (y-axis) in plasma cells from 98 subjects with MGUS, 104 with smoldering MM (SMM), and 544 newly diagnosed MM in the TT2 trial, as well as in 63 myeloma cell lines. The height of each bar indicates a sample's level of JUN (A) or EGR-1 (C) gene expression, as measured by Affymetrix microarray signal. Low JUN (B) and EGR-1 (D) expression levels were highly correlated with high-risk MM. JUN and EGR-1 expression levels, as measured by Affymetrix microarray signal, in 75 high-risk MM patients are significantly lower than in 469 low-risk MM patients. By the use of the optimal cutoffs, EFS (E, F, I, and J) and OS (G, H, K, and L), analyses were performed on JUN (E-H) or EGR-1 (I-L) mRNA expression levels, measured by Affymetrix microarray signal, in the TT2 and TT3 datasets.
We investigated the correlation of the length of event-free survival (EFS) and overall survival (OS) with expression levels of JUN or EGR-1 in patients enrolled in TT2 (n = 339) and TT3 (n = 205) clinical trials. Kaplan-Meier analyses showed that EFS and OS times were shorter in patients with low expression than in those with high expression of JUN (Figure 2E-H) or EGR-1 (Figure 2I-L). Thus, there appears to be a link between low levels of JUN or EGR-1 expression and clinical outcome in patients with MM.
JUN and EGR-1 are critical components in the apoptosis pathway induced by bortezomib
We noted a much more significant association of survival times and expression levels of JUN and EGR-1 in TT3 than in TT2 trial participants (Figure 2E-L). Because the drugs used in the TT3 regimen differ from those in the TT2 regimen only by addition of bortezomib, this observation led us to consider whether JUN and EGR-1 are critical components in the pathway-mediating response to this drug.
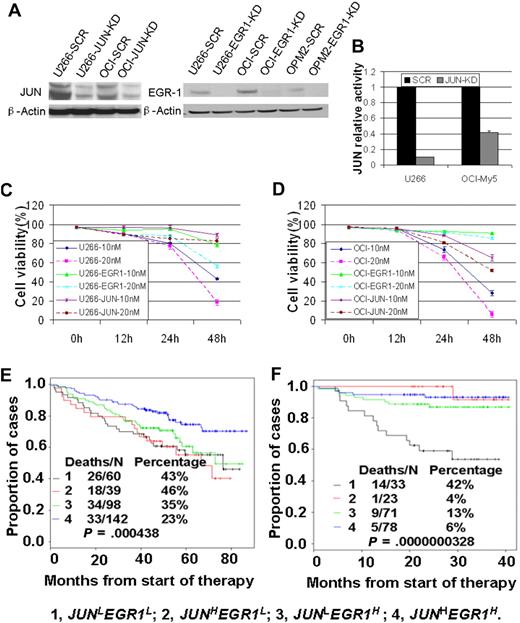
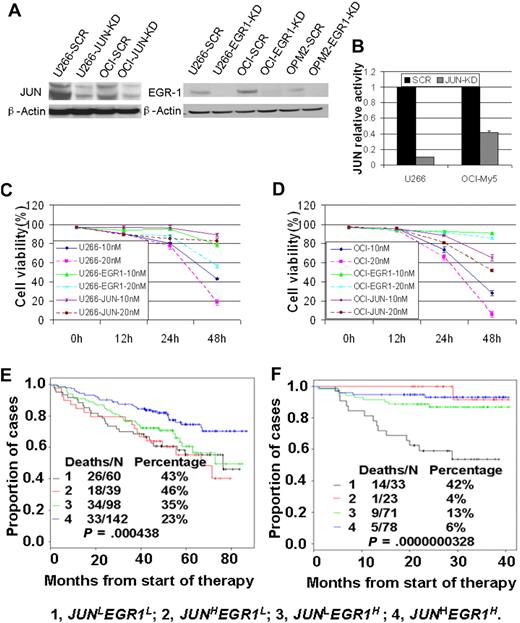
To examine the roles of JUN and EGR-1 expression in the response to bortezomib treatment, we first used siRNA-lentiviral transduction to silence JUN or EGR-1 expression in myeloma cell lines U266 and OCI-My5 (Figure 3A-B); silencing JUN or EGR-1 expression had no effect on cell proliferation or apoptosis (P > .05). We then treated unaltered cells of U266 and OCI-My5 myeloma cell lines and the corresponding JUN- or EGR-1–knockdown cell lines with 10nM or 20nM bortezomib and evaluated cell viability by trypan blue exclusion 48 hours after treatment. Bortezomib treatment induced massive apoptosis in the U266 and OCI-My5 cell lines (Figure 3C-D). In contrast, the JUN- or EGR-1–knockdown cell lines treated with bortezomib showed viability significantly better than the unaltered cell lines (P < .05; Figure 3C-D). These results strongly suggest that JUN and EGR-1are critical components in the apoptosis pathway induced by bortezomib.
JUN and EGR-1 are critical components in the apoptosis pathway induced by bortezomib. (A) Western blot demonstrated down-regulation of JUN expression in U266 and OCI-My5 myeloma cell lines transfected with JUN shRNA (JUN-KD) and EGR-1 down-regulation in U266, OCI-My5, and OPM2 cell lines transfected with EGR-1 shRNA (EGR-1-KD) compared with scrambled control shRNA (SCR). (B) JUN binding activity was evaluated in nuclear extracts of U266 and OCI-My5 JUN-KD cell lines by the use of an ELISA-based assay (expressed as relative value compared with each control). Each sample is expressed as the mean ± SEM of 2 duplicates. U266 (C) and OCI-My5 (D) cell lines, with and without shRNA-mediated JUN knockdown (JUN) or EGR-1 knockdown (EGR1), were cultured for 48 hours with 10nM and 20nM bortezomib. Cell viability was determined every 12 hours by trypan blue exclusion. Patients with newly diagnosed MM on TT2 (E) and TT3 (F) were divided into 4 groups based on the expression of JUN with EGR-1. Kaplan-Meier estimates on TT2 show 6-year actuarial probabilities of 43% death in cases with JUNLEGR1L, 46% death in cases with JUNHEGR1L, 35% death in cases with JUNLEGR1H, and 23% death with JUNHEGR1H. Kaplan-Meier estimates on TT3 show 3-year actuarial probabilities of 42% death in cases with JUNLEGR1L, 4% death in cases with JUNHEGR1L, 13% death in cases with JUNLEGR1H, and 6% death with JUNHEGR1H.
JUN and EGR-1 are critical components in the apoptosis pathway induced by bortezomib. (A) Western blot demonstrated down-regulation of JUN expression in U266 and OCI-My5 myeloma cell lines transfected with JUN shRNA (JUN-KD) and EGR-1 down-regulation in U266, OCI-My5, and OPM2 cell lines transfected with EGR-1 shRNA (EGR-1-KD) compared with scrambled control shRNA (SCR). (B) JUN binding activity was evaluated in nuclear extracts of U266 and OCI-My5 JUN-KD cell lines by the use of an ELISA-based assay (expressed as relative value compared with each control). Each sample is expressed as the mean ± SEM of 2 duplicates. U266 (C) and OCI-My5 (D) cell lines, with and without shRNA-mediated JUN knockdown (JUN) or EGR-1 knockdown (EGR1), were cultured for 48 hours with 10nM and 20nM bortezomib. Cell viability was determined every 12 hours by trypan blue exclusion. Patients with newly diagnosed MM on TT2 (E) and TT3 (F) were divided into 4 groups based on the expression of JUN with EGR-1. Kaplan-Meier estimates on TT2 show 6-year actuarial probabilities of 43% death in cases with JUNLEGR1L, 46% death in cases with JUNHEGR1L, 35% death in cases with JUNLEGR1H, and 23% death with JUNHEGR1H. Kaplan-Meier estimates on TT3 show 3-year actuarial probabilities of 42% death in cases with JUNLEGR1L, 4% death in cases with JUNHEGR1L, 13% death in cases with JUNLEGR1H, and 6% death with JUNHEGR1H.
To examine the clinical relevance of these findings, we divided the TT2 and TT3 participants into 4 groups on the basis of their expression of JUN and EGR-1: high JUN and EGR-1 expression (JUNHEGR1H), high JUN and low EGR-1 expression (JUNHEGR1L), low JUN and high EGR-1 expression (JUNLEGR1H), and low JUN and EGR-1 expression (JUNLEGR1L). In the TT2 trial, patients in the JUNHEGR1H group had OS superior to that of the other 3 groups (P < .005; Figure 3D), whereas no significant difference in OS was found among the other 3 groups. In the TT3 trial, patients in the JUNLEGR1L group had significantly decreased OS, compared with those with high expression of JUN and/or EGR-1 (Figure 3E). These results further suggest that low expression of JUN and/or EGR-1 plays a crucial role in high-risk and disease progression in MM. They also revealed that bortezomib improved the OS only in those patients with high expression of JUN or EGR-1.
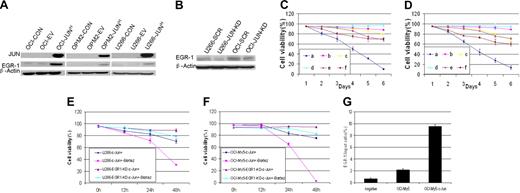
Overexpression of JUN in myeloma cell lines leads to profound growth inhibition and an apoptotic phenotype
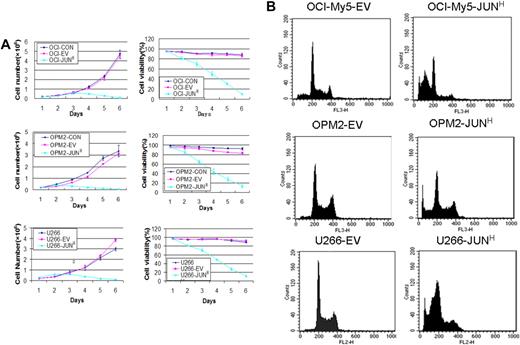
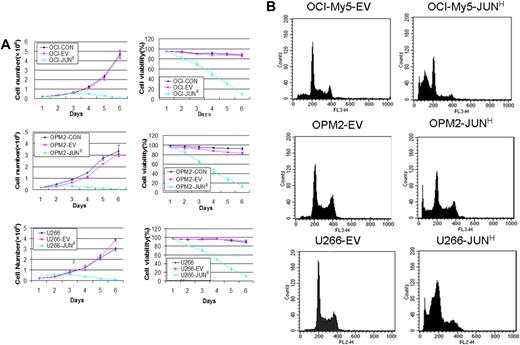
To identify JUN's function in myeloma cell growth and survival, we overexpressed JUN in the OCI-My5, OPM2, and U266 myeloma cell lines via lentiviral transduction. JUN overexpression decreased cell viability in the 3 cell lines within 48 hours. On day 5, almost all JUN-overexpressing cells had died, whereas the control cells continued to proliferate (P < .005; Figure 4A). JUN overexpression in these myeloma cell lines also had the effect of strongly inducing myeloma cell apoptosis, unlike control cells, as determined by flow cytometry 72 hours after transduction (Figure 4B). The percentages of sub–G0-phase DNA content were 56.2% plus or minus 2.6% versus 8.6% plus or minus 0.7% in OCI-My5–JUN versus OCI-My5–empty vector, 48.5% plus or minus 2.5% versus 5.4% plus or minus 0.36% in OPM2–JUN versus OPM2–empty vector, and 50.99% plus or minus 4.19% versus 4.03% plus or minus 0.25% in U266–JUN versus U266–empty vector (P < .05).
Effects of JUN overexpression on myeloma cell survival. (A) Effects on cell proliferation (left) and cell viability (right) in OCI-My5 (top), OPM2 (middle), and U266 (bottom) cells for JUN overexpression. Total number of cells and cell viability were evaluated daily by trypan blue exclusion. Error bars represent standard error of the mean for 2 independent experiments. (B) JUN overexpression induced apoptosis of OCI-My5, OPM2, and U266 cells as evaluated by flow cytometry performed 72 hours after lentiviral transfection with EV or JUN cDNA. Overexpression of JUN induced a dramatic increase in the percentages of cells with sub–G0-phase DNA content (indicative of apoptosis). CON indicates control; EV, empty vector; and JUNH, JUN overexpression.
Effects of JUN overexpression on myeloma cell survival. (A) Effects on cell proliferation (left) and cell viability (right) in OCI-My5 (top), OPM2 (middle), and U266 (bottom) cells for JUN overexpression. Total number of cells and cell viability were evaluated daily by trypan blue exclusion. Error bars represent standard error of the mean for 2 independent experiments. (B) JUN overexpression induced apoptosis of OCI-My5, OPM2, and U266 cells as evaluated by flow cytometry performed 72 hours after lentiviral transfection with EV or JUN cDNA. Overexpression of JUN induced a dramatic increase in the percentages of cells with sub–G0-phase DNA content (indicative of apoptosis). CON indicates control; EV, empty vector; and JUNH, JUN overexpression.
EGR-1 is the direct downstream target gene of JUN
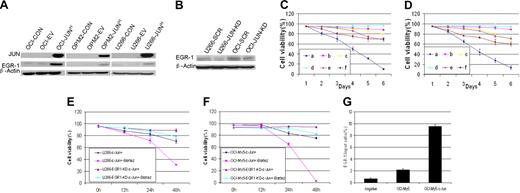
We next investigated the relationship in myeloma cells between JUN and EGR-1. Western blot analysis revealed that JUN overexpression resulted in up-regulation of EGR-1 protein (Figure 5A), whereas JUN knockdown resulted in down-regulation of EGR-1 protein expression (Figure 5B). This result supports our finding that overexpression of JUN in myeloma cells with EGR-1 knockdown resulted in cell viability that was better than that in cells without EGR-1 knockdown (Figure 5C-E). We also detected the cell viability of U266 and OCI-My5 EGR-1 knockdown JUN-overexpressing cells during 10nM bortezomib exposure and found that EGR-1 knockdown in JUN-overexpressing cells prevents cell apoptosis by bortezomib (P < .05; Figure 5E-F).Together, these results further support the possibility that JUN regulates EGR-1 expression.
EGR-1 is the direct downstream target gene of JUN. (A) Western blot analysis compared OCI-My5 (OCI), OPM2, and U266 myeloma cells when untransfected (CON), carrying empty vector (EV), or overexpressing JUN (JUNH). Results showed specificity of JUN overexpression and effects on regulation of EGR-1. β-Actin was used as a loading control. (B) Effect of nonspecific scrambled shRNA (SCR) and JUN shRNA (JUN-KD) in U266 and OCI-My5 (OCI) cells on regulation of EGR-1 expression was evaluated by Western blot. Effects on cell viability in OCI-My5 (C) and OPM2 (D) cells for JUN overexpression (a), silencing of EGR-1 expression (b), JUN overexpression in the case of EGR-1 knockdown (c), overexpression of Survivin (d), JUN overexpression in the case of Survivin overexpression (e), and exposure to the pan-caspase inhibitor Z-VAD-fmk (f). Cell viability was evaluated daily by trypan blue exclusion. Error bars represent SEM for 2 independent experiments. U266 (E) and OCI-My5 (F) EGR-1–knockdown JUN overexpression cells compared with U266 and OCI-My5 JUN overexpression cell lines during exposure to 10nM bortezomib (Bort). Cell viability was determined every 12 hours by trypan blue exclusion. Error bars represent standard error of the mean for 2 independent experiments. (G) JUN binding to the proximal EGR-1 promoter in OCI-My5 and JUN-overexpressing OCI-My5 cells (OCI-My5-JUNH) was analyzed by ChIP by the use of antibodies against JUN or no antibody (negative control); real-time PCR followed.
EGR-1 is the direct downstream target gene of JUN. (A) Western blot analysis compared OCI-My5 (OCI), OPM2, and U266 myeloma cells when untransfected (CON), carrying empty vector (EV), or overexpressing JUN (JUNH). Results showed specificity of JUN overexpression and effects on regulation of EGR-1. β-Actin was used as a loading control. (B) Effect of nonspecific scrambled shRNA (SCR) and JUN shRNA (JUN-KD) in U266 and OCI-My5 (OCI) cells on regulation of EGR-1 expression was evaluated by Western blot. Effects on cell viability in OCI-My5 (C) and OPM2 (D) cells for JUN overexpression (a), silencing of EGR-1 expression (b), JUN overexpression in the case of EGR-1 knockdown (c), overexpression of Survivin (d), JUN overexpression in the case of Survivin overexpression (e), and exposure to the pan-caspase inhibitor Z-VAD-fmk (f). Cell viability was evaluated daily by trypan blue exclusion. Error bars represent SEM for 2 independent experiments. U266 (E) and OCI-My5 (F) EGR-1–knockdown JUN overexpression cells compared with U266 and OCI-My5 JUN overexpression cell lines during exposure to 10nM bortezomib (Bort). Cell viability was determined every 12 hours by trypan blue exclusion. Error bars represent standard error of the mean for 2 independent experiments. (G) JUN binding to the proximal EGR-1 promoter in OCI-My5 and JUN-overexpressing OCI-My5 cells (OCI-My5-JUNH) was analyzed by ChIP by the use of antibodies against JUN or no antibody (negative control); real-time PCR followed.
To elucidate the mechanism by which JUN protein regulates EGR-1 activation, we performed ChIP experiments to determine whether JUN protein is associated with the proximal promoter of EGR-1 by using OCI-My5 as a representative myeloma cell line. Our analysis (in which we used ECR Browser software; ECR Software Corp) of a 2-kb fragment of the EGR-1 promoter identified the AP-1 binding site “ACTGACTGCGT” at 1572 bp site upstream of EGR-1 start site. Primers to this region were used in quantitative real-time reverse-transcription PCR to amplify and quantify the DNA fragment purified as a result of cross-linking and immunoprecipitation with JUN antibodies. In OCI-My5 cells, the amount of amplified DNA containing the putative AP-1 binding site was more than 3-fold greater than in negative control cells (Figure 5G). Furthermore, in JUN-overexpressing OCI-My5 cells, the target DNA was increased more than 16-fold (Figure 5G). Taken together, these data demonstrate that JUN up-regulates EGR-1 expression by binding its promoter and suggest that EGR-1 is the direct downstream target gene of the JUN transcription factor.
Overexpressed JUN interacts with EGR-1 to inhibit Survivin expression and induce apoptosis in myeloma cell lines
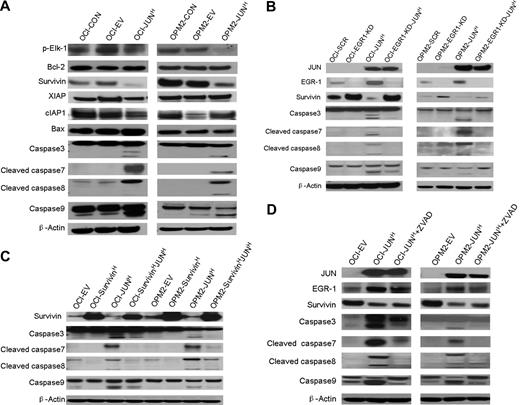
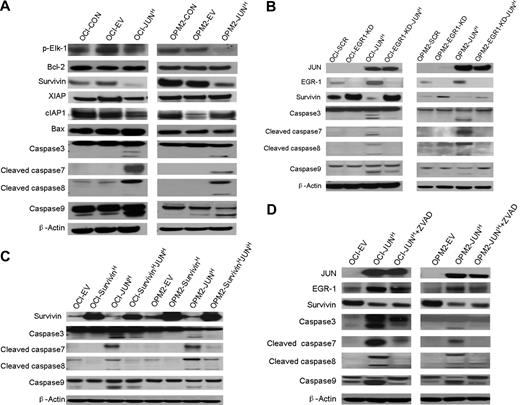
To further explore the precise mechanism by which the interaction of JUN with EGR-1 induces myeloma cell apoptosis, we used Western blots to detect phosphorylated Elk-1, antiapoptosis proteins Bcl-2 and IAP family proteins (Survivin, XIAP, and cIAP1), proapoptotic protein Bax, and proteins in the caspase signaling pathway. In OCI-My5 and OPM2 myeloma cell lines overexpressing JUN, Survivin was down-regulated and caspase-3, -7, -8, and -9 were activated; no significant alterations of Bcl-2, Bax, XIAP, cIAP1, and phosphorylated Elk-1 were observed (Figure 6A).These results support the idea that JUN down-regulates Survivin and triggers the caspase signaling pathway to induce cell apoptosis.
Overexpression of JUN induces apoptosis via an EGR-1–Survivin–caspase signaling pathway. (A) Western blot analysis compared OCI-My5 (OCI) and OPM-2 myeloma cells when untransfected (CON), carrying empty vector (EV), or overexpressing JUN (JUN). Results showed specificity of JUN overexpression and effects on regulation of Survivin, XIAP, cIAP1, phosphorylated Elk-1(p-Elk-1), Bcl-2, and Bax, and cleavage of caspase-3, -7, -8, and -9. β-Actin was used as a loading control. (B) Western blot analysis of OCI-My5 (OCI) and OPM2 myeloma cell lines with scrambled shRNA (SCR; control), EGR-1 shRNA (EGR1-KD), and/or JUN overexpression (JUNH). Silencing of EGR-1 activated Survivin expression and reduced Survivin inhibition and caspase cleavage by JUN overexpression. (C) Western blot analysis of OCI-My5 (OCI) and OPM2 myeloma cell lines with PWPI (EV; control), Survivin (SurvivinH), and/or JUN overexpression (JUNH). Overexpression of Survivin inhibited caspase cleavage triggered by JUN overexpression. (D) Western blot analysis of OCI-My5 (OCI) and OPM2 myeloma cell lines carrying empty vector or overexpressing JUN cultured in the absence or presence of pan-caspase inhibitor Z-VAD-fmk (ZVAD) shows effects on EGR-1 and Survivin levels and on cleavage of caspase-3, -7, -8, and -9. β-Actin was used as a loading control in all Western blots.
Overexpression of JUN induces apoptosis via an EGR-1–Survivin–caspase signaling pathway. (A) Western blot analysis compared OCI-My5 (OCI) and OPM-2 myeloma cells when untransfected (CON), carrying empty vector (EV), or overexpressing JUN (JUN). Results showed specificity of JUN overexpression and effects on regulation of Survivin, XIAP, cIAP1, phosphorylated Elk-1(p-Elk-1), Bcl-2, and Bax, and cleavage of caspase-3, -7, -8, and -9. β-Actin was used as a loading control. (B) Western blot analysis of OCI-My5 (OCI) and OPM2 myeloma cell lines with scrambled shRNA (SCR; control), EGR-1 shRNA (EGR1-KD), and/or JUN overexpression (JUNH). Silencing of EGR-1 activated Survivin expression and reduced Survivin inhibition and caspase cleavage by JUN overexpression. (C) Western blot analysis of OCI-My5 (OCI) and OPM2 myeloma cell lines with PWPI (EV; control), Survivin (SurvivinH), and/or JUN overexpression (JUNH). Overexpression of Survivin inhibited caspase cleavage triggered by JUN overexpression. (D) Western blot analysis of OCI-My5 (OCI) and OPM2 myeloma cell lines carrying empty vector or overexpressing JUN cultured in the absence or presence of pan-caspase inhibitor Z-VAD-fmk (ZVAD) shows effects on EGR-1 and Survivin levels and on cleavage of caspase-3, -7, -8, and -9. β-Actin was used as a loading control in all Western blots.
In myeloma cell lines with EGR-1 knockdown (shown as EGR-1-KD in Figure 6B), we observed up-regulation of Survivin. Moreover, overexpression of JUN markedly reduced inhibition of Survivin and activation of caspase-3, -7, -8, and -9 in EGR-1–knockdown myeloma cell lines, but not in myeloma cell lines with intact expression of EGR-1 (Figure 6B). To further identify the correlation between Survivin and caspase activation, we also overexpressed Survivin in OCI-My5 and OPM2 cell lines. Overexpression of Survivin markedly inhibited caspase activation (Figure 6C) and cell apoptosis (Figure 5B-C), which were triggered by JUN overexpression. Cleavage of caspase-3, -7, -8, and -9 was significantly reduced after incubating JUN-overexpressing myeloma cells with pan-caspase inhibitor Z-VAD-fmk, but regulation of EGR-1 and Survivin was almost unaffected (Figure 6C), which is consistent with our finding that JUN-overexpressing myeloma cells were more viable when treated with Z-VAD-fmk (Figure 5B-C). Taken together, these data revealed that JUN′s interaction with EGR-1 negatively regulated Survivin and subsequently strongly triggered cleavage of caspase-3, -7, -8, and -9.
Discussion
In the current study, we investigated the functional role of JUN in myeloma cell growth and survival. JUN is an important AP-1 transcription factor that promotes cell growth and mediates apoptosis in response to various stimuli. However, overexpression of JUN in myeloma cell lines led to profound growth inhibition and an apoptotic phenotype, a finding consistent with that of Podar et al,13 who showed that JUN inhibits myeloma cell growth and apoptosis via caspase-triggered c-Abl cleavage. Our study, however, revealed a novel mechanism whereby JUN modulates myeloma-cell apoptosis through interacting with EGR-1, which leads to down-regulation of Survivin and initiation of caspase signaling. In addition, our results suggest that low expression of JUN and EGR-1 in myeloma cells is a marker for poor survival and resistance to bortezomib treatment.
Low expression levels of JUN and EGR-1 appear to have important clinical implications for patients with MM and may be linked to high risk with shorter survival and disease progression. The present findings demonstrate that myeloma cells cocultured with osteoclasts and MSCs have markedly decreased expression of AP-1 transcription factor JUN as well as EGR-1, consistent with our recent data demonstrating similar effects with simultaneous decreases in the levels of AP-1 transcription factors.15 These data suggest that interactions between myeloma cells and bone marrow microenvironment result in low expression levels of JUN and EGR-1 and potentially contribute to myeloma cell survival and proliferation. Furthermore, low expression levels of JUN and EGR-1 were significantly lower in myeloma plasma cells from late-stage and high-risk disease than those in healthy plasma cells or cells from MGUS or low-risk MM, and patients with low JUN or EGR-1 expression levels had significantly shorter times of EFS and OS than those with high JUN or EGR-1 expression levels. Finally, our findings indicated that OS was longer for patients whose myeloma cells had high expression levels of both JUN and EGR-1 than for those with low expression levels of either individual gene (P < .001).
Our studies indicate that low expression of JUN and EGR-1 contributes to bortezomib resistance clinically and in vitro. Bortezomib, the first proteasome inhibitor approved for treating MM, has demonstrated significant antitumor activity as a single agent and as part of combination therapy, so understanding mechanisms of resistance to this drug is an important goal for advancing treatment of MM.35 The present study implied that expression levels of JUN or EGR-1 were more significantly associated with OS and EFS in patients treated with bortezomib (ie, those enrolled in TT3) than in patients who did not receive bortezomib (ie, those enrolled in TT2). This result is consistent with our in vitro findings that JUN- or EGR-1–knockdown cell lines were resistant to bortezomib treatment. Taken together, these data suggest that JUN and EGR-1 are critical components in the pathway mediating the response to bortezomib and that expression levels of these genes can be markers to identify bortezomib-responsive patients. Targeting the JUN and/or EGR-1 pathways potentially offers novel strategies for helping patients who are unresponsive to bortezomib, such as combining bortezomib with an agent that can up-regulate JUN and/or EGR-1 expression.
Our in vitro data suggest that EGR-1 is a target of JUN–EGR-1 protein and was up-regulated in JUN-overexpressing myeloma cells, and knockdown of EGR-1 in myeloma cells abrogated the effects of JUN overexpression on myeloma cell death and growth inhibition; further study confirmed that knockdown of EGR-1 in JUN-overexpressing myeloma cells prevents cell apoptosis by bortezomib. Furthermore, we demonstrated that JUN directly binds to the EGR-1 promoter, showing that JUN directly regulates EGR-1 transcription in myeloma cells. Hoffmann et al28 reported that JUN regulates mice EGR-1 via binding directly to the AP-1 element and via indirect recruitment of phosphorylated Elk-1 to the serum response elements and suggested that Elk-1, JUN, and EGR-1 form a transcription factor network. However, we found phosphorylated Elk-1 was not up-regulated in myeloma cell lines overexpressing JUN, which indicates JUN has no role of recruitment of Elk-1 to serum response elements at human EGR-1 promoter in myeloma cells. JUN regulates EGR-1 only via directly binding to the AP-1 site at EGR-1 promoter.
Because our findings indicate that expression of JUN and EGR-1 is important in myelomagenesis and MM treatment, we investigated their mechanisms of inducing myeloma cell apoptosis. The EGR-1 binding site (GCGGGGGCG) is present in the promoters of a variety of genes, and EGR-1 has been shown to stimulate synthesis of growth and differentiation factors via direct promoter activation.25,40 EGR-1 regulates radiation-induced cell apoptosis in solid tumors via p53, PTEN, Bcl-2, and Bax.25,40 In our study, EGR-1 was up-regulated when JUN was overexpressed in OCI-My5 myeloma cells (no p53 expression41 ) and OPM2 myeloma cells (no PTEN expression42 and mutated TP5343 ), which indicated that myeloma cell apoptosis induced by EGR-1 did not occur via p53 or PTEN. Inhibition of Bcl-2 and activation of Bax was not indicated in Western blot analyses of myeloma cell lines overexpressing JUN, which means that Bcl-2 and Bax are not involved in EGR-1's inducing of myeloma cell apoptosis. Myeloma cell apoptosis induced by EGR-1 is mediated by a different pathway.
The expression of Survivin, a member of IAP family proteins, was negatively regulated by EGR-1,44 which was supported by our data from silencing EGR-1 up-regulated Survivin expression that was inhibited by overexpressing JUN in myeloma cell lines. Survivin is highly expressed in various human cancers45-48 and plays an important role in suppressing apoptosis by interacting with caspase-3, -7, and -9 to inhibit their proapoptotic effects.49 Accordingly, we observed that overexpression of Survivin inhibited cell apoptosis and activation of caspase-3, -7, -8, and -9 triggered by overexpressing JUN in myeloma cell lines. No alteration of XIAP and cIAP1 protein in JUN overexpression myeloma cell lines further suggested that Survivin may be the specific IAP family protein involved in JUN overexpression inducing cell apoptosis. Pan-caspase inhibitor Z-VAD-fmk inhibited activation of these caspases but did not alter expression of EGR-1 or Survivin. Together, these data reveal that EGR-1 induces apoptosis via inhibition of Survivin and subsequent activation of caspase signaling.
In conclusion, these studies delineated the mechanistic roles of JUN and EGR-1 in MM microenvironment-mediated myeloma cell growth, survival, and resistance to bortezomib treatment, as well as disease progression. Low expression levels of the genes encoding JUN and/or EGR-1 are adverse prognostic markers for the efficacy of bortezomib therapy. In addition, we identified a novel mechanism by which overexpressed JUN induces myeloma cell apoptosis via interaction with EGR-1 to inhibit Survivin and subsequently activate caspase signaling to induce cell apoptosis. For patients with aggressive, bortezomib-resistant MM marked by low expression of JUN and EGR-1, the combination of bortezomib with an agent that can activate JUN/EGR-1 signaling pathways may represent a novel approach to successful therapy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Office of Grants and Scientific Publications at the University of Arkansas for Medical Sciences for editorial assistance during the preparation of the manuscript.
This work was supported by National Institutes of Health PO1 grant CA55819 (to B.B., J.D.S., and J.E.), National Institutes of Health RO1 CA113992 (to J.E.), the Fund to Cure Myeloma, the Nancy and Stephen Grand Philanthropic Fund, and the Multiple Myeloma Research Foundation (to I.E. and S.Y.). We thank Lei Shi, Yan Xiao, and Hongqing Zhu for assistance.
National Institutes of Health
Authorship
Contribution: F.Z., J.D.S., and S.W. designed the research; S.W., F.Z., J.D.S., and L.C. organized, analyzed, and interpreted the data; L.C., F.Z., X.W., W.X., I.E., J.E., and S.Y. performed the experiments; J.D.S., S.W., B.B., L.C., and Y.Z. drafted the manuscript; F.Z. and Y. Z. analyzed the clinical data; B.B. contributed to clinical samples; and B.B., J.E., and S.Y. provided critical suggestions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fenghuang Zhan, Division of Hematology/BMT/Myeloma Program, School of Medicine Huntsman Cancer Institute, University of Utah, 30 North 1900 East, Salt Lake City, UT 84132; e-mail: fenghuang.zhan@hsc.utah.edu; or John D. Shaughnessy Jr, PhD, Myeloma Institute for Research and Therapy, University of Arkansas for Medical Sciences, 4301 W Markham St, #776, Little Rock, AR 72205; e-mail: shaughnessyjohn@uams.edu.
References
Author notes
*L.C. and S.W. contributed equally to this work.