Abstract
Large granular lymphocyte (LGL) leukemia results from chronic expansion of cytotoxic T cells or natural killer (NK) cells. Apoptotic resistance resulting from constitutive activation of survival signaling pathways is a fundamental pathogenic mechanism. Recent network modeling analyses identified platelet-derived growth factor (PDGF) as a key master switch in controlling these survival pathways in T-cell LGL leukemia. Here we show that an autocrine PDGF regulatory loop mediates survival of leukemic LGLs of both T- and NK-cell origin. We found high levels of circulating PDGF-BB in platelet-poor plasma samples from LGL leukemia patients. Production of PDGF-BB by leukemic LGLs was demonstrated by immunocytochemical staining. Leukemic cells expressed much higher levels of PDGFR-β transcripts than purified normal CD8+ T cells or NK cells. We observed that phosphatidylinositol-3-kinase (PI3 kinase), Src family kinase (SFK), and downstream protein kinase B (PKB)/AKT pathways were constitutively activated in both T- and NK-LGL leukemia. Pharmacologic blockade of these pathways led to apoptosis of leukemic LGLs. Neutralizing antibody to PDGF-BB inhibited PKB/AKT phosphorylation induced by LGL leukemia sera. These results suggest that targeting of PDGF-BB, a pivotal regulator for the long-term survival of leukemic LGLs, may be an important therapeutic strategy.
Introduction
Large granular lymphocyte (LGL) leukemia is a lymphoproliferative disease of either CD3+ cytotoxic T lymphocytes (CTLs) or CD3− natural killer cells (NK cells). The majority of LGL patients with T-cell (CD3+, CD8+/CD57+) or NK-cell (CD3−, CD16+/CD56+) leukemia have a clinically indolent course.1,2 Leukemic LGLs of T-cell phenotype reflect polarized expansion of CD8+ terminal-effector memory cells.3 Expanded NK cells have an activated phenotype with dysregulated NK receptor expression.4,5 Fas resistance is an important biologic feature in leukemic LGLs of both T-cell and NK-cell type.3,6 Constitutive activation of survival signaling pathways is a central pathogenetic mechanism in LGL leukemia. Previously, phosphatidylinositol-3 (PI3) kinase activation and signal transducer and activator of transcription 3 up-regulation of Mcl-1 were shown to be important for survival of leukemic T-LGLs.7-9 More recently, molecular profiling of T-LGL leukemia revealed a survival role for constitutive sphingolipid signaling.10 Survival mechanisms in the NK type of LGL leukemia have been less extensively studied; however, a constitutively active retrovirus-associated DNA sequence (RAS)/mitogen-activated protein kinase (MEK)/extracellular signal-related kinase (ERK) survival pathway was identified.6 Given the complexity and interactive nature of signaling pathways, it is difficult to determine the importance of individual pathway components when studied in isolation. Using a network modeling approach, we found that the presence of interleukin-15 (IL-15) and platelet-derived growth factor (PDGF) is sufficient to reproduce all known deregulations in T-LGL leukemia.11 Work in this study focused on further examining the pivotal role of PDGF. We found that PDGF mediates survival of leukemic LGLs of both T- and NK-cell origin through an autocrine regulatory pathway.
Methods
Reagents
All chemicals were purchased from Sigma-Aldrich unless otherwise specified. Recombinant human (rh) PDGF-BB was purchased from ProSpec-TANY TechnoGene LTD; rhIL-2, from Promega Corporation; and human T-lymphotropic virus-I (HTLV-I)– and HTLV-II–infected plasma, from Zeptometrix. Antibodies and inhibitors were obtained from the following sources and used at the dilutions recommended by the manufacturers: anti–PDGFR-α (951) and anti–PDGFR-β (958) polyclonal antibodies, anti–phospho-Tyr monoclonal antibody (PY99), goat anti–mouse immunoglobulin G (IgG) antibody (Santa Cruz Biotechnology Inc); anti–PDGF-BB neutralizing antibody and anti-PDGF-BB antibody for immunocytochemistry/immunofluorescence (ICC/IF; Abcam Inc); anti–phospho-AKT and total AKT polyclonal antibodies, anti-MEK1/2, anti–phospho-MEK1/2, anti-ERK1/2, and phospho-ERK1/2 (Cell Signaling Technology); anti–phospho-Src (Tyr419) and anti-Src antibodies (Upstate Cell Signaling Solutions); β-actin monoclonal antibody (Sigma-Aldrich); mouse anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (Chemicon International); PI3K inhibitor LY294002 (Cell Signaling Technology); Src family kinase (SFK) inhibitor PP2 and PDGF receptor tyrosine kinase inhibitor AG1296 (Calbiochem-Novabiochem Corp).
Patient characteristics and preparation of PBMCs
All patients met the clinical criteria of T- or NK-LGL leukemia with increased numbers of CD3+, CD8+/CD57+ T lymphocytes or CD3−, CD16+/CD56+ NK cells in the peripheral blood. Patients were clinically stable and not on treatment at the time of sample acquisition (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Peripheral blood specimens from LGL leukemia patients were obtained and informed consents signed for sample collection according to the Declaration of Helsinki using a protocol approved by the Institutional Review Board of Penn State Hershey Cancer Institute. Not all of the investigational studies were performed in each patient, as outlined below. Buffy coats from age- and sex-matched normal donors were also obtained from the blood bank of the Milton S. Hershey Medical Center, Pennsylvania State University, College of Medicine. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque gradient separation, as described previously.3 Cell viability was determined by trypan blue exclusion assay with more than 95% viability in all the samples. CD3−/CD16+/CD56+ NK cells from normal donors and LGL leukemia patients were isolated by a negative selection process (Rosette Sep; StemCell Technologies), as described previously.12 Purified CD8+ cells from blood samples of normal controls or patients with T-LGL leukemia were isolated using Dynal CD8 antibody–positive isolation Kit (Invitrogen Life Technologies) following the manufacturer's protocols.13 The purity of freshly isolated CD8+ T cells and CD16+/CD56+ NK cells in each of the samples was determined by flow cytometry assay. The purity for NK cells was between 85% and 90%, and for T cells was more than 95%.
Cell culture
Culture of freshly isolated PBMCs and purified NK cells and CD8+ T lymphocytes was carried out using RPMI-1640 medium supplemented with 10% fetal bovine serum (both from Invitrogen). NKL cells, a leukemic LGL NK-cell line,14 were cultured in alpha-minimum essential medium (Invitrogen) supplemented with 20% fetal bovine serum, 1% sodium pyruvate, 1% nonessential amino acid, and rhIL-2 (100 IU/mL). All tissue culture media were further supplemented with glutamine (200 μM/L), penicillin (100 IU/mL), and streptomycin (100 μg/mL); cells were grown in 5% CO2 at 37°C.
PDGF-BB determination: ELISA
PDGF-BB protein levels in platelet-poor plasma samples from LGL leukemia patients and normal controls as well as HTLV-I– and HTLV-II–infected persons were determined using Human PDGF-BB Quantikine enzyme-linked immunosorbent assay (ELISA) Kit (R&D Systems Inc) following the manufacturer's protocol. Serial diluted rhPDGF-BB protein with concentrations ranging from 31.2 to 2000 pg/mL was used to generate a standard curve to calculate the PDGF-BB concentration in each sample. Samples were assayed in triplicate.
IP for PDGF receptor– and PDGF-BB–induced PDGF-RTK expression
Detection of PDGF receptor-α and receptor-β and induction of PDGF receptor tyrosine kinase (PDGF-RTK) phosphorylation were carried out in freshly isolated PBMCs from patients with either T- or NK-LGL leukemia or age/sex-matched normal controls, as described elsewhere.15 Briefly, 2 × 107 freshly isolated PBMCs were cultured with serum-free medium for 16 hours followed by recombinant human platelet growth factor-BB (rhPDGF-BB, 50 ng/mL) or vehicle (normal saline) treatment for 1 hour on ice and 10 minutes at 37°C. Cells were washed and lysed with cell lysis buffer (1% Nonidet P-40, 20mM/L, tris(hydroxymethyl)aminomethane-HCl, pH 7.5, 150mM/L NaCl, 10% glycerol, 1mM/L Na3VO4, 1% aprotinin [Bayer], and 1mM/L phenylmethylsulfonyl fluoride). Total protein from each clarified whole-cell lysate (250 μg) was incubated with anti–PDGFR-α or –PDGFR-β antibodies for 2 hours at 4°C followed by an overnight incubation with 100 μL of protein A/G Sepharose beads (Amersham Biosciences) at a 1:1 ratio with gentle rotation at 4°C. Immunoprecipitation (IP) samples were then washed with ice-cold lysis buffer 3 times to remove the unbound antibodies; proteins were separated by gel electrophoresis. Western immunoblotting assays were performed for PDGFR-α and -β and the phosphorylated PDGF RTK using 7% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis.
PDGF and PDGF receptor gene expression: real-time quantitative RT-PCR
Real-time reverse-transcription–polymerase chain reaction (RT-PCR) was performed using primer sets specific for PDGF-A and -B, PDGFR-α and -β, and an internal standard, 18S rRNA, in an ABI PRISM 7900 sequence detector (Applied Biosystems) as described elsewhere.16 Freshly purified CD8+ T cells or CD16+/CD56+ NK cells (5 × 106) from patients with either T-LGL (n = 5) or NK-LGL (n = 7) leukemia or from normal controls were subjected to total RNA extraction using TRIzoL LS Reagent (Invitrogen) following the manufacturer's directions. First-strand cDNA was synthesized from 2 μg of purified total RNA using random hexamers and Moloney murine leukemia virus reverse-transcription reagents (Invitrogen) in a total volume of 20 μL. A total of 1 μg of cDNA was applied in a 10-μL PCR mix using a QuantiTect SYBR Green PCR kit (QIAGEN). Amplification of triplicate cDNA template samples was then performed with denaturation for 15 minutes at 95°C, followed by 40 PCR cycles of denaturation at 94°C for 15 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds. A standard curve of cycle thresholds using serial dilutions of cDNA samples was established and used to calculate the relative abundance of the target gene between samples from patients and normal controls. Values were normalized to the relative amounts of 18S mRNA, which were obtained from a similar standard curve. The changes in fluorescence of SYBR green dye in every cycle were monitored by the ABI 7900 system software and the threshold cycle for each reaction was calculated. The relative amount of PCR products generated from each primer set was determined based on the threshold cycle or threshold cycle value.17 PCR analysis was performed on each cDNA sample at least twice. The following primers were used for the detection of PDGF isoform A and B, PDGFR-α and -β: PDGF-A sense, 5′-CCACACCTCCTCGCTGTAG-3′, antisense 5′-CAGCAGCCTGTGTGTTATC-3′; PDGF-B sense, 5′-GTGGCTTCTTTTCGTT-3′, antisense 5′-GAAAATGCAGGGTAGGA-3′; PDGFR-α sense, 5′-CTGGGTTTCCATCCTTGAG-3′, antisense 5′-TAGTAGGCTTCCTGCGTGG-3′; PDGFR-β sense, 5′-CATGGGGGTATGGTTTTGT-3′, antisense 5′-GTAAGGTGCCAACCTGCAA-3′; 18S rRNA-encoding DNA (rRNA) sense 5′-GTAACCCGTTGAACCCCATT-3′, 18S rRNA antisense 5′-CCATCCAATCGGTAGTAGCG-3′.
Immunofluorescence microscopy for PDGF-BB expression
PDGF-BB expression in purified CD8+ T cells and CD16+/CD56+ NK cells from patients with LGL leukemia (T-LGL, n = 3; NK-LGL, n = 3) and normal controls (n = 3) was detected by immunocytochemistry/immunofluorescence (ICC/IF) staining with polyclonal anti–PDGF-BB antibody and cyanin 5 (Cy5)–conjugated secondary antibody (Abcam). Briefly, 2 × 106 purified T or NK cells were seeded into poly-l-lysine–coated 3.5 cm in diameter, glass-bottom dishes (MatTek Co) and incubated for 30 minutes at room temperature. Adhered cells were then fixed with 3.7% paraformaldehyde phosphate-buffered saline at 37°C, and permeabilized in ice-cold methanol-acetone (1:1) for 10 minutes at −20°C. Cells were incubated with polyclonal anti–PDGF-BB antibody (1:200 dilution) at 4°C, and then with goat anti–rabbit IgG-Cy5 secondary antibody at 1:250 dilution in blocking solution. CD8+ T cells were further incubated with anti-CD8–fluorescein isothiocyanate (FITC) antibody (BD Biosciences) at 1:200 dilution for an additional 2 hours at room temperature. Nuclei counterstaining for all the samples was carried out by incubating with 4′-6-diamidino-2-phenylindole (DAPI; Invitrogen) at 100 μg/mL concentration for 10 minutes at room temperature. Slides were mounted with Aqua Poly/Mount (Polysciences Inc). Cells were visualized using Leica TCS SP2 AOBS confocal microscopy (Leica Microsystems) according to the manufacturer's directions. CD8 surface marker on T cells was visualized by FITC green fluorescent staining on the cell membrane. PDGF-BB was visualized by Cy5 red fluorescent staining in the cytoplasm compartment. Nonspecific staining was not detected with secondary antibody alone.
Kinase assays
PBMCs from patients with T-LGL leukemia (n = 6) and normal controls (n = 6) were lysed in IP buffer (50mM/L N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 10mM/L NaCl, 2mM/L EDTA, 0.5% Nonidet P-40, 10% glycerol, 0.1mM/L phenylmethylsulphonyl fluoride, 0.1mM/L Na3VO4, 0.5 mL of NaF, 5 μg/mL leupeptin, 0.1mM/L dithiothreitol). Total protein from each sample (50 μg) was immunoprecipitated with 2 μg of Src antibody (Upstate Biotechnology) for 2 hours. Protein-G agarose beads (50 μL) were added and incubation continued at 4°C overnight. Beads were washed 3× in IP buffer and 1× in kinase buffer (100mM/L N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 20mM/L MgCl2, 10mM/L MnCl2, 0.1M/L dithiothreitol). The reaction was started by adding 10 μL of reaction cocktail containing kinase buffer, 1 μL of acid-denatured enolase (Sigma-Aldrich), 1 μL of 100μM/L cold adenosine triphosphate, and 1.2 μL of [32P] adenosine triphosphate, and incubated at 27°C for 30 minutes. The samples were electrophoresed by SDS-PAGE on a mini gel (Bio-Rad). The gel was dried and exposed to x-ray film.
MTT assays
A set of experiments was conducted to test whether rhPDGF-BB or serum containing PDGF-BB mediated cell growth/proliferation of NKL cells. Briefly, cells were treated with rhPDGF-BB (2.5 ng/mL) in the presence or absence of PP2, LY294002, or the combination of both compounds at indicated concentrations or with 10 μL of pooled sera from patients with either T-LGL (n = 3) or NK-LGL (n = 3) leukemia, or from normal controls (n = 3). Serum samples were pretreated with either polyclonal anti–PDGF-BB neutralizing antibody (Abcam) or anti–rabbit IgG isotype control antibody (Santa Cruz Biotechnology Inc) for 2 hours at 10-ng/mL concentration. The efficacy of PDGF-BB neutralizing antibody was pretested by examining rhPDGF-BB–mediated cell proliferation in NKL cells. Cell viability/proliferation was determined by reading the plates at 490-nm wavelength in Synergy HT Multi-Detection Microplate Reader (Bio-TEK). All samples were assayed in triplicate and each experiment was repeated at least twice.
Apoptosis assay
Apoptosis was determined in PBMC samples from patients with LGL leukemia (T-LGL, n = 5; NK-LGL, n = 4) and normal controls (n = 4) by 2-color flow cytometry with annexin-V and 7-amino-actinomycin D (BD Pharmingen Transduction Laboratories) staining using 5 × 105 cells per sample. Cell viability was also confirmed by trypan blue exclusion. The percentage of specific apoptosis was calculated using the following formula: Apoptosis (%) = (% annexin-V–FITC positive in assay well − % annexin-V–FITC positive in the dimethyl sulfoxide [DMSO] control well) × 100/(100 − % annexin-V–FITC positive in the DMSO control well).
Western blot analysis
Western blot analyses of phosphorylated protein kinase B (PKB)/AKT, MEK1/2, ERK1/2, and SFK expression, PDGFR-α and -β, PDGF-BB protein expression, and PDGF-RTK autophosphorylation were performed on whole-cell lysates or samples immunoprecipitated with specific antibodies from patients with LGL leukemia or normal controls. Blots were washed and developed with enhanced chemiluminescence (ECL-plus; Amersham) following the manufacturer's instructions. Semiquantitative densitometry analysis was performed using ChemDox XR gel documentary system (Bio-Rad) to determine the relative concentration of proteins on the Western blot membranes.
Statistical analysis
All data are expressed as mean plus or minus SEM. Paired Student t test (2-tail paired) and 2-way analysis of variance test were used to determine the statistical significance, and P value of .05 or less was considered statistically significant.
Results
PDGF-BB expression in LGL leukemia
PDGF gene expression by real-time RT-PCR.
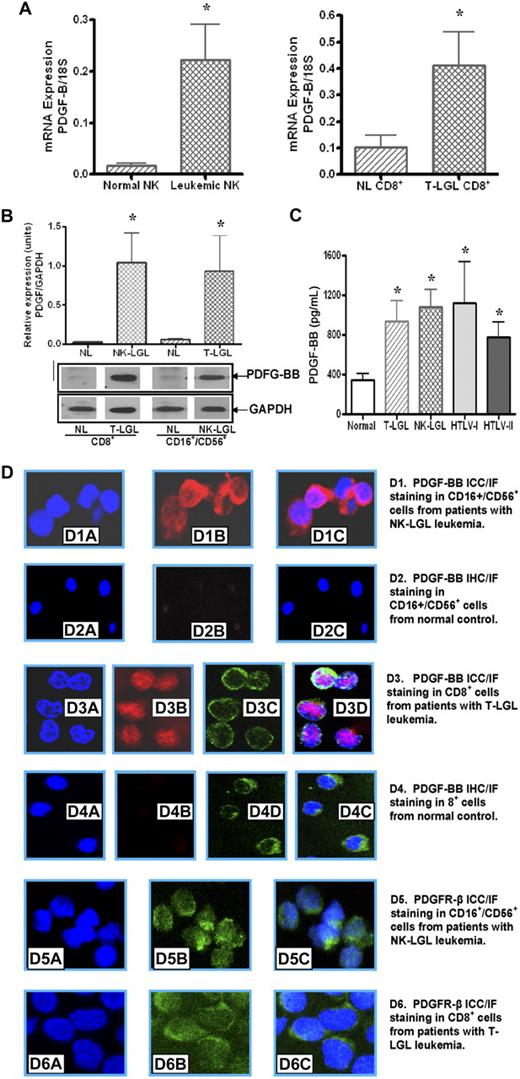
PDGF-B mRNA expression levels from purified NK cells of NK-LGL leukemia patients (n = 7) were significantly higher than levels observed in purified normal NK cells (n = 7) as shown in Figure 1A, (0.24 ± 0.96 vs 0.018 ± 0.006 mRNA copies/18S, NK-LGL vs normal NK, P < .05). Similarly, elevated PDGF-B gene expression in CD8+ cells from T-LGL leukemia patients (n = 5) was observed compared with the level in CD8+ T cells from normal controls (n = 5) as shown in Figure 1A (0.41 ± 0.11 vs 0.104 ± 0.04 mRNA copies/18S, LGL vs normal, P < .02). No statistical difference in PDGF-A isoform gene expression was observed in CD8+ T cells or CD16+/CD56+ NK cells from LGL leukemia patients compared with purified cells from normal controls (data not shown).
Leukemic LGLs express PDGF-BB and PDGFR-β. (A) PDGF-BB mRNA expression levels in NK cells from patients with NK-LGL leukemia and normal controls, and in T cells from patients with T-LGL leukemia and normal controls, were determined by real-time quantitative RT-PCR (expressed as the ratio of PDGF-BB mRNA copies/18S control mRNA copies; *P < .05). (B) Western blot analysis was performed for PDGF-BB production in cells lysates of CD16+/CD56+ cells from patients with NK-LGL leukemia or CD8+ cells from patients with T-LGL leukemia as well as in their counterpart cells from normal controls. GAPDH detection was used to confirm equal loading of total protein in each sample. Densitometry was performed on these Western blot results to determine the average level of PDGF-BB protein expression. *P < .05. (C) PDGF-BB protein expression in plasma samples from 10 normal controls, 9 NK-LGL leukemia patients, 10 T-LGL leukemia patients, 4 HTLV-I–infected persons, and 4 HTLV-II–infected persons was determined by ELISA; *P < .05 compared with plasma sample levels from normal controls. (D) PDGF-BB protein ICC/IF staining as visualized by confocal microscopy in LGL leukemia cells compared with normal T or NK cells. Nuclei staining was visualized by DAPI (blue); PDGF-BB, by Cy5 fluorescent staining in the cytoplasm compartment (red). CD8 surface marker on T cells was visualized by FITC-fluorescent staining on the cell membrane (green). (D1-2) NK cells from a patient with NK-LGL leukemia and a normal control. Nuclei staining (D1A,2A), PDGF-BB staining (Dib-iib), and merged image (Dic-iic). (D3-4) CD8+ cells from a patient with T-LGL leukemia and a normal control. Nuclei staining (D3A-4A), PDGF-BB staining (D3B,4B), CD8 surface marker (D3C,4C), and merged image (D3D,4D). (D5-6) PDGFR-β staining on NK cells and T cells from a patient with NK-LGL (D5) and T-LGL leukemia (D6), respectively. Nuclei staining: D5A, D6A; PDGFR-β: D5B, D6B; and merged images: D5C, D6C.
Leukemic LGLs express PDGF-BB and PDGFR-β. (A) PDGF-BB mRNA expression levels in NK cells from patients with NK-LGL leukemia and normal controls, and in T cells from patients with T-LGL leukemia and normal controls, were determined by real-time quantitative RT-PCR (expressed as the ratio of PDGF-BB mRNA copies/18S control mRNA copies; *P < .05). (B) Western blot analysis was performed for PDGF-BB production in cells lysates of CD16+/CD56+ cells from patients with NK-LGL leukemia or CD8+ cells from patients with T-LGL leukemia as well as in their counterpart cells from normal controls. GAPDH detection was used to confirm equal loading of total protein in each sample. Densitometry was performed on these Western blot results to determine the average level of PDGF-BB protein expression. *P < .05. (C) PDGF-BB protein expression in plasma samples from 10 normal controls, 9 NK-LGL leukemia patients, 10 T-LGL leukemia patients, 4 HTLV-I–infected persons, and 4 HTLV-II–infected persons was determined by ELISA; *P < .05 compared with plasma sample levels from normal controls. (D) PDGF-BB protein ICC/IF staining as visualized by confocal microscopy in LGL leukemia cells compared with normal T or NK cells. Nuclei staining was visualized by DAPI (blue); PDGF-BB, by Cy5 fluorescent staining in the cytoplasm compartment (red). CD8 surface marker on T cells was visualized by FITC-fluorescent staining on the cell membrane (green). (D1-2) NK cells from a patient with NK-LGL leukemia and a normal control. Nuclei staining (D1A,2A), PDGF-BB staining (Dib-iib), and merged image (Dic-iic). (D3-4) CD8+ cells from a patient with T-LGL leukemia and a normal control. Nuclei staining (D3A-4A), PDGF-BB staining (D3B,4B), CD8 surface marker (D3C,4C), and merged image (D3D,4D). (D5-6) PDGFR-β staining on NK cells and T cells from a patient with NK-LGL (D5) and T-LGL leukemia (D6), respectively. Nuclei staining: D5A, D6A; PDGFR-β: D5B, D6B; and merged images: D5C, D6C.
PBGF-BB protein expression.
We found high levels of expression of PDGF-BB protein as detected by Western blot in purified LGL leukemia samples of either T-cell (n = 3) or NK-cell (n = 3) origin. In contrast, normal purified CD8+ cells or NK cells expressed little, if any, PDGF protein (Figure 1B).
ELISA detection of PDGF-BB.
Previously, we found increased levels of PDGF-BB in sera from T-LGL leukemia patients.11 It is known that PDGF is released during clotting; therefore, values measured in serum do not reflect levels of circulating PDGF. To determine circulating levels of PDGF-BB, platelet-poor plasma samples from patients with T-LGL leukemia (n = 10) and NK-LGL leukemia (n = 9), normal controls (n = 10), as well as HTLV-I–infected (n = 4) and HTLV-II–infected (n = 4) persons were examined in this study. As expected, increased levels of PDGF-BB in plasma samples from HTLV-I– and HTLV-II–infected persons were detected (Figure 1C). We also found nearly 3-fold higher levels of PDGF-BB in plasma samples from both T- and NK-types of LGL leukemia compared with normal controls (plasma PDGF-BB level: T-LGL: 938 ± 250, NK-LGL: 1118 ± 231, HTLV-I: 1124 ± 471, and HTLV-II: 911 ± 237 vs normal control: 342 ± 69 pg/mL, P ≤ .05). There was no statistical difference in PDGF-BB concentrations between T-LGL and NK-LGL leukemia.
ICC/IF staining for PDGF-BB.
ICC/IF staining was used to investigate the specific PDGF-BB production at protein level in LGL leukemia cells. As shown in Figure 1D, strong positive staining (red) for PDGF-BB in cytoplasmic compartments was observed in both CD16+/CD56+ NK cells (Figure 1D1B-C) and CD8+ T cells (Figure 1D3B,D) from LGL leukemia patients. The counterpart cells from normal controls showed negative staining for PDGF-BB molecules (Figure 1D2B for normal NK cells and D4B for normal CD8+ T cells). DAPI staining (blue) was applied to visualize nucleus (Figure 1D1A-4A). CD8 surface marker staining was used to identify purified CD8+ cells from patients with T-LGL leukemia and normal controls (green in Figure 1D3C-4C) using FITC-conjugated anti-CD8 antibody. These results indicated that leukemic T and NK cells from patients with LGL leukemia are responsible for PDGF-BB production.
PDGF receptor expression
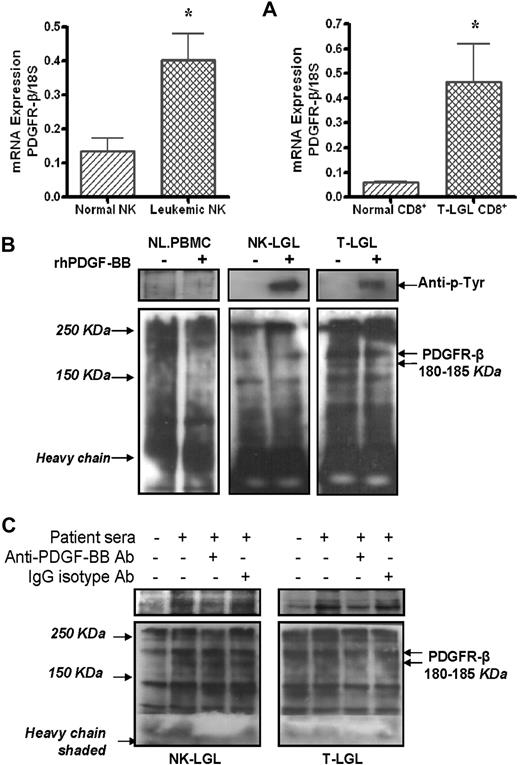
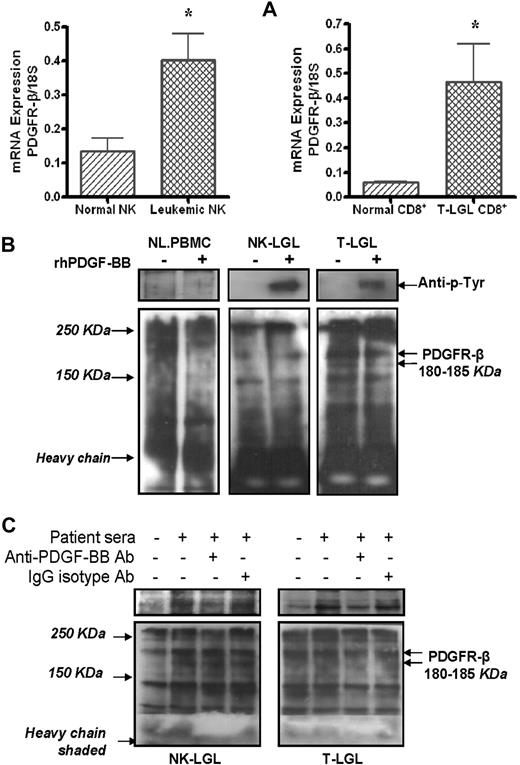
Using real-time quantitative RT-PCR, we showed that PDGFR-β mRNA levels were significantly higher in samples from purified leukemic NK (n = 7) or T cells (n = 5) than the levels expressed in their purified normal counterparts (Figure 2A, 0.41 ± 0.08 vs 0.13 ± 0.06 mRNA copies/18S, NK-LGL vs normal NK, P < .02, and 0.47 ± 0.12 vs 0.06 ± 0.004 mRNA copies/18S, T-LGL CD8+ vs normal CD8+, P < .01). Using Western blot assay, we found high levels of PDGFR-β protein expression in PBMCs from patients with either T-LGL (n = 3) or NK-LGL (n = 3) leukemia, whereas PDFGR-β was nearly undetectable in PBMCs from normal controls (Figure 2B). Using immunocytochemistry/immunofluorescence staining, we confirmed direct expression of PDFG-Rβ (green) on freshly isolated CD16+/CD56+ NK cells and CD8+ T cells from patients with LGL leukemia as shown in Figure 1D5B and C and D6B and C. In addition to expression of PDGF-Rβ, we were able to show PDGF-RTK autophosphorylation in PBMCs from patients with LGL leukemia of both T and NK type in the presence of rhPDGF-BB or LGL leukemia sera (Figure 2B-C). PDGF-RTK autophosphorylation in presence of LGL leukemic sera was inhibited by addition of an anti–human PDGF-BB neutralizing antibody (Figure 2C).
PDGFR-β expression and downstream activation in LGL leukemia. (A) PDGFR-β mRNA expression levels in CD3−/CD16+/CD56+ NK cells from either 7 patients with NK-LGL or 7 normal controls, and in CD8+ T cells from either 5 patients with T-LGL leukemia or 5 normal controls, were determined by real-time quantitative RT-PCR (expressed as the ratio of PDGFR-β mRNA copies/18S control mRNA copies, *P < .05). (B) Western blot assay for PDGFR-β and autophosphorylated PDGF-β-RTK expression in immunoprecipitation samples in PBMCs from a representative patient with T-LGL or NK-LGL leukemia as well as normal controls in presence or absence of rhPDGF-BB. (C) Western blot assay for PDGFR-β and autophosphorylated PDGF-β-RTK expression in immunoprecipitation samples in PBMCs from a representative patient with T-LGL or NK-LGL leukemia in the presence of pooled patient sera with or without anti–PDGF-BB neutralizing antibody. Rabbit IgG was used for isotype control. The gels in top panels indicate autophosphorylated PDGF-β-RTK expression in the same membranes used for PDGF-βR detection (bottom panel) reblotted using anti–p-Tyr antibody (PY99).
PDGFR-β expression and downstream activation in LGL leukemia. (A) PDGFR-β mRNA expression levels in CD3−/CD16+/CD56+ NK cells from either 7 patients with NK-LGL or 7 normal controls, and in CD8+ T cells from either 5 patients with T-LGL leukemia or 5 normal controls, were determined by real-time quantitative RT-PCR (expressed as the ratio of PDGFR-β mRNA copies/18S control mRNA copies, *P < .05). (B) Western blot assay for PDGFR-β and autophosphorylated PDGF-β-RTK expression in immunoprecipitation samples in PBMCs from a representative patient with T-LGL or NK-LGL leukemia as well as normal controls in presence or absence of rhPDGF-BB. (C) Western blot assay for PDGFR-β and autophosphorylated PDGF-β-RTK expression in immunoprecipitation samples in PBMCs from a representative patient with T-LGL or NK-LGL leukemia in the presence of pooled patient sera with or without anti–PDGF-BB neutralizing antibody. Rabbit IgG was used for isotype control. The gels in top panels indicate autophosphorylated PDGF-β-RTK expression in the same membranes used for PDGF-βR detection (bottom panel) reblotted using anti–p-Tyr antibody (PY99).
PDGF-BB–mediated PKB/AKT and ERK1/2 phosphorylation in LGL leukemia cells
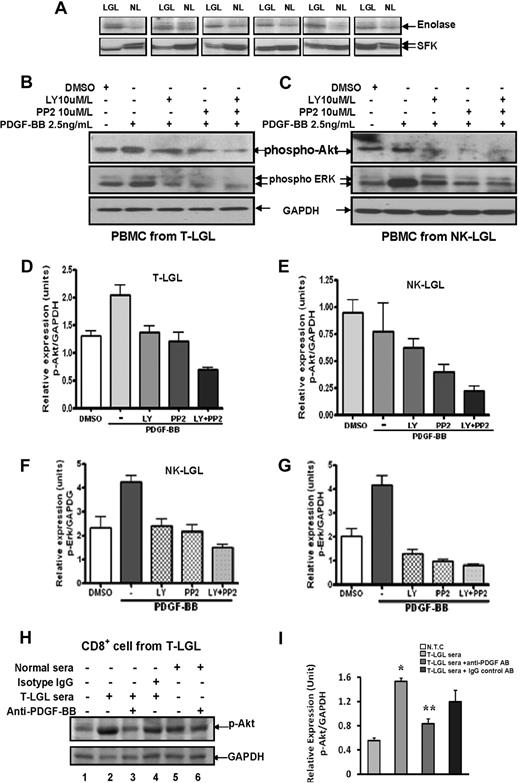
A previous study has demonstrated constitutive activation of AKT dependent on upstream activity of SFK in leukemic LGLs of T-cell origin.8 Using a kinase assay, we demonstrated directly increased SFK activity in leukemic T-LGLs (Figure 3A). We confirmed constitutive activation of AKT/ERK in these cells and extended this finding to leukemic LGLs of NK origin (Figure 3B-C). PDGF-RTK autophosphorylation has been shown to mediate PI3K and SFK activation through SH2 and SH3 domain interactions, which in turn, mediate the activation of multiple downstream activation pathways.18,19 Therefore, we hypothesized that constitutive expression of PDGF-BB by LGL leukemia cells may be responsible for activation of PKB/AKT pathways. To confirm our hypothesis, rhPDGF-BB–induced AKT and ERK1/2 phosphorylation in PBMCs from patients with LGL leukemia of both subtypes was determined by Western blot assay. As shown in a representative experiment (Figure 3B), rhPDGF-BB mediated elevation of both phospho-AKT and phospho-ERK1/2 expression in PBMCs from T-LGL leukemia patients. SFK inhibitor PP2 and PI3K inhibitor LY294002 dramatically down-regulated PDGF-BB–induced phosphorylation of AKT and ERK, and dual inhibition showed maximum inhibitory effects in both T and NK leukemic LGLs (Figure 3B-C). Quantitation of p-AKT from these experiments by densitometry analysis is summarized in Figure 3D-E; and p-ERK, in Figure 3F-G. These results suggested that PDGF-β-RTK autophosphorylation causes activation of both PI3K and SFK pathways in leukemic LGLs and subsequent phosphorylation of MAPK/ERK.20,21 To confirm that PDGF-BB isoform was sustaining activation through PKB/AKT pathway in leukemic LGLs, purified CD8+ T cells from 3 patients with T-LGL leukemia were studied. We observed that 10% pooled sera from 3 leukemic T-LGL patients induced a further increase in phospho-AKT (Ser473) expression compared with the samples receiving 10% normal sera. Two-hour preincubation of anti–human PDGF-BB neutralizing antibody significantly inhibited the elevation of phospho-AKT induced by LGL leukemia patient sera. Samples receiving 10% sera from normal controls with/without anti–human PDGF-BB neutralizing antibody showed only mild effects in mediating PKB/AKT phosphorylation (Figure 3H-I).
PDGF-BB mediates downstream target AKT/ERK pathway activation via PI3K and SFK pathways in leukemic LGLs. (A) Western blot assay was carried out for SFK protein expression in PBMCs from 6 patients with T-LGL leukemia (LGL) and 6 normal controls (NL). The gels in the bottom panel indicate SFK expression of 60 KDa molecular weight. Kinase assay (top panel) was performed in IP samples to determine SFK activities (enolase). (B-C) Phospho-AKT and phospho-ERK (p44/42 MAPK) expression was determined in PBMC lysates from a representative patient with T-LGL leukemia (B) and a patient with NK-LGL leukemia(C) using Western blot assay. Western blot assay for GAPDH expression was performed to confirm equal loading of total protein in each lane. (D-G) Densitometry analysis was performed on Western blot results from 3 patients with T-LGL leukemia (D,F) and 3 patients with NK-LGL leukemia (E,G) to determine the average level of phospho-AKT expression (D-E) or phospho-ERK expression (F-G). Data expressed as relative expression units of p-AKT/GAPDH or p-ERK/GAPDH ratios. (H) Western blot assay was performed for phospho-AKT protein expression in lysates of CD8+ cells from a representative patient with T-LGL leukemia. Cells received different treatments as indicated. (Lane 1) Medium only. (Lane 2) Ten percent pooled sera from 3 patients with T-LGL leukemia. (Lanes 3,6) Ten percent pooled sera from either T-LGL leukemia patients or normal controls that received 2-hour preincubation with anti–PDGF-BB neutralizing antibody. (Lane 4) Ten percent pooled sera from patients with T-LGL leukemia that received 2-hour preincubation with IgG isotype control antibody. (Lane 5) Ten percent pooled sera from 3 normal controls. Western blot analysis for GAPDH was performed to confirm equal loading of total protein in each lane. (I) Densitometry was performed for Western blot results from 3 different experiments to determine the average level of phospho-AKT protein expression. *P < .03, no treatment control (NTC) versus 10% T-LGL patient sera treatment; **P < .05, treatment with 10% patient sera versus treatment with 10% patient sera preincubated with anti–PDGF-BB neutralizing antibody.
PDGF-BB mediates downstream target AKT/ERK pathway activation via PI3K and SFK pathways in leukemic LGLs. (A) Western blot assay was carried out for SFK protein expression in PBMCs from 6 patients with T-LGL leukemia (LGL) and 6 normal controls (NL). The gels in the bottom panel indicate SFK expression of 60 KDa molecular weight. Kinase assay (top panel) was performed in IP samples to determine SFK activities (enolase). (B-C) Phospho-AKT and phospho-ERK (p44/42 MAPK) expression was determined in PBMC lysates from a representative patient with T-LGL leukemia (B) and a patient with NK-LGL leukemia(C) using Western blot assay. Western blot assay for GAPDH expression was performed to confirm equal loading of total protein in each lane. (D-G) Densitometry analysis was performed on Western blot results from 3 patients with T-LGL leukemia (D,F) and 3 patients with NK-LGL leukemia (E,G) to determine the average level of phospho-AKT expression (D-E) or phospho-ERK expression (F-G). Data expressed as relative expression units of p-AKT/GAPDH or p-ERK/GAPDH ratios. (H) Western blot assay was performed for phospho-AKT protein expression in lysates of CD8+ cells from a representative patient with T-LGL leukemia. Cells received different treatments as indicated. (Lane 1) Medium only. (Lane 2) Ten percent pooled sera from 3 patients with T-LGL leukemia. (Lanes 3,6) Ten percent pooled sera from either T-LGL leukemia patients or normal controls that received 2-hour preincubation with anti–PDGF-BB neutralizing antibody. (Lane 4) Ten percent pooled sera from patients with T-LGL leukemia that received 2-hour preincubation with IgG isotype control antibody. (Lane 5) Ten percent pooled sera from 3 normal controls. Western blot analysis for GAPDH was performed to confirm equal loading of total protein in each lane. (I) Densitometry was performed for Western blot results from 3 different experiments to determine the average level of phospho-AKT protein expression. *P < .03, no treatment control (NTC) versus 10% T-LGL patient sera treatment; **P < .05, treatment with 10% patient sera versus treatment with 10% patient sera preincubated with anti–PDGF-BB neutralizing antibody.
Stimulation of NK leukemia cell line growth by PDGF or LGL patient sera
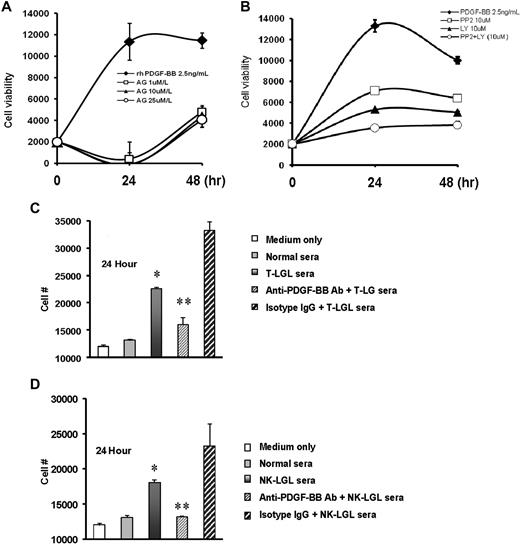
We found that treatment of NKL cells with PDGF-BB led to a 6-fold increase in cell numbers at 24 hours. This effect could be completely blocked with addition of AG1296, a PDGF-β-RTK inhibitor at a concentration of 1μM/L (Figure 4A). Inhibition of the PDGF-BB effect was also seen when NKL cells were treated with the PI3K inhibitor, LY129004, or with the SFK inhibitor, PP2 (Figure 4B). We were also interested in determining whether the high levels of PDGF-BB in serum samples from patients with leukemic LGLs could stimulate growth of NKL in vitro. As shown in Figure 4C-D, the presence of pooled sera from either T- or NK-LGL leukemia patients significantly increased proliferation of NKL cells compared with either culture medium or normal human serum. This effect was significantly blocked by addition of neutralizing antibody to PDGF-BB.
Inhibition of PDGF or LGL patient sera stimulation of NK leukemia cell line growth by blockade of PI3K/SFK or neutralizing antibody to PDGF-BB, respectively. (A-B) NKL cells were treated with rhPDGF-BB in presence of AG1296 at different concentrations (A), or in presence or absence of LY294002, PP2, or the combination of these 2 compounds (B); cell proliferation was measured by MTT assay. (C-D) NKL cells were treated with 10% pooled sera from patients with T-LGL leukemia (C) or from patients with NK-LGL leukemia (D) for 24 hours; cell proliferation was determined by MTT assay. Ten percent pooled sera sample from normal donors was used as controls. *P < .01 for cell viability in samples receiving 10% normal sera treatment versus samples receiving 10% LGL patient sera treatment; **P < .03 for cell viability in the samples receiving 10% patient sera treatment versus samples receiving treatment of 10% patient sera preincubated with anti–PDGF-BB neutralizing antibody. Rabbit IgG antibody was used as isotype control.
Inhibition of PDGF or LGL patient sera stimulation of NK leukemia cell line growth by blockade of PI3K/SFK or neutralizing antibody to PDGF-BB, respectively. (A-B) NKL cells were treated with rhPDGF-BB in presence of AG1296 at different concentrations (A), or in presence or absence of LY294002, PP2, or the combination of these 2 compounds (B); cell proliferation was measured by MTT assay. (C-D) NKL cells were treated with 10% pooled sera from patients with T-LGL leukemia (C) or from patients with NK-LGL leukemia (D) for 24 hours; cell proliferation was determined by MTT assay. Ten percent pooled sera sample from normal donors was used as controls. *P < .01 for cell viability in samples receiving 10% normal sera treatment versus samples receiving 10% LGL patient sera treatment; **P < .03 for cell viability in the samples receiving 10% patient sera treatment versus samples receiving treatment of 10% patient sera preincubated with anti–PDGF-BB neutralizing antibody. Rabbit IgG antibody was used as isotype control.
PI3K and SFK pathway inhibition induces apoptosis in PBMCs from patients with LGL leukemia
A previous study demonstrated induction of apoptosis via inhibition of PI3K signaling in leukemic LGLs of T-cell origin.8 In addition to confirming these results, we also wanted to investigate whether such survival signaling was responsible for the long-term survival of leukemic NK-LGLs. Therefore, PI3K and SFK inhibition with LY294002 and PP2, respectively, were tested for efficacy in mediating apoptosis in PBMCs from patients with T- or NK-LGL leukemia. Both LY294002 and PP2 induced apoptosis in PBMCs from leukemic LGLs of T- and NK-cell subtypes in a time- and dose-dependent manner; moreover, the combination showed additive effects in induction of apoptosis (Figure 5A-B). These findings were confirmed in a larger sample of patients with T-LGLs (n = 5) and NK-LGLs (n = 4) using a dose of 25μM (Figure 5C).
Inhibition of PI3K and SFK pathways induces apoptosis in PBMCs from patients with LGL leukemia. (A-B) Freshly isolated PBMCs from a representative patient with NK-LGL leukemia (A) or a patient with T-LGL leukemia (B) were treated with LY129004 (PI3K inhibitor) or PP2 (SFK inhibitor) at doses of 10, 25, or 50μM/L, respectively, and with the combination of both inhibitors at doses of 10 and 25μM/L concentrations for 24 or 48 hours, respectively. DMSO served as vehicle control. (C) PBMCs from 5 patients with T-LGL leukemia (patients 1-5), from 4 patients with NK-LGL leukemia (patients 6-9), and from 4 normal controls (NLs 10-13) were treated with LY129004 or SFK PP2 at a concentration of 25μM, or with the combination of LY and PP2 at 25μM for 24 hours. Apoptosis was determined by flow cytometry assay with annexin-V–FITC/7-amino-actinomycin D staining.
Inhibition of PI3K and SFK pathways induces apoptosis in PBMCs from patients with LGL leukemia. (A-B) Freshly isolated PBMCs from a representative patient with NK-LGL leukemia (A) or a patient with T-LGL leukemia (B) were treated with LY129004 (PI3K inhibitor) or PP2 (SFK inhibitor) at doses of 10, 25, or 50μM/L, respectively, and with the combination of both inhibitors at doses of 10 and 25μM/L concentrations for 24 or 48 hours, respectively. DMSO served as vehicle control. (C) PBMCs from 5 patients with T-LGL leukemia (patients 1-5), from 4 patients with NK-LGL leukemia (patients 6-9), and from 4 normal controls (NLs 10-13) were treated with LY129004 or SFK PP2 at a concentration of 25μM, or with the combination of LY and PP2 at 25μM for 24 hours. Apoptosis was determined by flow cytometry assay with annexin-V–FITC/7-amino-actinomycin D staining.
Discussion
Dysregulated apoptosis appears to be a fundamental mechanism underlying the pathogenesis of LGL leukemia of both T- and NK-cell origin. Leukemic LGLs correspond to activated cytotoxic cells that have escaped Fas-mediated activation-induced cell death.3,6 Mechanisms of apoptotic resistance have been studied more extensively in T-LGL leukemia than in the NK type of LGL leukemia. Gene expression signature and pathway-based microarray analyses showed profound dysregulation of apoptotic genes, suggesting uncoupling of activation and apoptotic pathways as a mechanism for failure of activation-induced cell death in leukemic LGLs.10 Increased numbers of circulating leukemic LGLs then result from activation of multiple survival pathways. Constitutive activation of JAK2/signal transducer and activator of transcription 3/Mcl-1, RAS/MAPK, and SFK/PI3K/AKT and sphingolipid signaling pathways have been demonstrated in leukemic LGLs.7-10 Using a network theory approach, we recently showed that IL-15 and PDGF are master switches responsible for turning on and controlling cross-talk among these survival pathways.11
Here we demonstrate the autocrine nature of PDGF-mediated survival of both T and NK leukemic LGLs and further define downstream components involved in this process. PDGF is a major mitogen for connective tissue cells and certain other cell types.18,22 It is a dimeric molecule consisting of disulfide-bonded, structurally similar A- and B-polypeptide chains that combine to homodimers and heterodimers.19 PDGF isoforms exert their cellular effects by binding to and activating 2 structurally related protein tyrosine kinase receptors (PDGF-RTK), denoted receptor-α and receptor-β.19,22-24 Normally, both human T and NK cells express high-affinity PDGF receptors.23,25-29 We demonstrated increased expression of PDGFR-β gene transcripts in leukemic LGLs compared with normal purified CD8+ T cells or NK cells. Increased protein expression was also detected. Direct detection of PDGFR-β on leukemic LGLs was also observed.
Excessive production of PDGF molecules is involved in autocrine and paracrine growth stimulation of human tumors that express PDGF receptors.19,28,30 We found increased expression of PDGF-B gene transcripts and PDGF-BB protein in purified leukemic cells compared with their normal counterparts. Previously we had demonstrated increased serum levels of PDGF-BB in patients with T-LGL leukemia.11 To more accurately determine circulating levels of PDGF, we examined platelet-poor plasma samples by ELISA. Plasma levels of PDGF-BB were significantly elevated in patients with either T- or NK-type of LGL leukemia, similar to levels seen in HTLV-I– or HTLV-II–infected persons. Using immunocytochemistry, we demonstrated that leukemic LGLs were a source of PDGF-BB production. These results support an autocrine PDGF-BB regulatory loop in LGL leukemia. A key question remains: what initiated leukemic LGLs to make PDGF? Leukemic LGLs appear to be antigen-driven cytotoxic cells. Recent evidence implicates human cytomegalovirus as the inciting antigen in the rare CD4+ subset of LGL leukemia.31 The cytomegalovirus-induced gene expression signature was characterized by inflammatory/immune response and apoptotic resistance, similar to features described for the more common CD8+ LGL leukemia. It is of interest that lymphocyte production of PDGF-BB had been observed previously only in the setting of HTLV-I or -II infection.28,32 Although LGL leukemia patients are not infected with these viruses, we have found seroreactivity to the BA21 protein of HTLV-I envelope in patients with either T-cell or NK-cell form of the disease.33,34 These data taken together with documentation of a common PDGF-dependent survival pathway suggest a similar, perhaps viral, etiology of both T- and NK-LGL leukemia.
PDGF receptor tyrosine kinase contains several cellular signaling transduction domains, such as SH2 and SH3 domains and PTB domain.19 Autophosphorylation of tyrosine residues located outside the kinase domain creates docking sites for interactions with many SH2 and SH3 domain containing molecules such as PI3-kinase regulatory unit p85, SFK, phospholipase C-r, and a GTPase activation protein for RAS, as well as other adaptor proteins and nuclear transcription factors.18,19,35 A previous study has documented the importance of SFK/PI3K/AKT activation in pathogenesis of LGL leukemia of T-cell type.8 We confirmed these results and also extended this finding to leukemic NK cells. Importantly, we demonstrated a central role of PDGF in this activation pathway. We found autophosphorylation of PDGF-RTK in PBMCs of LGL leukemia patients in presence of LGL leukemia sera. We also observed that exogenous rhPDGF-BB mediated PDGF-β-RTK autophosphorylation and its downstream PKB/AKT (Thr473), SFK (Tyr419), as well as MEK1/2 and ERK1/2 phosphorylation in PBMCs from patients with leukemic LGLs of both T and NK subtypes. The biologic importance of overexpression of PDGF-BB in LGL leukemia was demonstrated by showing the effectiveness of a specific anti–human PDGF-BB neutralizing antibody in inhibition of PKB/AKT phosphorylation in purified CD8+ cells from all 3 tested patients with T-LGL leukemia. PDGF-BB also provided an important proliferative signal for the NKL cell line. PDGF-BB signaling, therefore, may also be important in the pathogenesis of acute NK-LGL leukemia, which is characterized by high cell turnover and an aggressive clinical course.
We also found that simultaneous inhibition of both pathways with PP2 and LY294002 mediated additive effects in induction of apoptosis in PBMCs from patients with LGL leukemia of both T and NK subtypes. Dual inhibition mediated a profound down-regulation of PKB/AKT and ERK phosphorylation compared with the effects seen by individual pathway inhibition. These results suggest that targeting of downstream components of PDGF signaling may be a potential therapeutic strategy for LGL leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Lynn F. Ruiz, research study coordinator and LGL registrar. We also thank the Penn State Cancer Institute Flow Cytometry Core Facility and Confocal Imaging Core Facility at Penn State Hershey Cancer Institute for acquisition and analysis data. The following reagent was obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: recombinant human interleukin-2, catalog no. 136 from Dr Maurice Gately, Hoffman-LaRoche Inc.
This work was supported by National Institutes of Health grant R01 CA112112.
National Institutes of Health
Authorship
Contribution: J.Y., X.L., and T.P.L. designed research, analyzed data, and wrote the paper; J.Y., X.L., S.B.N., R.Z., K.B., L.K.R., and R.I. performed research; and K.T.B. and N.R.J. prepared blood samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jun Yang, Penn State University, College of Medicine, Penn State Hershey Cancer Institute, Experimental Therapeutics-CH74, 500 University Dr, Hershey, PA 17033; e-mail: jyang@psu.edu.