Abstract
Dyskeratosis congenita (DC) is a rare inherited form of bone marrow failure (BMF) caused by mutations in telomere maintaining genes including TERC and TERT. Here we studied the prevalence of TERC and TERT gene mutations and of telomere shortening in an unselected population of patients with BMF at our medical center and in a selected group of patients referred from outside institutions. Less than 5% of patients with BMF had pathogenic mutations in TERC or TERT. In patients with BMF, pathogenic TERC or TERT gene mutations were invariably associated with marked telomere shortening (≪ 1st percentile) in peripheral blood mononuclear cells (PBMCs). In asymptomatic family members, however, telomere length was not a reliable predictor for the presence or absence of a TERC or TERT gene mutation. Telomere shortening was not pathognomonic of DC, as approximately 30% of patients with BMF due to other causes had PBMC telomere lengths at the 1st percentile or lower. We conclude that in the setting of BMF, measurement of telomere length is a sensitive but nonspecific screening method for DC. In the absence of BMF, telomere length measurements should be interpreted with caution.
Introduction
Dyskeratosis congenita (DC) is a rare inherited bone marrow failure syndrome (IBMFS). Patients with DC typically present with progressive bone marrow failure (BMF) and the classic triad of mucocutaneous features, including abnormal pigmentation, dystrophic nail changes, and leukoplakia of the oral mucosa.1-3 Disease penetrance is highly variable, ranging from hardly detectable, to severe forms causing death in early childhood, as seen in Hoyeraal Hreidarson (HH) syndrome. With the identification of 6 genes that when mutated can cause DC (DKC1, TERC, TERT, NOP10, NHP2, and TINF2)4-9 and the availability of genetic testing, it has become increasingly evident that the classic mucocutaneous features are only present in a small proportion of patients with DC, suggesting that a diagnosis based on the presence of these manifestations alone is likely to overlook a significant proportion of patients with DC.10 Screening for pathogenic mutations is expensive, time-consuming, and, in approximately half of patients, inconclusive. Thus, using the methods currently available, mutation screening does not seem to be a suitable screening test for the diagnosis of DC. However, all genes implicated in DC are involved in telomere maintenance, suggesting that dysfunctional and excessively short telomeres are the common denominator in this disease and that short and dysfunctional telomeres play an important role in the pathogenesis of disease in patients with DC
Telomeres are complex DNA-protein structures at the end of chromosomes.11,12 In most eukaryotes including humans, telomeric DNA is composed of guanine-rich DNA repeat sequences.13 Telomeres shorten with each cell division.14 When telomeres become critically short, a DNA damage response is activated, causing cell cycle arrest or cell death.15 In humans, telomerase-based telomere elongation is the major mechanism that counteracts this process of continuous telomere shortening.16,17 However, in humans after birth, telomerase expression is confined to germ cells, stem cells and their immediate progeny, activated T cells, and monocytes. Notably, most cancer cells express telomerase activity.15,18 Despite telomerase expression in stem cells, human telomeres shorten with age.19,20 In peripheral white blood cells rapid telomere shortening occurs within the first year of life, followed by a more gradual decline over time.21 Although there is considerable variation among people of the same age, the relative telomere length is at least in part inherited.22 Telomerase is a ribonucleoprotein complex consisting of the catalytic subunit (TERT), its RNA component TERC, and the 4 H/ACA RNA associated proteins, dyskerin, NHP2, NOP10, and GAR1.23,24 Notably, 5 of the 6 telomerase components have been found to be mutated in patients with DC. The 6th gene mutated in DC is TINF2 encoding TIN2, a protein component of the telomere ribonucleoprotein complex shelterin.9 These findings suggest that in patients with DC, telomere elongation and maintenance are impaired. Indeed, patients with DC have very short telomeres.25-27 We were interested in determining the frequency of TERC and TERT gene mutations in patients with BMF, whether the identified mutations were associated with short telomeres, and whether telomere measurement could be used to identify people with DC. For this purpose, we screened the TERC and TERT genes for mutations and measured telomere length in peripheral blood mononuclear cells (PBMCs) in an unselected population of patients with BMF at Washington University Medical Center (WUMC) and in a selected group of patients referred from outside institutions between 2001 and 2007. We found that patients with BMF due to DC diagnosed either by the presence of a pathogenic mutation or by the presence of classic mucocutaneous features have very short telomeres. However, short telomeres were also identified in other patients with BMF. Although there was a trend, short telomere length was not a significant predictor for the mutation status in families with DC.
Methods
Patient characteristics
A total of 247 patients with BMF were enrolled through our ongoing study investigating the molecular mechanism of BMF (http://bmf.im.wustl.edu). The selection and characteristics of these patients are summarized in Table 1. The Human Research Protection Offices at WUMC and collaborating centers approved the study. Informed consent was obtained in accordance with the Declaration of Helsinki from all study participants or through their legal guardians before participation. Inclusion criteria included an unselected population of patients, children and adults, who were evaluated or treated at WUMC because of BMF, defined as cytopenia for at least 3 months, in at least 1 cell lineage (hemoglobin ‘Hb’ ≤ 11 g/dL, with white blood cell ‘WBC’ count < 3.5 × 103/μL, polymorphonuclear ‘PMN’ cells < 1.5 × 103/μL, platelets ≤ 100 × 103/μL) and a hypocellular bone marrow. At WUMC, children and adult patients were enrolled as they presented and therefore represent a relatively unbiased patient population from a tertiary referral center. To enrich our patient population for those with DC or suspected DC and to include patients with other IBMFS, we also enrolled patients through collaboration with institutions outside Washington University (referred). The diagnosis of IBMFS was made according to previously published criteria including clinical presentation, laboratory testing, and/or the presence of pathogenic mutations.3,28-30 Peripheral blood was obtained for the isolation of genomic DNA and for the analysis of telomere length. Due to limited or inappropriate samples, telomere measurements or part of the mutation analysis could not be performed in selected samples. The numbers of patients analyzed for each assay are indicated. In those in whom a mutation was identified, family members were invited to participate in the study. Telomere length measurement and mutation analysis were performed in all participating family members.
Isolation and amplification of DNA
Genomic DNA was extracted from peripheral blood leukocytes with the DNA blood mini kit (QIAGEN, Valencia, CA). Phi29-based whole-genome amplification was performed on genomic DNA in selected samples where DNA was limited (QIAGEN Repli-G service).31
Mutation analysis
Mutation screening was performed on genomic DNA or amplified DNA. Gene sequences from GenBank and Ensembl were used as reference for assembly and primer construction. The 151-bp 5′ flanking and exonic regions of the TERT gene, including the exon/intron boundaries, were amplified by polymerase chain reaction (PCR) to generate amplicons for direct nucleotide sequencing. An amplicon of 1190 bp extending from 433 bp 5′ flanking to 306 bp downstream of the TERC gene was amplified for the sequencing of TERC. Primers used have been published previously32,33 ; additional primers and primers used for Pyrosequencing (Biotage AB, Charlottesville, VA) are summarized in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The sequencing of TERT was performed on an ABI3730 automated sequencer (Applied Biosystems, Foster City, CA); sequencing of TERC, DKC1, and TINF2 exon 6 were done manually.
For robotic sequencing, an automated analysis report that identifies sequence alterations, including single nucleotide changes, insertions, and deletions, was generated by using PolyPhred and PolyScan software. These reported alterations were visually inspected on the individual forward and reverse chromatographs to exclude false positive reads. Identified sequence variants were verified by separate PCR direct sequencing or by restriction endonuclease digestion.
The TERT H412Y sequence variant was typed in 276 cancer-free controls. All controls were > 65 years of age with no history of cancer. Control DNAs were made available by the Siteman Cancer Center Hereditary Cancer Core (http://www.siteman.wustl.edu/internal.aspx?id = 2570). The CT variant was detected by Pyrosequencing using the manufacturers' reagent, protocols, and software to sequence and analyze the data.
Detection of deletion at the 5′ or 3′ end of TERC
Quantitative PCR was used to detect gene deletion at the 5′ end or 3′ end of TERC. The primer sequences used for the detection of 5′ end deletion were F: 5′-GAGAGCCGCGAGAGTCAG-3′, R: 5′-GCCTACGCCGTTCTCAGTT-3′. The primer sequences used for 3′ end deletion were F: 5′-GAAGAGGAACGGAGCGAGT-3′, R: 5′-GAAGAGGAACGGAGCGAGT-3′. The β-globin gene (β-G) served as the reference gene in the determination of the copy number of TERC F: 5′-GCTTCTGACACAACTGTGTTCACTAGC-3′, R: 5′-CACCAACTTCATCCACGTTCACC-3′.34 Aliquots of 20-ng genomic DNA were assayed with a SYBR Green core reagent kit (Applied Biosystems). All PCRs were performed on the Prism 7500F Sequence Detection System (Applied Biosystems). Individual copy number of TERC at the 5′ end or 3′ end was calculated as the ratio of TERC/β-G of each sample relative to that of 2 patients with DC who have been described previously by us to carry a deletion of 316 bp at the 5′ end or a deletion of 74 bp at the 3′ end.
Functional analysis of TERT and TERC variants
Nonsynonymous mutations identified in TERT and TERC were introduced in a p3.1 + TERT plasmid or a pUC TERC plasmid by using a QuickChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) as previously described.32 Four micrograms of wild-type (WT) or mutant TERT (TERC) plasmid was transfected into WI-38 VA-13 fibroblasts at 80% confluence in the presence of equal amount of WT TERC (TERT) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Thirty-six hours after transfection, the cells were collected, lysed, and the protein concentration in the resulting cell lysates was measured by using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Hercules, CA). Telomerase activity in the serially diluted cell lysates was determined using quantitative PCR-based telomeric repeat amplification protocol (TRAP) assay.
Telomere length measurement by flow cytometric fluorescence in situ hybridization
Telomere lengths in PBMCs isolated by Ficoll-Hypaque gradient centrifugation were measured as previously described,35 using fluorescein isothiocyanate (FITC)–conjugated (C3TA2)3 peptide nucleic acid (PNA; Applied Biosystems) probe with a flow cytometer (Beckman Coulter, Fullerton, CA). Relative telomere length (RTL) was determined by comparing isolated PBMC with a control cell line (GM03671C; Coriell Institute of Medical Research, Camden, NJ), a tetraploid cell line that served as an internal control and was assigned a telomere length of 100%. Telomere lengths of 234 healthy people (ranging in age from 1 day to 94 years old) were used to determine the distribution of age-dependent telomere lengths. Short telomeres were defined as a telomere length less than or equal to the 1st percentile of the normal telomere length distribution.
Statistical analysis
All data analyses were performed using the SAS computer software (version 9.1; SAS Institute, Cary, NC). SAS mixed model was used to assess the association between age and telomere length, separately among unaffected family members from families ascertained through affected probands who were TERC, TERT, or DKC1 mutation carriers, and among control families. Familial dependences were accounted for by using sandwich estimator method implemented in SAS mixed procedure. We used the same procedure with the least mean square approach to assess mean differences of telomere length across the 3 groups of mutation carriers after correction for the effect of age. This approach was also used to assess TERC or TERT gene mutations in predicting shortened telomeres among unaffected family members from ascertained families with affected probands after correction for the effect of age. Differences of telomerase activities among TERT and TERC mutation variants were tested using one-way analysis of variance (ANOVA) followed by post hoc Bonferroni analysis. One-sided Fisher exact probability test was used to assess the association between the C1234T (H412Y) mutation and the disease status. A P value less than .05 after Bonferroni correction for multiple tests was used to flag statistical significance.
Results
Patients with TERC and TERT gene mutations may present with clinical symptoms indistinguishable from patients with aplastic anemia.10 We therefore sought to determine whether mutations in TERT or TERC are common in patients with BMF and whether telomere measurement would be a sensitive screening method to identify patients with DC. We determined telomere length and screened for TERC and TERT gene mutations in adults and children who were diagnosed or treated because of BMF at WUMC and collaborating institutions (referred).
Mutation analysis
Results from the mutation analysis are summarized in Table 2.
TERC gene.
The TERC gene was screened for mutations in 200 patients (WUMC 133, referred 67). Figure S1A and Table S2 summarize the sequence variants identified. This group included 2 unrelated patients in whom we previously identified large TERC gene deletions.5,36 We have previously identified the G58A and G228A transitions within the TERC gene as rare polymorphisms in the healthy African American population.33,37,38 In addition, we identified 2 novel TERC RNA mutations. The T100A mutation affected a highly conserved polyuridine track in the P2b TERC pseudoknot domain and is predicted to inhibit pseudoknot formation.39 Indeed, telomerase activity after transfection into WI-38 VA-13 cells was severely impaired (P < .0001, Figure 1). The mutation was identified in 2 sisters with clinical manifestations of DC, including presentation with hypoplastic myelodysplastic syndrome (MDS), reticular skin pigmentation, pulmonary fibrosis, carcinoma of the tongue, and a family history of pulmonary fibrosis, myelodysplasia, and cancer (sFigure S2A,B). The C35T TERC RNA mutation was identified in a patient with hypoplastic MDS. In vitro reconstitution assays using WI-38 VA-13 cells revealed a 76% reduction in telomerase activity (P = .003, Figure 1). The patient's brother, his nephew, and 6 of his 7 children inherited the same mutation; 3 of them showed a mild macrocytic anemia as the only clinical manifestation, whereas the other 5 mutation carriers in this family had normal peripheral blood values and appeared healthy (Figure S2C,D).
In vitro telomerase activity of the TERC RNA and TERT variants in WI-38 VA-13 cells. WI-38 VA-13 cells were transfected with a plasmid expressing the mutant or WT TERT cDNA sequences and a plasmid expressing the respective WT or mutant or TERC RNA. Telomerase activity was determined using a quantitative PCR-based TRAP assay. Activity is shown in comparison to the activity obtained after transfection with WT (= 1.0). Values above each column are the average of triplicate experiments. Error bars represent SD, * indicates the difference between the variant, and WT is statistically significant at the level of .05. In vitro telomerase activities of the TERT mutants H412Y, P704S, Y846C, and H876Q have been reported previously.32
In vitro telomerase activity of the TERC RNA and TERT variants in WI-38 VA-13 cells. WI-38 VA-13 cells were transfected with a plasmid expressing the mutant or WT TERT cDNA sequences and a plasmid expressing the respective WT or mutant or TERC RNA. Telomerase activity was determined using a quantitative PCR-based TRAP assay. Activity is shown in comparison to the activity obtained after transfection with WT (= 1.0). Values above each column are the average of triplicate experiments. Error bars represent SD, * indicates the difference between the variant, and WT is statistically significant at the level of .05. In vitro telomerase activities of the TERT mutants H412Y, P704S, Y846C, and H876Q have been reported previously.32
TERT gene.
The TERT gene was screened for mutations in 199 patients (WUMC 130, referred 69). Figure S1B and Table S3 summarize the sequence variants identified. Four nonsynonymous sequence variants in the TERT gene were found more than once in unrelated families ‘G835A (A279T), C1234T (H412Y), C2110T (P704S), and G3184A (A1062T)’. Three (G835A (A279T), C1234T (H412Y), G3184A (A1062T)) have been previously identified in patients with aplastic anemia and pulmonary fibrosis.6,40,41 Two (G835A (A279T) and G3184A (A1062T)) have been reported also in healthy control subjects with an allelic frequency similar to the frequency identified in our patient population and found by others in patients with BMF or pulmonary fibrosis.6,40,41 We therefore concluded that G835A (A279T) and G3184A (A1062T) are rare single nucleotide polymorphisms (SNPs) that are unlikely to cause or contribute to the development of BMF. The C1234T (H412Y) sequence variant was identified in 4 patients, 1 diagnosed with MDS and one with Diamond Blackfan anemia (DBA). After transfection into WI-38 VA-13 cells, the C1234T (H412Y) sequence variant was associated with a reduced telomerase activity (Figure 1).6,32 The variant has previously been reported to be pathogenic in 2 patients with aplastic anemia (AA), as it was not found in 528 control subjects.6 However, C1234T (H412Y) is represented in the CEPH SNP database (rs34094720). To further determine the pathogenicity of this variant, we analyzed an additional 274 healthy people. Because of the variable onset of BMF in patients with autosomal dominant DC, we selected people who were healthy at the age of 65 years or older. Three subjects carried the C1234T (H412Y) variant (allele frequency 0.005). Although the frequency of this variant was higher in our BMF patient population, this was not statistically significant (P = .22). Thus both the G835A (A279T) and the C1234T (H412Y) sequence variants were not considered to be pathogenic. Whether carriers of the C1234T (H412Y) variant have an increased risk for the development of BMF or of other DC-associated diseases, including pulmonary fibrosis, liver fibrosis, osteoporosis, MDS, and acute myeloid leukemia (AML) requires further studies including large numbers of patients and controls. In contrast, the C2110T (P704S) variant has never been identified before in healthy controls, was not present in the SNP database, and was associated in vitro with reduced telomerase activity (Figure 1).32 The variant was identified in 2 unrelated patients. One patient heterozygous for the C2110T (P704S) mutation had a history of recurrent AA after antithymocyte globulin (ATG) and cyclosporine therapy. The other was homozygous for this mutation and presented with the mucocutaneous features characteristic of DC.32 We therefore concluded that the C2110T (P704S) mutation is pathogenic. We identified 4 additional TERT gene mutations in 3 families, one of whom we have reported on previously.32 One patient with therapy-resistant, progressive AA was a compound heterozygote for the A2537G (Y846C) and C2628G (H876Q) mutations; her parents, who were heterozygous for the mutations, had no clinical manifestations. A novel C2147T (A716V) mutation was identified in one child with severe pancytopenia and a family history significant for AA and lung disease. Finally, a C3043T (C1015R) mutation was found in a 62-year-old woman with therapy-refractory AA. All 4 TERT gene mutations caused a significant reduction in telomerase activity after transfection into WI-38 VA-13 cells (Figure 1), which is consistent with the finding that these mutations are likely to be pathogenic.
DKC1 gene.
DKC1 mutations usually are associated with the classic mucocutaneous manifestations when signs of BMF become obvious.42 Indeed, mucocutaneous manifestations often precede signs of BMF in X-linked DC.43 Furthermore, no DKC1 mutations were found in a previous study of 200 adult patients with AA.6 We therefore decided to screen the DKC1 gene only in: (1) patients presenting with mucocutaneous features (n = 5); (2) patients with signs of HH (n = 3); and (3) male patients with BMF, no other identified IBMFS, and a telomere length at the 1st percentile (n = 11) or lower. Six DKC1 mutations were identified in 7 patients (Table 2). One of the mutations was known to be the cause of disease in the affected patient before participation in our study. In 6 patients, the mutation was unknown before study enrollment. The A353V mutation was identified in 2 unrelated patients. In 1, the mutation had occurred de novo, and in the other, the maternal grandfather and 1 sibling had also been diagnosed with DC. All mutations identified have previously been described in unrelated patients with DC. No DKC1 mutation was identified in patients with BMF and short telomeres in the absence of typical mucocutaneous manifestations or clinical signs of HH.
TINF2 exon 6.
Recently mutations in exon 6 of TINF2 have been identified in patients with DC and short telomeres.9,44 We therefore screened all patients with telomeres less than or equal to the 1st percentile for mutations in TINF2 exon 6 (n = 49). None of our patients with short telomeres carried one of the 3 known TINF2 mutations (K280E, R282H, or R282S).
Patients with BMF, short telomeres (≤ 1st percentile), and no mutation in the TERC, TERT, DKC1, or TINF2 genes.
No mutations in TERC, TERT, DKC1, or TINF2 and telomere length less than or equal to the 1st percentile were detected in one patient with clinical features consistent with HH, 2 patients with MDS, 4 patients with paroxysmal nocturnal hemoglobinuria (PNH), 13 patients with AA, 5 patients with DBA, and one patient with familial platelet disorder (FPD).
Telomere length measurements
We determined the normal age-dependent telomere length distribution in PBMC from 234 healthy control subjects between the ages of 1 day and 94 years (Figure 2A). Figure 2B shows the telomere length of all study participants with BMF in whom a telomere measurement was obtained (n = 160). The number of patients with BMF and short telomeres (≤ 1st percentile of normal) is summarized in Table 3. Forty-nine patients with BMF (30.6%) were found to have short telomeres. Nineteen patients with BMF carried the diagnosis of DC at the time of enrollment because they presented either clinical characteristic features or a known mutation in one of the DC-associated genes. All patients with DC and BMF showed a telomere length far below the 1st percentile (Figure 2B). As we previously reported in a large family with autosomal dominant DC,26 patients with DC and BMF all had a similar telomere length and did not show an age-dependent decline in telomere length (β = −0.0011, P = .95) as seen in the healthy control population (β = −0.066, P < .001). This is consistent with our hypothesis that the telomere length in patients with DC and BMF represents the minimal telomere length in mononuclear cells able to contribute to the peripheral blood cell pool.26 The lack of age-dependent telomere shortening in patients with DC was confirmed by Alter and colleagues in the patient population studied at the National Institutes of Health (NIH).27 We did not find a significant difference in telomere length whether the mutation was caused by a DKC1, TERT, or TERC gene mutation (P > .05). Five (14.7%) patients with DBA (Figure 3D), 1 (20%) with Shwachman-Diamond syndrome (SDS; Figure 3E), 3 (17.6%) with MDS (Figure 2F), 4 (16%) with PNH (Figure 2G), 16 (28.1%) with AA (Figure 2H), and one patient with FDP (Figure 2I) also had telomere length less than or equal to the 1st percentile. In one patient with MDS, a TERC gene mutation was identified (arrow in Figure 2F). Five TERT gene mutations were identified in 5 patients originally diagnosed with AA, and in 3, telomere length measurements were available (arrows in Figure 3H). Patients with AA or MDS and a TERC or TERT gene mutation showed a telomere length similar to those identified in patients with DC and BMF. Patients with AA and the G835A (A279T) or C1234T (H412Y) TERT gene variants, in contrast, showed a telomere length within the normal telomere length distribution consistent with our interpretation that these variants were not responsible for disease in these patients (stippled arrows in Figure 2H).
Telomere lengths in PBMC from patients with BMF. Telomere lengths in PBMC were measured by flow cytometric fluorescence in situ hybridization. (A) Telomere length in 234 healthy control subjects between the ages of 1 day and 94 years. The 1st, 5th, 25th, 50th, 75th, 95th, 99th percentiles of healthy controls are shown. (B) Telomere length in 160 patients with BMF from whom telomere measurements were obtained. (C) Telomere length in patients enrolled in the study with the diagnosis of DC. The red squares highlight patients with DC, while gray circles represent the remaining patients with BMF. (D) Telomere lengths in patients with DBA (highlighted in blue). (E) Telomere length in patients with SDS (highlighted in pink). (F) Telomere length in patients with MDS (highlighted in green). Arrows indicate patients in whom a pathogenic C35T TERC gene mutation was identified. (G) Telomere length in patients with PNH (highlighted in brown). (H) Telomere length in patients with aplastic anemia not otherwise classified (AA, highlighted in purple). Arrows indicate patients in whom a pathogenic TERT gene mutation was identified (from left to right; Y846C/H876Q, A716V, A716V). Stippled arrows indicate patients with the variant H412Y. (I) Telomere length in patients with other forms of IBMFS (Pearson syndrome, Fanconi anemia, and FPD, from left to right, highlighted in gold).
Telomere lengths in PBMC from patients with BMF. Telomere lengths in PBMC were measured by flow cytometric fluorescence in situ hybridization. (A) Telomere length in 234 healthy control subjects between the ages of 1 day and 94 years. The 1st, 5th, 25th, 50th, 75th, 95th, 99th percentiles of healthy controls are shown. (B) Telomere length in 160 patients with BMF from whom telomere measurements were obtained. (C) Telomere length in patients enrolled in the study with the diagnosis of DC. The red squares highlight patients with DC, while gray circles represent the remaining patients with BMF. (D) Telomere lengths in patients with DBA (highlighted in blue). (E) Telomere length in patients with SDS (highlighted in pink). (F) Telomere length in patients with MDS (highlighted in green). Arrows indicate patients in whom a pathogenic C35T TERC gene mutation was identified. (G) Telomere length in patients with PNH (highlighted in brown). (H) Telomere length in patients with aplastic anemia not otherwise classified (AA, highlighted in purple). Arrows indicate patients in whom a pathogenic TERT gene mutation was identified (from left to right; Y846C/H876Q, A716V, A716V). Stippled arrows indicate patients with the variant H412Y. (I) Telomere length in patients with other forms of IBMFS (Pearson syndrome, Fanconi anemia, and FPD, from left to right, highlighted in gold).
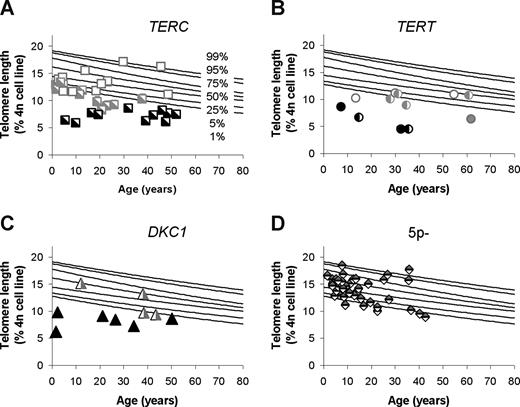
Telomere lengths in patients with DC and first-degree family members. (A) Patients with DC due to a TERC gene mutation. (B) Patients with DC due to a TERT gene mutation. (C) Patients with DC due to a DKC1 gene mutation. People with signs of BMF are shown in black; family members without BMF are shown in gray. Heterozygous mutation carriers are indicated with a half-filled symbol; filled symbol indicates homozygosity (hemizygosity/compound heterozygosity) for the mutation. (D) Telomere length in 41 subjects with a de novo TERT gene deletion due to a chromosome deletion syndrome that includes the TERT gene (5p- syndrome).35
Telomere lengths in patients with DC and first-degree family members. (A) Patients with DC due to a TERC gene mutation. (B) Patients with DC due to a TERT gene mutation. (C) Patients with DC due to a DKC1 gene mutation. People with signs of BMF are shown in black; family members without BMF are shown in gray. Heterozygous mutation carriers are indicated with a half-filled symbol; filled symbol indicates homozygosity (hemizygosity/compound heterozygosity) for the mutation. (D) Telomere length in 41 subjects with a de novo TERT gene deletion due to a chromosome deletion syndrome that includes the TERT gene (5p- syndrome).35
To further investigate the heredity of the disease-associated mutations and of telomere lengths, we next studied family members of patients with BMF in whom a TERC, TERT, or DKC1 mutation was identified. Figure 3 shows the telomere length in patients with BMF and their first-degree family members due to mutations in TERC (Figure 3A), TERT (Figure 3B), or DKC1 (Figure 3C). Telomere analysis in families with mutations in TERC, TERT, or DKC1 clearly show that, although in a patient population with BMF, short telomeres are a very sensitive indicator of patients with BMF due to DC, mutations in these genes were not always associated with disease or short telomeres. Furthermore, short telomeres were also found in some family members who have not inherited the mutation. Statistical analysis revealed that, although mutation carriers tended to have shorter telomere length than nonmutation carriers (10.77 vs 13.11), the difference was not statistically significant (P = .109).
Discussion
Patients with autosomal dominant DC caused by TERC or TERT gene mutations may present with clinical manifestations of BMF indistinguishable from patients with idiopathic AA. In our study, we were interested in determining the frequency of TERC and TERT mutations in patients with BMF, whether the identified mutations were associated with short telomeres, and whether the measurement of telomeres may be used to identify people with DC or mutation carriers in affected families. We found that a small number of patients with BMF have mutations in the TERC or TERT genes. In our patient population enrolled at WUMC, one of 133 BMF patients investigated carried a TERC gene mutation (0.8%), and 3 of 130 carried a TERT gene mutation (2.3%). Among patients referred to us from outside institutions, 3 of 67 carried a TERC mutation (4.5%), and 2 of 69 carried a TERT gene mutation (2.9%), reflecting an enrollment bias toward patients known or suspected to have BMF due to DC. These frequencies are comparable with those reported in a previous study from patients enrolled at the NIH.6 In patients with BMF, the presence of a pathogenic TERC or TERT gene mutation was associated with very short telomeres (well below the 1st percentile of telomere length found in healthy controls). The difference from normal was particularly pronounced in young people below the age of 40 years (Figure 2B). After the age of 50 years, due to age-dependent telomere shortening in the normal population, the difference of telomere lengths between healthy controls and patients with BMF and DC was less striking. There was no significant difference between the telomere lengths of patients with BMF due to TERC, TERT, or DKC1 mutations underlining the role of short telomeres in the pathogenesis of BMF in these people.
We identified a total of 4 different pathogenic TERC and 5 different pathogenic TERT gene mutations. Four patients with pathogenic TERC or TERT gene mutations were diagnosed with DC (n = 2) or suspected to have DC (n = 2) before enrollment, 4 were thought to have idiopathic AA, and one was diagnosed with MDS. The determination of pathogenicity of a particular mutation can be difficult. This is particularly true for TERC and TERT gene mutations because of the phenomenon of disease anticipation and because many of the mutations are hypomorphic, impairing but not abolishing telomerase activity. A statistically significant increase in the prevalence of the mutation in patients with disease, the segregation with clinical manifestations of the disease in the affected family, and a decreased telomerase activity in the in vitro assay are helpful supportive criteria in determining the pathogenicity of a sequence variant. In our study, we found that all patients with BMF and pathogenic TERC or TERT gene mutations had very short telomeres. In contrast, in patients with BMF and variants considered not to be pathogenic ‘G835A (A279T), C1234T (H412Y), or G3184A (A1062T)’, telomere lengths were within the normal distribution. Thus, in patients with BMF and a TERC or TERT sequence variant, short telomeres (≤ 1st percentile) seem to be a sine qua non criterion for the variant to be considered as pathogenic (responsible for BMF in the proband).
Telomere lengths less than or equal to the 1st percentile were not specific for BMF patients with TERC, TERT, or DKC1 mutations but were also observed in patients with DBA, SDS, PNH, and in patients with MDS or AA not otherwise classified. Replicative telomere erosion of a reduced number of stem cells or oxidative DNA damage preferentially occurring at the telomeres may account for short telomeres in some of these patients.45 Alternatively, in particular in patients with a telomere length similar to patients with DC, mutations in other genes associated with telomere maintenance may be responsible for the short telomeres and possibly for BMF.
Finally, we were interested in determining if telomere length measurements may be used to identify mutation carriers in families with DC. Family members of patients with DC due to DKC1 mutations have to be evaluated separately, because in female carriers circulating blood cells usually only express the normal allele. It is therefore interesting that, in peripheral mononuclear cells from female carriers, the telomere lengths were highly variable (Figure 3C). The clinical significance of a mutation and of telomere lengths in female DKC1 mutation carriers will be the subject of future studies. Telomere length analysis in families with TERC or TERT gene mutations demonstrated that mutation carriers with clinical signs of BMF had the shortest telomeres. However, telomere length was not a significant predictor for the presence or absence of a mutation in family members without BMF. Telomere length below the 1st percentile was observed in family members who did not inherit the mutation, suggesting these people have inherited the short telomeres, but not the disease-causing mutation. Likewise, normal telomeres were found in family members carrying the disease-causing mutation, suggesting these people have inherited a normal telomere length, and the mutation alone does not sufficiently shortened the telomeres in one generation to cause disease or excessively short telomeres. Alternatively, modifier genes may compensate for the loss in telomere length in these people. We have also shown previously, in our study of 41 subjects with a TERT gene deletion due to large chromosomal deletion, that telomere length measurements do not necessarily identify TERT gene mutation carriers. The majority (85%) of subjects missing one copy of TERT had a telomere length within the normal distribution (5p- syndrome, Figure 3D).35
In summary, our analysis shows that TERC and TERT gene mutations may account for BMF in a small but distinctive population of patients. The measurement of very short telomeres in patients with BMF is a sensitive albeit nonspecific method to identify these patients. In other words, in a patient with BMF, a normal telomere length excludes that he/she has BMF due to DC (high sensitivity). In contrast, the finding of short telomeres in a patient with BMF does not necessarily indicate that he/she has DC (low specificity). This agrees with a previous report in a more selected population of patients with inherited bone marrow failure syndromes.27 However, in contrast to that report, our results show that in the absence of BMF, telomere length measurement does not predict the presence or absence of a mutation in healthy family members. This is important to consider when telomere measurement is performed as a substitute for mutation analysis during the selection of a human leukocyte antigen (HLA)-matched sibling bone marrow donor or for the identification of mutation carriers in an affected family. Whether a sibling donor with no mutation but short telomeres or a sibling donor with a mutation but normal telomere length qualifies as a potential bone marrow donor remains to be determined. In the meantime, the absence of a genetic mutation and normal telomere length is probably the preferred scenario. Future long-term studies will determine the clinical importance of pathogenic mutations in the absence of short telomeres and the clinical significance of short telomeres in the absence of a pathogenic mutation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to all patients and their physicians for participating in our studies (http://bmf.im.wustl.edu). We are grateful to Paul Goodfellow and Diane Struckhoff from the Hereditary Cancer Core for the cancer-free controls and pyrosequencing and for their help in the enrollment of patients. We thank the Tissue Procurement Core for DNA extraction and sample storage, and the Genome Sequencing Center, in particular Rachel Maupin and Seth D. Crosby, for high-throughput sequencing and help in the data analysis.
Our studies are supported by National Institutes of Health grants CA105312 (to M.B. and D.B.W.) and CA106995 (to P.J.M.). The Hereditary Cancer and Tissue Procurement Cores are part of the Alvin J. Siteman Cancer Center (National Cancer Institute) Cancer Center Support Grant CA91842.
National Institutes of Health
Authorship
Contribution: M.B., H.Y.D., D.B.W., and P.J.M. designed the study; H.Y.D. and E.P. performed the experiments; H.Y.D. and M.B. analyzed the data; H.Y.D. and P.A. did the statistical analysis; M.B., D.B.W., R.T.M., U.M.R., D.C., A.S., A.V., J.M.L., R.K.G., and F.G. enrolled patients; J.I. coordinated enrollment of patients and families; H.Y. and M.B. wrote the paper; and P.J.M. and D.B.W. contributed to the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Monica Bessler, MD, PhD, Washington University School of Medicine, 660 South Euclid Avenue, Box 8125, St Louis, MO 63110; e-mail: mbessler@dom.wustl.edu.