Abstract
CCAAT enhancer-binding protein-epsilon (C/EBP-ϵ) is required for the terminal differentiation of neutrophils and eosinophils. Human C/EBP-ϵ is expressed as 4 isoforms (32, 30, 27, and 14 kDa) through differential RNA splicing, and alternative promoters and translational start sites. The C/EBP-ϵ32/30 isoforms are transcriptional activators, whereas C/EBP-ϵ27 interacts with and represses GATA-1 transactivation of eosinophil promoters. C/EBP-ϵ14 contains only DNA-binding and -dimerization domains and may function as a dominant-negative regulator. To define functional activities for these C/EBP-ϵ isoforms in myelopoiesis, human CD34+ progenitors were transduced with internal ribosomal entry site–enhanced green fluorescent protein retroviral vectors encoding the 32/30, 27, and 14-kDa isoforms, purified by fluorescence-activated cell sorter, and analyzed in colony-forming assays and suspension cultures. Progenitors transduced with C/EBP-ϵ32/30 default exclusively to eosinophil differentiation and gene expression, independent of interleukin-5, and regardless of inclusion of cytokines to induce other lineages. In contrast, the putative repressor C/EBP-ϵ27 isoform strongly inhibits eosinophil differentiation and gene expression, including GATA-1, promoting granulocyte (neutrophil)-macrophage differen-tiation. The C/EBP-ϵ14 repressor isoform strongly inhibits eosinophil development and gene expression, promoting erythroid differentiation, an effect enhanced by erythropoietin. Thus, C/EBP-ϵ isoforms can reprogram myeloid lineage commitment and differentiation consistent with their predicted activities based on activator and repressor domains and in vitro functional activities.
Introduction
Evidence from studies of both avian and human systems indicates that hematopoietic progenitor cell commitment and differentiation to the eosinophil lineage are transcriptionally regulated.1,2 The eosinophil developmental program requires the combinatorial activities of transcription factors that include GATA-1,3 CCAAT enhancer-binding protein-α (C/EBP-α),4 PU.1,2,5,6 and C/EBP-ϵ.7 C/EBP-ϵ, a member of the bZIP C/EBP family of transcription factors that includes C/EBP-α, -β, -γ, -δ, and -ζ,8 is preferentially expressed in granulocytes and required for the promyelocyte to myelocyte transition of terminally differentiating neutrophils and eosinophils.7,9,10 C/EBP-ϵ–null mice lack mature functional granulocytes (both neutrophils and eosinophils), and the mice die by 3 to 5 months of age because of opportunistic infections.7,11 Similarly, patients with a frame-shift mutation in the C/EBP-ϵ gene suffer from specific granulocyte deficiency disease, characterized by recurrent pyogenic infections, defective neutrophil chemotaxis, and bactericidal activity, and lack of neutrophil secondary granule proteins.12 Eosinophils in these patients also lack secondary granule proteins and are functionally impaired.13,14
In the mouse, C/EBP-ϵ is expressed as 2 activator isoforms of approximately 36 kDa and 34 kDa,15 whereas in humans, 4 distinct isoforms are expressed as proteins of 32, 30, 27, and 14 kDa through alternative splicing, differential promoter usage, and translational start sites.9,10,16,17 The human C/EBP-ϵ isoforms are all identical in sequence at their carboxyl terminus, which encodes the basic DNA-binding and leucine zipper (bZIP) domains.9,10 Human neutrophils and eosinophils express all 4 C/EBP-ϵ isoforms,2,9 with the highest levels of expression reported during the promyelocyte to myelocyte transition in both neutrophilic cell lines9,10,18,19 and neutrophil progenitors purified from human bone marrow.20 Whether eosinophil progenitors express all 4 C/EBP-ϵ isoforms has not been evaluated, but we previously reported that human eosinophil myeloblast (AML14) and myelocyte (AML14.3D10) cell lines express all the isoforms, whereas mature blood eosinophils express high levels of mainly the 14-kDa isoform.2 Although the C/EBP-ϵ32 and C/EBP-ϵ30 isoforms are weak transcriptional activators that interact with and require coactivators for full functional activity,7,21 the activities and combinatorial roles of the shorter 27-kDa and 14-kDa isoforms are unclear.10 Our transactivation studies in cell lines suggest they may function as transcriptional repressors of GATA-1 (C/EBP-ϵ27) or other C/EBPs (C/EBP-ϵ14).2,22
The full-length human C/EBP-ϵ32 and shorter ϵ30 isoforms contain well-defined transactivation, repression, DNA binding, and dimerization domains (Figure 1),10,16,23 but their function as transcriptional activators of myeloid promoters requires transcriptional cofactors, particularly c-myb.21 Cotransfection of c-myb with C/EBP-ϵ32 or C/EBP-ϵ30 in CV-1 cells cooperatively transactivates both the mim-1 and neutrophil-elastase promoters.21
Functional domains and sequence alignments of the C/EBP-ϵ isoforms. Functional domains and predicted transcriptional activities of the 4 C/EBP-ϵ isoforms (32, 30, 27, and 14 kDa), based on mutagenesis and transactivation studies of the human and murine full-length isoforms16,23,49,50 and shorter human isoforms,2,22 are shown schematically in panel A. Alignments of their amino acid sequences and locations of the various transactivation, repression, and bZIP domains are shown in panel B. All 4 isoforms are identical at their carboxyl terminus, which encodes the RDII repressor, basic DNA binding, and bZIP dimerization domains. The activator isoform C/EBP-ϵ32, a 281-amino acid protein, contains 2 transcriptional activation domains (TADI, TADII), 2 repressor domains (RDI, RDII), and the DNA-binding domain composed of the basic region (BR) and leucine zipper (LZ). C/EBP-ϵ30, a 250-amino acid protein, is derived from an alternative translation start site 100 bp (33 amino acids) downstream of the start site for C/EBP-ϵ32. C/EBP-ϵ27, a 253-amino acid protein derived through alternative RNA splicing, contains a unique 68-amino acid N-terminal repression domain we have designated as RD27.22 The shortest isoform, C/EBP-ϵ14, a 130-amino acid protein, consists mainly of the basic DNA binding and leucine zipper domains, the RDII repressor domain, with no transactivation domain. The RDI domain contains a highly conserved “VKEEP” sumoylation consensus sequence (boxed in panel B), through which sumoylation increases the transcriptional activity of the murine activator isoforms.23,49,50
Functional domains and sequence alignments of the C/EBP-ϵ isoforms. Functional domains and predicted transcriptional activities of the 4 C/EBP-ϵ isoforms (32, 30, 27, and 14 kDa), based on mutagenesis and transactivation studies of the human and murine full-length isoforms16,23,49,50 and shorter human isoforms,2,22 are shown schematically in panel A. Alignments of their amino acid sequences and locations of the various transactivation, repression, and bZIP domains are shown in panel B. All 4 isoforms are identical at their carboxyl terminus, which encodes the RDII repressor, basic DNA binding, and bZIP dimerization domains. The activator isoform C/EBP-ϵ32, a 281-amino acid protein, contains 2 transcriptional activation domains (TADI, TADII), 2 repressor domains (RDI, RDII), and the DNA-binding domain composed of the basic region (BR) and leucine zipper (LZ). C/EBP-ϵ30, a 250-amino acid protein, is derived from an alternative translation start site 100 bp (33 amino acids) downstream of the start site for C/EBP-ϵ32. C/EBP-ϵ27, a 253-amino acid protein derived through alternative RNA splicing, contains a unique 68-amino acid N-terminal repression domain we have designated as RD27.22 The shortest isoform, C/EBP-ϵ14, a 130-amino acid protein, consists mainly of the basic DNA binding and leucine zipper domains, the RDII repressor domain, with no transactivation domain. The RDI domain contains a highly conserved “VKEEP” sumoylation consensus sequence (boxed in panel B), through which sumoylation increases the transcriptional activity of the murine activator isoforms.23,49,50
The C/EBP-ϵ27 isoform contains a unique N-terminal (RD27) and RDI repressor domain (Figure 1),16 both of which we showed contribute to its inhibition of GATA-1 activity.2,22 GATA-1 is essential for the development of hematopoietic lineages, including erythrocytes, megakaryocytes, mast cells, and eosinophils, and GATA-1–null mice show impaired development of all these lineages at various levels of differentiation.3,24-27 Enforced retroviral expression of GATA-1 in hematopoietic progenitors leads to exclusive development of eosinophils in the presence or absence of interleukin 5 (IL-5),3 and transgenic deletion of a high affinity palindromic double GATA site in the HS-2 region of the murine GATA-1 promoter leads to a lineage-specific eosinophil deficiency.25 Our prior studies showed that C/EBP-ϵ27 (but not the other C/EBP-ϵ isoforms) specifically antagonizes GATA-1 transactivation of a hallmark eosinophil-specific promoter, the major basic protein-1 (MBP1) P2 promoter,2 and transduction of an eosinophil myelocyte cell line (AML14.3D10) with an HIV Tat–C/EBP-ϵ27 fusion protein potently inhibits endogenous MBP1 gene expression.22 The AML14.3D10 line expresses all of the C/EBP-ϵ isoforms and GATA-1, and coimmunoprecipitations with antibodies to GATA-1 or C/EBP-ϵ showed that C/EBP-ϵ27 physically interacts with GATA-1.2 C/EBP-ϵ27 is therefore hypothesized to be a potent repressor of GATA-1 activity, potentially impacting GATA-1–dependent eosinophil and possibly erythroid development and gene transcription.
The C/EBP-ϵ14 isoform contains only DNA-binding and bZIP-dimerization domains (Figure 1). It is hypothesized to function as a dominant negative repressor either as a heterodimer with other C/EBP family members (eg, C/EBP-ϵ32/30, C/EBP-α, or C/EBP-β) or by direct competition as a homodimer for C/EBP sites in granulocyte promoters.8 Our reports showing that C/EBP-ϵ14 inhibits transactivation of the eosinophil MBP1-P2 promoter by C/EBP-α and C/EBP-β in a dose-dependent manner2 and inhibits endogenous MBP1 gene transcription in AML14.3D10 eosinophils transduced with an HIV Tat–C/EBP-ϵ14 fusion protein22 support a repressor role for this isoform.
We provide novel evidence demonstrating that these human C/EBP-ϵ isoforms have distinct functions in myelopoiesis and are capable of altering myeloid lineage commitment and terminal differentiation when ectopically expressed in cord blood–derived human CD34+ hematopoietic progenitors.
Methods
Construction of retroviral expression vectors
C/EBP-ϵ32/30 was amplified from a pcDNA3 expression vector using a 5′ primer containing a NotI site and 3′ primer containing a BamHI site. The amplicon was digested and ligated into the pGCDnSam internal ribosome entry site–enhanced green fluorescent protein (IRES-eGFP) retroviral vector containing respective cohesive termini. The C/EBP-ϵ27 and C/EBP-ϵ14 cDNAs in the pcDNA3 vector were digested using HindIII, ends filled in with Klenow enzyme, and the inserts of 759 bp and 461 bp, respectively, excised using BamHI. The PGCDnSam IRES-eGFP vector was digested using NotI, blunted by filling in with Klenow enzyme, and digested with BamHI. Ligation of the inserts into the retroviral vector was performed using the Roche Rapid Ligation Kit (Roche Diagnostics, Mannheim, Germany). The C/EBP-ϵ isoform cDNA inserts were verified by restriction mapping and DNA sequencing.
Stable cell lines for production of retrovirus
The GP293 cell line was cotransfected with the C/EBP-ϵ isoform retroviral vectors along with VSV-G env gene (Clontech, Mountain View, CA) using the calcium phosphate method. The viral supernatants were used to transduce the mouse fibroblast retroviral packaging line PG13 (ATCC, Manassas, VA). Stably GFP+ transduced PG13 cells were sorted by fluorescence-activated cell sorter (FACS), and cells with an mean fluorescence intensity of more than or equal to 104 were frozen in aliquots at early passage. These PG13 cell lines were acclimatized to grow in Iscove modified Dulbecco medium, and expression of the C/EBP-ϵ isoforms was confirmed by Western blotting (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Retroviral transduction of CD34+ cells
The C/EBP-ϵ isoform PG13 stable lines were thawed and plated such that fresh viral supernatant could be collected for each CD34+ cell transduction. Non–tissue-culture treated 6-well plates (BD Biosciences, San Jose, CA) were coated with 8 μg/cm2 retronectin (Takara, Kyoto, Japan) according to the manufacturer's instructions. Plates were preloaded with filtered viral supernatant for 2 hours at room temperature. Cord blood (CB) CD34+ cells (> 97% pure) purchased from AllCells (Emeryville, CA) were prestimulated for 18 hours at 106 cells/mL in presence of 100 ng/mL recombinant human stem cell factor (rh-SCF), recombinant human fms-like tyrosine kinase 3 ligand (rh-Flt-3L), and recombinant human thrombopoietin (rh-TPO; R&D Systems, Minneapolis, MN). The cells were transduced repetitively 4 times in retronectin-coated plates every 16 hours in the presence of 50 ng/mL rh-SCF, rh-Flt-3L, and rh-TPO. After 72 hours, the cells were stained with anti–CD34-phycoerythrin (PE)–conjugated antibody or IgG1 isotype control (BD PharMingen, San Diego, CA) and sorted by FACS (MoFlo; Dako North America, Carpinteria, CA) based on dual expression of CD34 and GFP. These studies have Institutional Review Board approval from the University of Illinois at Chicago for obtaining and purifying CD34+ hematopoietic progenitors anonymously from umbilical cord blood.
CD34+ cell suspension cultures
CB CD34+ cells (AllCells), nontransduced or transduced with the C/EBP-ϵ isoforms or control retroviral vectors, were cultured in suspension at 0.3 × 106 cells/mL in Iscove modified Dulbecco medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 50 μM β-mercaptoethanol, 10 U/mL penicillin, 10 μg/mL streptomycin, and 2 mM l-glutamine. The CD34+ cells were differentiated toward eosinophils using SCF (50 ng/mL), Flt3-L (50 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF; 0.1 nM, only in experiments using nontransduced cells), IL-3, and IL-5 (0.1 nM) for the first 3 days. Thereafter, cells were cultured in IL-3 and IL-5 (0.1 nM) only, counted every 3 to 4 days, and maintained at 0.5 × 106 cells/mL. Double CD34+GFP+ transduced cells were cultured in IL-3 and IL-5 (0.1 nM) for 14 days, with fresh media containing cytokines added every 3 to 4 days.28 Cytokines were purchased from R&D Systems. Overexpression of the C/EBP-ϵ isoforms was confirmed by Western blotting (Figure S1B). Cytocentrifuge slides were stained with May-Grünwald Giemsa or Fast Green/Neutral Red for performance of differential cell counts (Figure S2).
Semiquantitative RT-PCR
For temporal assessments of C/EBP-ϵ isoform mRNA expression, total RNA was prepared using TRIzol reagent (Invitrogen) every 3 to 4 days from CD34+ cells differentiated toward eosinophils. The RNA was reverse-transcribed to cDNA using a Superscript cDNA Synthesis Kit (Invitrogen). Primer pairs for the C/EBP-ϵ isoforms were synthesized based on previous designs9 (Table S1). The optimal cycles for each primer pair were first determined using RNA from the HL-60 cell line, which expresses all 4 isoforms.2,9 All samples were analyzed in a predetermined linear dose-response range. Expression of β-2–microglobulin (β2M) was used as the control for equalizing cDNA inputs and normalizing the results for quantitation.29,30 The polymerase chain reaction (PCR) amplifications were done using α-32P-dCTP, reactions analyzed on native 4% or 6% polyacrylamide gel electrophoresis gels, and the dried gels analyzed by PhosphorImager (GE Healthcare, Little Chalfont, United Kingdom) using their ImageQuant software.
Quantitative real-time PCR
Quantitative reverse-transcriptase PCR (RT-Q-PCR) was performed using an iCycler iQ5 system (Bio-Rad, Hercules, CA) with iQSYBR supermix. All primer pairs are available in Table S1. Melting curves were performed to select primer sets that produced single peaks at high efficiencies. For all eosinophil-specific genes, cDNA from AML14.3D10 eosinophil myelocytes was used to generate standard curves for quantitation; for erythroid genes, GATA-1 and β-globin, cDNA from the erythroleukemia line K562 was used. For all primer sets, β2M was the internal control, and results are expressed as the ratio to β2M.
Hematopoietic colony assays
Double CD34+GFP+ cells transduced with the C/EBP-ϵ or control retroviral vectors were used to perform colony assays in Collagen Cult (StemCell Technologies, Vancouver, BC). The C/EBP-ϵ isoform or empty GFP vector-transduced cells were plated in duplicate or triplicate (750 CD34+GFP+ cells/chamber) and cultured using different cytokine cocktails to induce (1) mixed myeloid, (2) mixed myeloid plus erythroid, (3) eosinophil, (4) neutrophil, or (5) erythroid differentiation (Table 1). The slides were incubated under humidified conditions in a 5% CO2 incubator at 37°C for 14 days, dehydrated, fixed, and stained according to manufacturer's instructions. The transduced cells remain more than 95% GFP+ for the duration of these 14-day colony assays (Figure S3).
Cytokine cocktails for hematopoietic colony assays
| Hematopoietic lineage(s) . | Cytokine cocktail . |
|---|---|
| Mixed myeloid* | SCF, IL-3, GM-CSF, IL-5 |
| Mixed myeloid plus erythroid* | SCF, IL-3, GM-CSF, IL-5, EPO |
| Eosinophil† | SCF, IL-3, IL-5 |
| Neutrophil† | SCF, IL-3, G-CSF |
| Erythroid† | SCF, IL-3, EPO |
| Hematopoietic lineage(s) . | Cytokine cocktail . |
|---|---|
| Mixed myeloid* | SCF, IL-3, GM-CSF, IL-5 |
| Mixed myeloid plus erythroid* | SCF, IL-3, GM-CSF, IL-5, EPO |
| Eosinophil† | SCF, IL-3, IL-5 |
| Neutrophil† | SCF, IL-3, G-CSF |
| Erythroid† | SCF, IL-3, EPO |
SCF, IL-3, IL-5, GM-CSF (50 ng/mL), and EPO (5 U/mL).
SCF, IL-5 (50 ng/mL), G-CSF, IL-3 (10 ng/mL), and EPO (3 U/mL).
Histochemical and enzymatic staining
Differential colony counts were performed using May-Grünwald Giemsa (Sigma-Aldrich, St Louis, MO) (all myeloid colonies), benzidine (Acros Organics, Fairlawn, NJ) staining for erythroid colonies, Fast Green/Neutral Red (Fluka Chemical, Ronkonkoma, NY) for eosinophil colonies, naphthol AS-D chloroacetate esterase staining (Sigma-Aldrich) for granulocyte colonies, and α-naphthyl acetate esterase staining (Sigma-Aldrich) for monocyte colonies, all performed according to the manufacturer's suggestions.
Results
Expression of the C/EBP-ϵ isoforms during eosinophilopoiesis
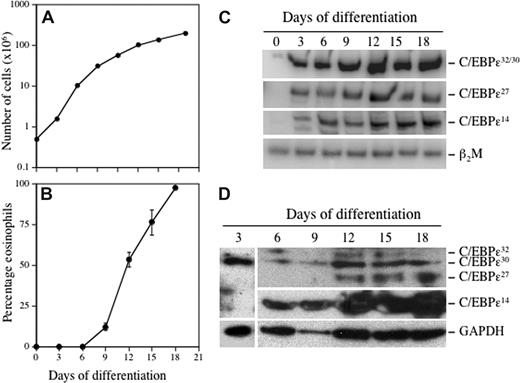
To determine whether there are temporal, developmentally regulated changes in the expression of the C/EBP-ϵ isoforms during eosinophil differentiation, CB CD34+ progenitors were cultured in suspension with the lineage-specific cytokine IL-5 (Figure 2). Eosinophil differentiation was monitored by staining cells with May-Grünwald/Giemsa, or Fast Green/Neutral Red that selectively stains mature secondary granules only in eosinophils (Figure S2). By day 21, more than 95% of cells had differentiated to eosinophils (Figure 2B). A low level of C/EBP-ϵ32/30 mRNA was detectable in undifferentiated CD34+ cells (day 0; Figure 2C), and this isoform was the most strongly induced of the 3 isoforms studied, increasing steadily through days 9 to 12, corresponding to the promyelocyte to myelocyte transition. The C/EBP-ϵ27 isoform mRNA was initially detected on day 3, with expression increasing and reaching a plateau somewhat later (days 12-18) than the 32-kDa isoform (Figure 2C). Expression of C/EBP-ϵ14 began on day 3, increasing steadily throughout the 18-day period (Figure 2C). Thus, all 3 C/EBP-ϵ isoform mRNAs are expressed concurrently during eosinophil differentiation, albeit at markedly different levels, with increasing levels of the repressor (C/EBP-ϵ27 C/EBP-ϵ14) isoforms relative to the activator (C/EBP-ϵ32) isoform as cells terminally differentiate. At the protein level (Figure 2D), expression of the C/EBP-ϵ32/30 activator, but not ϵ27 or ϵ14 repressor, isoforms was detected after 3 days of eosinophil differentiation, peaking around day 12, whereas expression of the C/EBP-ϵ27 repressor isoform began later on day 12 and continued to increase through day 18. The C/EBP-ϵ14 isoform was first expressed at day 6 and increased significantly throughout the duration of eosinophil differentiation. Of note, differentiating eosinophils expressed more of the 30-kDa than the 32-kDa activator isoform. These results indicate that the C/EBP-ϵ activator and repressor isoforms are expressed with different kinetics and levels in a developmentally regulated manner during eosinophil differentiation.
Temporal changes in expression of the C/EBP-ϵ isoforms during eosinophilopoiesis. CD34+ progenitors were differentiated to the eosinophil lineage by suspension culture in SCF, IL-3, IL-5, GM-CSF, and Flt3-L for 3 days, followed by only IL-3 and IL-5 thereafter. The cells were maintained at 0.5 × 106 cells/mL, total and eosinophil counts determined every 3 to 4 days, and total RNA for RT-PCR and total protein for Western blotting prepared from 1 × 106 cells. Cell proliferation (A) and the percentage of eosinophils (B) developing in the cultures based on differential cell counts using Fast Green/Neutral Red staining to distinguish secondary granule formation is shown. Semiquantitative RT-PCR was performed using α-32P-dCTP and C/EBP-ϵ isoform selective primers, with β2M amplified as the internal control for mRNA (cDNA) input (C). C/EBP-ϵ isoform protein expression was analyzed by Western blotting of whole cell lysates using a combination of anti–C/EBP-ϵ C-terminal (C-22; SC-158) and N-terminal (H-75; SC-25770) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), compared with GAPDH expression as the loading control (D). Representative results from 2 independent experiments are shown.
Temporal changes in expression of the C/EBP-ϵ isoforms during eosinophilopoiesis. CD34+ progenitors were differentiated to the eosinophil lineage by suspension culture in SCF, IL-3, IL-5, GM-CSF, and Flt3-L for 3 days, followed by only IL-3 and IL-5 thereafter. The cells were maintained at 0.5 × 106 cells/mL, total and eosinophil counts determined every 3 to 4 days, and total RNA for RT-PCR and total protein for Western blotting prepared from 1 × 106 cells. Cell proliferation (A) and the percentage of eosinophils (B) developing in the cultures based on differential cell counts using Fast Green/Neutral Red staining to distinguish secondary granule formation is shown. Semiquantitative RT-PCR was performed using α-32P-dCTP and C/EBP-ϵ isoform selective primers, with β2M amplified as the internal control for mRNA (cDNA) input (C). C/EBP-ϵ isoform protein expression was analyzed by Western blotting of whole cell lysates using a combination of anti–C/EBP-ϵ C-terminal (C-22; SC-158) and N-terminal (H-75; SC-25770) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), compared with GAPDH expression as the loading control (D). Representative results from 2 independent experiments are shown.
C/EBP-ϵ repressor isoforms alter erythroid and granulocyte-macrophage differentiation
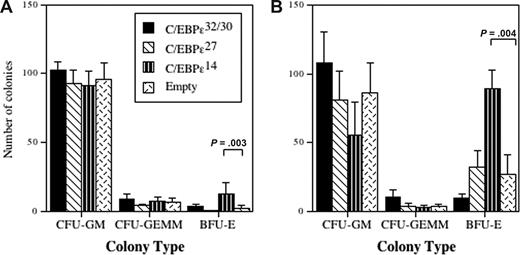
Both C/EBP-ϵ27 and C/EBP-ϵ14 are hypothesized to function as repressor isoforms based on their transactivation activities in heterologous cell lines,2 but their functions, especially in granulocyte development, have not been determined. We transduced CD34+ progenitors with IRES-eGFP retroviral vectors encoding each isoform, the transduced cells identified by GFP expression, and the CD34+GFP+ cells sorted and cultured in Collagen Cult media to assess their effects on mixed myeloid colony formation (Figure 3). Two different culture conditions were evaluated in which transduced cells were induced to differentiate toward mixed myeloid lineages except the erythroid lineage (Figure 3A) or toward both mixed myeloid and erythroid lineages (Figure 3B). In the absence of erythropoietin (EPO), there was increased erythroid colony formation (BFU-E) in cells transduced with the C/EBP-ϵ14 isoform compared with vector control (Figure 3A). Although C/EBP-ϵ27 decreased erythroid colonies in the absence of EPO, the difference from the vector control was not statistically significant. In contrast, when cells were driven to both mixed myeloid and erythroid lineages, there was a significant increase in erythroid colonies for cells transduced with the C/EBP-ϵ14 isoform (Figure 3B), but any inhibitory effect of C/EBP-ϵ27 was abrogated by the addition of EPO to drive erythroid differentiation. Under these multilineage conditions, C/EBPe32/30-transduced cells gave rise to equivalent numbers of GM colonies as the vector control. In contrast, enforced expression of C/EBP-ϵ14 favored differentiation toward the erythroid lineage, consistent with its suggested role as a negative regulator of C/EBPϵ32/30 and/or other C/EBPs required for granulocyte-macrophage development.2
Effects of the C/EBP-ϵ isoforms on myeloid and erythroid lineage colony formation. CD34+ CB progenitors were transduced with retroviral vectors encoding the C/EBP-ϵ isoforms and after 72 hours of transduction, CD34+GFP+ cells were sorted by FACS and cultured in semisolid Collagen Cult (StemCell Technologies) colony assays using cytokines to drive differentiation of mixed myeloid lineages excluding erythroid (A: culture in SCF, GM-CSF, IL-3, and IL-5), or both mixed myeloid and erythroid lineages (B: culture with SCF, GM-CSF, IL-3, IL-5, and EPO). The mean (± SEM) number of granulocyte-macrophage colony-forming unit (CFU-GM), granulocyte-erythrocyte-macrophage-megakaryocyte colony-forming unit (CFU-GEMM), and BFU-E colonies that developed from 750 plated CD34+GFP+ transduced progenitors is plotted for 3 or 4 independent experiments performed in duplicate or triplicate. Statistically significant differences of interest are shown (brackets and P values) for comparisons using 1-way analysis of variance (ANOVA) and least significant difference (LSD).
Effects of the C/EBP-ϵ isoforms on myeloid and erythroid lineage colony formation. CD34+ CB progenitors were transduced with retroviral vectors encoding the C/EBP-ϵ isoforms and after 72 hours of transduction, CD34+GFP+ cells were sorted by FACS and cultured in semisolid Collagen Cult (StemCell Technologies) colony assays using cytokines to drive differentiation of mixed myeloid lineages excluding erythroid (A: culture in SCF, GM-CSF, IL-3, and IL-5), or both mixed myeloid and erythroid lineages (B: culture with SCF, GM-CSF, IL-3, IL-5, and EPO). The mean (± SEM) number of granulocyte-macrophage colony-forming unit (CFU-GM), granulocyte-erythrocyte-macrophage-megakaryocyte colony-forming unit (CFU-GEMM), and BFU-E colonies that developed from 750 plated CD34+GFP+ transduced progenitors is plotted for 3 or 4 independent experiments performed in duplicate or triplicate. Statistically significant differences of interest are shown (brackets and P values) for comparisons using 1-way analysis of variance (ANOVA) and least significant difference (LSD).
Effects of the C/EBP-ϵ activator and repressor isoforms on granulocyte vs erythroid development
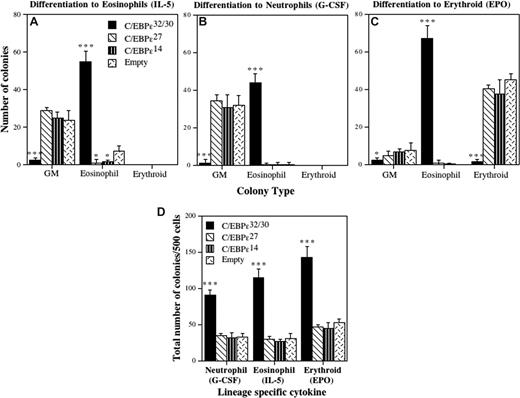
C/EBP-ϵ is required for the terminal differentiation of both eosinophils and neutrophils.7 To determine the activities of the individual C/EBP-ϵ isoforms in this process, CD34+GFP+ progenitors transduced with each isoform were plated in cytokine conditions favoring eosinophil (Figure 4A), neutrophil (Figure 4B), or erythroid (Figure 4C) differentiation, and colony counts performed after 14 days. For C/EBPϵ32/30, greater than 90% of the transduced cells differentiated toward eosinophils (Figure 4A). This marked induction of eosinophil colony formation was at the expense of GM colonies, including both neutrophil and neutrophil-macrophage colonies. The C/EBP-ϵ27 and C/EBP-ϵ14 isoforms both significantly inhibited eosinophil colony formation compared with control vector-transduced cells. Surprisingly, for transduced CD34+ progenitors induced to the neutrophil lineage (Figure 4B), the C/EBP-ϵ32/30 isoforms still induced greater than 90% eosinophil colonies despite the addition of granulocyte colony-stimulating factor (G-CSF). In contrast, for C/EBP-ϵ27 and C/EBP-ϵ14, there were no differences in the number of GM colonies that developed in G-CSF. For transduced cells differentiated toward the erythroid lineage (Figure 4C), greater than 90% of cells transduced with C/EBP-ϵ32/30 again developed into eosinophils, despite the presence of EPO, whereas C/EBP-ϵ27 and C/EBP-ϵ14 transduced cells both differentiated to erythroid colonies, with no difference in the number of BFU-E.
Effects of the C/EBP-ϵ isoforms on eosinophil, erythroid, and neutrophil differentiation. CD34+ CB progenitors were transduced with retroviral vectors encoding each of the C/EBP-ϵ isoforms. After 72 hours of transduction, CD34+GFP+ cells were sorted by FACS, plated in Collagen Cult colony assays, and the cells induced to differentiate toward the eosinophil (A: culture in SCF, IL-3, and IL-5), neutrophil (B: culture in SCF, IL-3, and G-CSF), and erythroid (C: culture in SCF, IL-3, and EPO) lineages. The mean (± SD) number of GM, eosinophil, and erythroid colonies that developed from 750 plated CD34+GFP+ transduced progenitors is plotted for 3 independent experiments performed in duplicate or triplicate. The effect of the C/EBP-ϵ isoforms on plating efficiency is shown in panel D as the total number of hematopoietic colonies (myeloid + erythroid) developed from 500 plated CD34+GFP+ progenitors for each of the cytokine cocktails used to induce neutrophil (G-CSF), eosinophil (IL-5), and erythroid (EPO) colony formation. Statistically significant differences are shown for comparisons using 1-way ANOVA and LSD (*P ≤ .05, ***P ≤ .001).
Effects of the C/EBP-ϵ isoforms on eosinophil, erythroid, and neutrophil differentiation. CD34+ CB progenitors were transduced with retroviral vectors encoding each of the C/EBP-ϵ isoforms. After 72 hours of transduction, CD34+GFP+ cells were sorted by FACS, plated in Collagen Cult colony assays, and the cells induced to differentiate toward the eosinophil (A: culture in SCF, IL-3, and IL-5), neutrophil (B: culture in SCF, IL-3, and G-CSF), and erythroid (C: culture in SCF, IL-3, and EPO) lineages. The mean (± SD) number of GM, eosinophil, and erythroid colonies that developed from 750 plated CD34+GFP+ transduced progenitors is plotted for 3 independent experiments performed in duplicate or triplicate. The effect of the C/EBP-ϵ isoforms on plating efficiency is shown in panel D as the total number of hematopoietic colonies (myeloid + erythroid) developed from 500 plated CD34+GFP+ progenitors for each of the cytokine cocktails used to induce neutrophil (G-CSF), eosinophil (IL-5), and erythroid (EPO) colony formation. Statistically significant differences are shown for comparisons using 1-way ANOVA and LSD (*P ≤ .05, ***P ≤ .001).
To determine whether transduction of CD34+ progenitors with the C/EBP-ϵ isoforms affected plating efficiency, we enumerated the total numbers of myeloid colonies (Figure 4D). Under all 3 cytokine conditions for differentiation toward eosinophil, neutrophil, and erythroid lineages (Figure 4A–C), the plating efficiency for progenitors transduced with C/EBP-ϵ32/30 (which develop exclusively to the eosinophil lineage) was significantly increased (∼2- to 3-fold) compared with C/EBP-ϵ27, C/EBP-ϵ14, and vector control (Figure 4D), indicating that C/EBP-ϵ32/30 increases the commitment, survival, and terminal differentiation of these cells toward eosinophils. Although C/EBP-ϵ32/30 increased plating efficiency for the transduced cells under all cytokine conditions, it did not increase eosinophil colony size (data not shown), indicating that it did not increase eosinophil progenitor cell proliferation. Likewise, the repressor C/EBP-ϵ27 and C/EBP-ϵ14 isoforms neither decreased nor increased myeloid or erythroid colony size (data not shown). Overall, these results demonstrate that enforced expression of the C/EBP-ϵ32/30 activator isoforms defaults progenitor cell differentiation to the eosinophil lineage at the expense of all other myeloid lineages, regardless of the presence of neutrophil or erythroid-specific cytokines, and maintains their proliferative capacity, whereas C/EBP-ϵ27 and C/EBP-ϵ14 isoforms both antagonize eosinophil differentiation, consistent with their in vitro and hypothesized repressor activities (Figure 1).
The C/EBP-ϵ32/30 activator isoforms induce rapid eosinophil differentiation independent of IL-5
Results thus far show that CD34+ progenitors transduced with the C/EBP-ϵ32/30 isoforms default to the eosinophil lineage despite the presence of other myeloid lineage-specific cytokines (Figure 4B,C), suggesting that IL-5 may be unnecessary. To determine whether IL-5 is required, we performed colony assays comparing the ability of C/EBP-ϵ32/30 and control vector-transduced CD34+GFP+ progenitors to differentiate to eosinophil or granulocyte-macrophage colonies in the presence vs absence of IL-5 (Figure 5). In the absence of IL-5, enforced expression of C/EBP-ϵ32/30 significantly increased the percentage of eosinophil colonies to approximately 100% (Figure 5A), as identified by their content of eosinophilic myelocytes containing Fast Green–positive secondary granules (Figure S4), and equivalent to the percentage of eosinophil colonies in the presence of IL-5 (Figure 5B), indicating that C/EBP-ϵ32/30 induces eosinophil differentiation independent of signaling by IL-5. Of note, in the absence of IL-5, enforced expression of C/EBP-ϵ32/30 also induced the development of more eosinophil colonies (∼75%) as early as 7 days relative to vector control transduced cells (Figure S5). Comparable results were obtained for C/EBP-ϵ32/30 transduced CD34+ progenitors cultured in suspension, which contained significantly increased numbers of eosinophilic myelocytes containing Fast Green–positive secondary granules at both 7 and 14 days of culture compared with vector control transduced cells (Figure S6). Because CD34+ progenitors do not normally develop into clearly discernable eosinophilic myelocytes until approximately 9 to 12 days of culture (eg, in Figures 2B, S2), these results indicate that C/EBP-ϵ32/30 not only supports the survival and proliferation of eosinophil progenitors but increases their rate of terminal differentiation.
C/EBP-ϵ32/30 enhancement of eosinophil differentiation does not require IL-5. CD34+ CB progenitors were transduced 3 times over a period of 72 hours with the retroviral vector encoding the C/EBP-ϵ32 activator isoform or empty vector control. CD34+GFP+ cells were sorted by FACS, and either plated in Collagen Cult colony assay media containing SCF and IL-3, and the cells allowed to differentiate for 14 days without (A) or with (B) IL-5 added to drive eosinophil colony formation. Eosinophil and GM colonies were enumerated using staining with Fast Green/Neutral Red and May-Grünwald Giemsa, respectively. The mean (± SD) number of CFU-Eos and CFU-GM colonies that developed from 750 CD34+GFP+ transduced progenitors is plotted for triplicate determinations (A,B).
C/EBP-ϵ32/30 enhancement of eosinophil differentiation does not require IL-5. CD34+ CB progenitors were transduced 3 times over a period of 72 hours with the retroviral vector encoding the C/EBP-ϵ32 activator isoform or empty vector control. CD34+GFP+ cells were sorted by FACS, and either plated in Collagen Cult colony assay media containing SCF and IL-3, and the cells allowed to differentiate for 14 days without (A) or with (B) IL-5 added to drive eosinophil colony formation. Eosinophil and GM colonies were enumerated using staining with Fast Green/Neutral Red and May-Grünwald Giemsa, respectively. The mean (± SD) number of CFU-Eos and CFU-GM colonies that developed from 750 CD34+GFP+ transduced progenitors is plotted for triplicate determinations (A,B).
C/EBP-ϵ27 and C/EBP-ϵ14 antagonize IL-5–induced eosinophil differentiation
To further characterize the activities of the C/EBP-ϵ isoforms on eosinophil differentiation and gene expression, the CD34+GFP+ transduced progenitors were differentiated to eosinophils with IL-5 in suspension cultures for 17 days (Figure S7). There was a 5-fold increase in the number of eosinophils in the C/EBP-ϵ32/30 transduced cells compared with vector control. In contrast, there were 5-fold and 3-fold decreases in eosinophils that developed from progenitors transduced with C/EBP-ϵ27 and C/EBP-ϵ14, respectively, consistent with their inhibition of eosinophil colony formation (Figure 4A). Similar to results from the colony assays, the proliferation rate of transduced CD34+ progenitors was not affected by any of the C/EBP-ϵ activator or repressor isoforms (data not shown).
The C/EBP-ϵ isoforms differentially activate or repress eosinophil gene expression
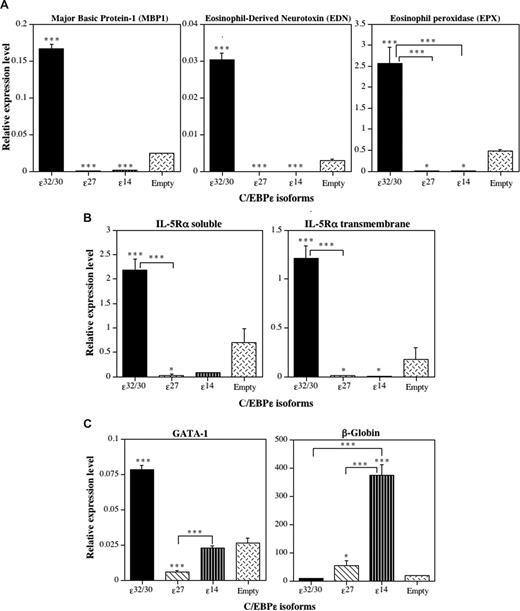
To determine whether the induction or repression of eosinophil differentiation by the C/EBP-ϵ isoforms is accompanied by activation or repression of eosinophil-specific gene transcription, we performed RT-Q-PCR analyses of RNA obtained from C/EBP-ϵ isoform-transduced CD34+GFP+ progenitors after 14 days in suspension culture with IL-3 plus IL-5 (Figure 6). Expression of mRNAs encoding a panel of C/EBP-regulated eosinophil genes, including the eosinophil-specific secondary granule proteins MBP1, eosinophil-derived neurotoxin (EDN), and eosinophil peroxidase (EPX), was analyzed by RT-Q-PCR, along with soluble and transmembrane isoforms of the IL-5–binding α-subunit of the high-affinity IL-5 receptor (IL-5Rα). Because we reported that C/EBP-ϵ27 antagonizes the activity of GATA-1,2 and the GATA-1 promoter contains a high-affinity “eosinophil-specific” autoregulatory GATA-1–binding site,25 we also assessed the effects of the C/EBP-ϵ isoforms on GATA-1 mRNA expression. Because C/EBP-ϵ14 promoted erythroid differentiation over granulocyte-macrophage development in the context of mixed-lineage choices, we also assessed β-globin expression as an erythroid marker. C/EBP-ϵ32/30 strongly induced the expression of mRNAs for all of the eosinophil secondary granule proteins analyzed (Figure 6A), the IL-5Rα subunit (Figure 6B), and the expression of GATA-1 itself (Figure 6C), consistent with its induction of eosinophil differentiation. In contrast, C/EBP-ϵ27 and C/EBP-ϵ14 both significantly inhibited expression of all eosinophil-specific secondary granule and IL-5Rα mRNAs analyzed (Figure 6A,B). Importantly, the C/EBP-ϵ27 isoform also significantly inhibited the expression of GATA-1 (Figure 6C), consistent with its potent inhibition of GATA-1 transactivation of eosinophil-specific promoters2,22 and GATA-1 autoregulatory activity in the eosinophil lineage.25 The C/EBP-ϵ14 isoform was also a potent inducer of β-globin gene expression (Figure 6C), consistent with its promotion of erythroid differentiation.
The C/EBP-ϵ isoforms differentially induce or inhibit eosinophil gene transcription. Analysis by real-time RT-Q-PCR. CD34+ CB cells were transduced 3 times with the retroviral vectors encoding the 3 C/EBP-ϵ isoforms and empty retroviral vector control over a period of 72 hours, double CD34+GFP+ progenitors sorted by FACS, grown for 14 days in suspension cultures supplemented with IL-3 + IL-5 to drive eosinophil differentiation (as in Figure 6), and total RNA prepared and reverse-transcribed to cDNA. The expression of genes encoding eosinophil secondary granule proteins, including major basic protein-1 (MBP1), eosinophil-derived neurotoxin/ribonuclease-2 (EDN, RNS2), and eosinophil peroxidase (EPX) (A), the soluble and transmembrane alternative RNA splice forms of the eosinophil-specific IL-5 receptor α (IL-5Rα) subunit (B), and GATA-1 and β-globin (C) were analyzed by RT-Q-PCR. The mean cDNA expression levels relative to the expression of the β2M input control in each sample amplified at the same time are plotted (± SD) for 3 independent experiments analyzed in triplicate. Statistically significant differences are shown for comparisons between means using 1-way ANOVA and LSD (*P ≤ .05, ***P ≤ .001).
The C/EBP-ϵ isoforms differentially induce or inhibit eosinophil gene transcription. Analysis by real-time RT-Q-PCR. CD34+ CB cells were transduced 3 times with the retroviral vectors encoding the 3 C/EBP-ϵ isoforms and empty retroviral vector control over a period of 72 hours, double CD34+GFP+ progenitors sorted by FACS, grown for 14 days in suspension cultures supplemented with IL-3 + IL-5 to drive eosinophil differentiation (as in Figure 6), and total RNA prepared and reverse-transcribed to cDNA. The expression of genes encoding eosinophil secondary granule proteins, including major basic protein-1 (MBP1), eosinophil-derived neurotoxin/ribonuclease-2 (EDN, RNS2), and eosinophil peroxidase (EPX) (A), the soluble and transmembrane alternative RNA splice forms of the eosinophil-specific IL-5 receptor α (IL-5Rα) subunit (B), and GATA-1 and β-globin (C) were analyzed by RT-Q-PCR. The mean cDNA expression levels relative to the expression of the β2M input control in each sample amplified at the same time are plotted (± SD) for 3 independent experiments analyzed in triplicate. Statistically significant differences are shown for comparisons between means using 1-way ANOVA and LSD (*P ≤ .05, ***P ≤ .001).
Ectopic overexpression of C/EBP-ϵ32/30 inhibits expression of the C/EBP-ϵ14 repressor
In analyses of C/EBP-ϵ32/30 overexpression in transduced CD34+ progenitors, we noted strongly inhibited expression of the endogenous C/EBP-ϵ14 repressor isoform (Figure 7). C/EBP-ϵ32/30 transduced CD34+GFP+ sorted cells, cultured for 14 days in IL-5 to induce eosinophil differentiation, showed a nearly complete loss of expression of the endogenous C/EBP-ϵ14 repressor isoform at both the mRNA (Figure 7A) and protein (Figure 7B) levels. These findings suggest that the expression level of the C/EBP-ϵ32/30 activator isoform may regulate alternative splicing to the C/EBP-ϵ14 repressor isoform, highlighting a potential mechanism contributing to the differentiation of C/EBP-ϵ32/30–transduced CD34+ progenitors exclusively to eosinophils.
Transduction of CD34+ progenitors with the C/EBP-ϵ32/30 activator isoforms blocks expression of the C/EBP-ϵ14 repressor isoform. CD34+ cells were transduced for 72 hours with the C/EBP-ϵ32/30 (lane 1) or empty GFP (lane 2) retroviral vectors and grown in suspension cultures supplemented with IL-5 to drive eosinophil differentiation. Total protein and RNA was prepared at 14 days from 106 cells lysed in TRIzol (Invitrogen) and analyzed by Western blotting (A) for expression of the C/EBP-ϵ32/30 and ϵ14 isoforms, or semiquantitative RT-PCR (B) for expression of the C/EBP-ϵ14 repressor isoform. Western blotting for GAPDH was used to control for equal protein loading (A), and PCR for β2M was used for comparison of cDNA inputs (B). Vertical lines have been inserted in panel A to indicate repositioned gel lanes.
Transduction of CD34+ progenitors with the C/EBP-ϵ32/30 activator isoforms blocks expression of the C/EBP-ϵ14 repressor isoform. CD34+ cells were transduced for 72 hours with the C/EBP-ϵ32/30 (lane 1) or empty GFP (lane 2) retroviral vectors and grown in suspension cultures supplemented with IL-5 to drive eosinophil differentiation. Total protein and RNA was prepared at 14 days from 106 cells lysed in TRIzol (Invitrogen) and analyzed by Western blotting (A) for expression of the C/EBP-ϵ32/30 and ϵ14 isoforms, or semiquantitative RT-PCR (B) for expression of the C/EBP-ϵ14 repressor isoform. Western blotting for GAPDH was used to control for equal protein loading (A), and PCR for β2M was used for comparison of cDNA inputs (B). Vertical lines have been inserted in panel A to indicate repositioned gel lanes.
Discussion
The human C/EBP-ϵ gene is expressed as 4 distinct isoforms through differential mRNA splicing, alternative translational start sites, and differential promoter usage (Figure 1A).9 The full-length isoform of 843 bp encodes a protein of approximately 34 kDa routinely referred to as C/EBP-ϵ32 in prior publications.10 The shorter C/EBP-ϵ30 activator isoform is generated through an alternative translational start site 100 bp downstream of the first ATG (Figures 1B, S1). Only C/EBP-ϵ27, which is generated by alternative RNA splicing, contains a unique N-terminal sequence (repressor domain) of 68 amino acids (RD27, Figure 1) shown in our prior studies to inhibit GATA-1 activity for the eosinophil-specific MBP1-P2 promoter.2,22 The shortest alternatively spliced isoform, C/EBP-ϵ14, consists mainly of the bZIP dimerization and basic DNA-binding domains (Figure 1) and was predicted to function as a dominant negative repressor of C/EBPs in a manner similar to the shorter isoforms of C/EBP-α31 and C/EBP-β (LIP),32 and can inhibit the activity of both C/EBP-α and C/EBP-β in transactivation assays.2
The current studies are the first to characterize individual functional activities in myeloid development for the human C/EBP-ϵ activator and repressor isoforms. Surprisingly, overexpression of the C/EBP-ϵ32/30 activator isoforms in CD34+ HSCs is capable of driving them exclusively to the eosinophil lineage at the expense of the neutrophil and other myeloid lineages in a cytokine (IL-5)–independent manner, regardless of the presence of other lineage-specific cytokines. In contrast, the C/EBP-ϵ27 and C/EBP-ϵ14 repressor isoforms are both capable of inhibiting CD34+ cell differentiation to the eosinophil lineage, even in the presence of IL-5 to drive eosinophil development. As demonstrated by RT-Q-PCR analyses, these findings are reflected in the induction (by C/EBP-ϵ32/30) or repression (by C/EBP-ϵ27 and C/EBP-ϵ14) of eosinophil-specific genes encoding secondary granule proteins and the IL-5 receptor, genes that uniquely define the eosinophil lineage. Under conditions allowing mixed lineage choices, the “dominant-negative” C/EBP-ϵ14 isoform is capable of inhibiting differentiation toward the granulocyte-macrophage lineages while promoting erythropoiesis, as demonstrated by marked increases in erythroid colonies and induction of GATA-1 and β-globin mRNA expression. The ability of the C/EBP-ϵ14 repressor isoform to drive progenitors to the erythroid lineage is consistent with results in zebrafish in which enforced ectopic expression of C/EBP-α dominant-negative isoforms, through deletion mutations of C/EBP-α (zD420), which mimic human dominant-negative mutations, induced primitive erythropoiesis with no discernible effect on granulopoiesis.33 C/EBP-ϵ14 may increase erythroid progenitor proliferation and differentiation by increasing the expansion of GATA-1–expressing committed erythroid progenitors, and because GATA-1 expression is autoregulated, may serve to expand this cell population.
Enforced overexpression of the C/EBP-ϵ32/30 activator isoforms in the context of endogenous expression of all the C/EBP-ϵ isoforms maintained the proliferation and potently induced the differentiation of CD34+ progenitors to the eosinophil lineage in the absence of IL-5, regardless of whether cytokines were added to drive the development of other lineages. These results suggest that early expression of C/EBPϵ32/30 in hematopoietic progenitors defaults their program exclusively to the eosinophil lineage. Importantly, it implies that subtle changes in the balance between the activator vs repressor isoforms of C/EBP-ϵ, eg, the increased expression of C/EBP-ϵ27 and C/EBP-ϵ14 relative to C/EBP-ϵ32/30 observed during eosinophil terminal differentiation (Figure 2D), may impact the outcome of their combinatorial activities in terms of gene transcription and granulocyte (eosinophil vs neutrophil) terminal differentiation. As well, our finding that eosinophils can differentiate from C/EBP-ϵ32/30–transduced CD34+ progenitors in the absence of IL-5 is consistent with findings for both IL-5 and IL-5Rα knockout (null) mice, in which basal eosinophilopoiesis proceeds normally in the bone marrow, but the mice fail to develop significant blood or tissue eosinophilia in response to allergic stimuli or parasitic infections.34,35 These results support the concept that basal eosinophilopoiesis occurs in a transcription factor-dependent but IL-5–independent manner and that IL-5 signaling may only be required for blood and tissue eosinophilia in response to innate or Th2-mediated allergic or antiparasite immune responses.36,37
Eosinophils are unique in their commitment and terminal differentiation from CMP (common myeloid progenitor) or GMP (granulocyte-macrophage progenitor) as they require combinatorial expression of transcription factors that include C/EBP-α, PU.1, GATA-1, and C/EBP-ϵ.1 Their developmental program is clearly different from that of neutrophils because GATA-1 is an absolute requirement for eosinophil development3 and a pivotal difference between these 2 granulocyte lineages.1,2 However, the role of C/EBP-ϵ has largely been considered ancillary to C/EBP-α, which regulates its expression in granulocyte development.38 All 4 human C/EBP-ϵ isoforms are expressed early and concurrently during granulopoiesis in the bone marrow,20 including during eosinophilopoiesis (this article). However, both the timing and expression levels of the activator and repressor isoforms vary significantly during IL-5–induced eosinophil differentiation (Figure 2), such that their expression ratios and combinatorial interactions may serve to finely regulate one another's transcriptional activities, the activities of other C/EBPs, for example, C/EBP-α or C/EBP-β (C/EBP-ϵ14), or GATA-1 (C/EBP-ϵ27),2 or serve to down-regulate secondary granule protein gene expression during terminal granulocyte differentiation.39 Consistent with the latter, expression of the C/EBP-ϵ27 and -ϵ14 repressor isoforms increases significantly during eosinophil terminal differentiation (Figure 2), and we previously reported that mature blood eosinophils continue to express very high levels of mainly the C/EBP-ϵ14 repressor isoform.2
There are several possible mechanisms by which overexpression of the C/EBP-ϵ32/30 isoforms in CD34+ progenitors exclusively induces eosinophil differentiation. First, our results (Figure 2) indicate that developing eosinophil progenitors express the C/EBP-ϵ30 > C/EBP-ϵ32 activator isoform. One possibility is that the ratio of the ϵ32 to ϵ30 activator isoforms differentially specifies neutrophil (ϵ32 > ϵ30) vs eosinophil (ϵ30 > ϵ32) gene expression and development. Whether and how eosinophil progenitors preferentially use the downstream translational start site for expression of the ϵ30 > ϵ32 isoform requires further investigation. Second, overexpression of the C/EBPϵ32/30 isoforms in this study was at a much earlier time point (in undifferentiated CD34+ progenitors) relative to their endogenous expression mainly at the promyelocyte to myelocyte transition. Overexpression of these activator isoforms early in CD34+ hematopoietic progenitors, when low levels of other key transcription factors such as GATA-1 are also expressed and required for eosinophil, but not neutrophil, differentiation (which requires switching off GATA-1), may also impact the ability of the C/EBP-ϵ32/30 isoforms to drive eosinophil development exclusively at the expense of neutrophil differentiation. A third possibility is that CD34+ progenitors transduced with the C/EBP-ϵ32/30 isoforms showed nearly complete inhibition of endogenous C/EBP-ϵ14 repressor expression, both protein and mRNA (Figure 7), an effect we have reproduced in an eosinophil-committed cell line (AML14) stably transduced with the same C/EBP-ϵ32/30 retroviral vector (data not shown). Inhibited expression of the “dominant negative” C/EBP-ϵ14 repressor could free up C/EBP sites for the C/EBP-ϵ32/30 activator isoforms, driving eosinophil gene expression and terminal differentiation. C/EBP-ϵ32/30 has been shown to physically and functionally interact with and require c-myb for the activation of granulocyte genes,21 and c-myb was recently reported to affect pre-mRNA 5′-splice site selection through interaction with the spliceosome.40 The importance of c-myb to eosinophil development is highlighted by a report in which a c-myb point mutation (M303V) in the mouse leads to ablation of the eosinophil lineage.41 Thus, interaction of c-myb with C/EBP-ϵ32/30 might lead to inhibition of pre-mRNA splicing to the C/EBP-ϵ14 isoform, further contributing to C/EBP-ϵ32/30 induction of the eosinophil differentiation program.
The C/EBP-ϵ32 activator isoform is suggested to play a role in taking proliferating myeloid progenitors out of cell cycle, inducing them to terminally differentiate.42,43 However, our enforced overexpression of C/EBP-ϵ32/30 in CD34+ hematopoietic progenitors did not induce cell-cycle arrest because they continued to proliferate at the same rate throughout the duration of their differentiation to eosinophil myelocyte and metamyelocyte stages in suspension culture. As well, we anticipated that CD34+ cells transduced with C/EBP-ϵ32/30 would drop out of cell cycle, terminally differentiate, and undergo apoptosis, inhibiting significant eosinophil colony formation. To the contrary, C/EBP-ϵ32/30 increased both the commitment and survival of eosinophil progenitors as evidenced by increased plating efficiency, numbers of eosinophil colonies, and continued proliferation in suspension cultures. Actively cycling HSCs express low levels of p27,44-47 and C/EBP-ϵ32 was reported to induce cell-cycle arrest and differentiation by interacting with p27 and inhibiting E2F regulated gene transcription,43 and in myeloid cell lines by up-regulating p27 with concomitant down-regulation of cdk4/6 and cyclin D2/A/E, a process that requires the N terminus of C/EBP-ϵ.42 Ectopic expression of C/EBP-ϵ32/30 early in HSCs or committed progenitors may enable their continued proliferation before inducing terminal differentiation exclusively to eosinophils. The lack of an antiproliferative effect of C/EBP-ϵ32/30 in early hematopoietic progenitors may therefore be context dependent, similar to C/EBP-β, which was shown to enable continued proliferation of granulocyte (neutrophil) progenitors before inducing their terminal differentiation as part of emergency granulopoiesis.48
In conclusion, enforced ectopic expression of the C/EBP-ϵ isoforms in CD34+ hematopoietic progenitors was able to reprogram their myeloid lineage choice decisions and terminal differentiation in a manner consistent with their predicted activator (C/EBP-ϵ32/30) vs repressor (C/EBP-ϵ27 and C/EBP-ϵ14) roles, and their interactions with other hematopoietic transcription factors, such as GATA-1 or other C/EBP family members. Studies defining their combinatorial interactions during granulopoiesis in hematopoietic progenitors should further inform our understanding of their roles in eosinophil versus neutrophil development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Monica Stankiewicz for her initial studies in the laboratory that first identified the repressor activities of C/EBP-ϵ27 for GATA-1 and mapped the repressor domains22 ; Drs Kleanthis Xanthopoulos and Julie Lekstrom-Himes (National Institutes of Health ‘NIH’) for providing the C/EBP-ϵ isoform cDNAs in the pcDNA3 expression vector; Dr Atsushi Iwama (University of Tokyo, Tokyo, Japan) for providing the retroviral bicistronic vector pGCDnSam IRES eGFP; Dr Nissim Hay (University of Illinois at Chicago ‘UIC’) for providing the GP293 cell line; Dr Bettina Moser, Dr Tiffany Sharma, and Laura Periera for assistance with real-time PCR; Dr Amittha Wickrema for the benzidine staining protocol; Jewell Graves and Dr Karen Hagen at the UIC Research Resource Center for performing the flow cytometry and sorting of CD34+/GFP+ transduced cells; Dr Miranda Buitenhuis for helpful discussions and provision of cytokine conditions for culturing transduced CD34+ cells to eosinophils; and the technical support staff at Stem Cell Technologies for assistance in establishing the Collagen Cult hematopoietic colony assay method.
This work was supported by the NIH (grant AI33043; S.J.A.). R.B. was supported in part by an Institutional T32 training grant (Training Program in Signal Transduction and Cellular Endocrinology, T32 DK07739) from the NIDDK/NIH (S.J.A.).
National Institutes of Health
Authorship
Contribution: R.B. designed and performed all of the research, graphed and analyzed the data, and wrote the paper; this work was performed by R.B. in partial fulfillment of the requirement for her PhD; J.D. designed and characterized PCR primers for the IL-5Rα and C/EBP-ϵ isoforms, performed initial experiments on expression in CD34+ progenitors, provided assistance in methods development, and critically reviewed the manuscript; A.K.S. provided assistance in experimental design, generation of stable cell lines expressing retroviral vectors, FACS purification of transduced cells, and critical review of the manuscript; I.G. provided assistance in experimental design, statistical analyses, and critical review of the manuscript; and S.J.A. supervised the conceptualization, design, and performance of all of the research, methods, data analysis, and the writing, editing, critical review, and submission of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven J. Ackerman, Department of Biochemistry and Molecular Genetics (M/C 669), University of Illinois at Chicago, Room 2074 MBRB, 900 S Ashland Avenue, Chicago, IL 60607; e-mail: sackerma@uic.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal