Abstract
Chromosomal translocations involving the Mixed Lineage Leukemia (MLL) gene produce chimeric proteins that cause abnormal expression of a subset of HOX genes and leukemia development. Here, we show that MLL normally regulates expression of mir-196b, a hematopoietic microRNA located within the HoxA cluster, in a pattern similar to that of the surrounding 5′ Hox genes, Hoxa9 and Hoxa10, during embryonic stem (ES) cell differentiation. Within the hematopoietic lineage, mir-196b is most abundant in short-term hematopoietic stem cells and is down-regulated in more differentiated hematopoietic cells. Leukemogenic MLL fusion proteins cause overexpression of mir-196b, while treatment of MLL-AF9 transformed bone marrow cells with mir-196–specific antagomir abrogates their replating potential in methylcellulose. This demonstrates that mir-196b function is necessary for MLL fusion-mediated immortalization. Furthermore, overexpression of mir-196b was found specifically in patients with MLL associated leukemias as determined from analysis of 55 primary leukemia samples. Overexpression of mir-196b in bone marrow progenitor cells leads to increased proliferative capacity and survival, as well as a partial block in differentiation. Our results suggest a mechanism whereby increased expression of mir-196b by MLL fusion proteins significantly contributes to leukemia development.
Introduction
The Mixed Lineage Leukemia (MLL) gene is commonly involved in chromosome translocations that cause leukemia.1,2 MLL-associated leukemias can be myeloid, lymphoid or biphenotypic, depending on the partner gene to which MLL is fused.3 There have been more than 60 different MLL fusion partners isolated to date and, in most cases, overexpression of a subset of HOX genes is a hallmark of the disease.4 HOX genes are transcription factors that play an important role during development and hematopoiesis.5,6 Humans have 13 paralogous groups of HOX genes clustered on 4 different chromosomes. Expression of HOX genes is spatially and temporally regulated with 3′ genes expressed earlier and having a more anterior boundary of expression.5 Similarly, expression of HOX genes is tightly regulated during hematopoiesis. Genes located at the 3′ end of the cluster are down-regulated as CD34+ cells become lineage-specific progenitors while 5′ genes, like HOXA10, are turned off only after cells progress to the more differentiated CD34− stage.7 MLL regulates expression of some of the HOX genes at the chromatin level by binding to the promoters and recruiting various transcriptional regulators.8-10 However, what happens at the molecular level in the presence of the leukemogenic fusion proteins to cause HOX overexpression is still poorly understood.
Among 6800 genes analyzed by expression microarrays, overexpression of HOXA9 was the most correlative marker of poor prognosis in acute myeloid leukemia patients.11 Immortalization of bone marrow progenitors by the MLL fusion protein MLL-ENL is dependent on the presence of Hoxa9 and Hoxa7genes.12 Like other HOX genes, the HOXA9 locus gives rise to several alternatively spliced transcripts.13-15 The precise role of each one of these transcripts is still unclear, but some of them do not code for proteins and are more likely to have a strictly regulatory role.
MicroRNAs (miRNAs) are small RNA transcripts (∼ 22 nt) that function in posttranscriptional regulation of gene expression. The mature miRNAs are processed from much longer primary precursor molecules through a multistep process that involves several proteins. MiRNA precursors are transcribed by RNA polymerase II and initial processing is performed in the nucleus by Drosha. Subsequent transport into the cytoplasm and additional processing by Dicer and helicase give rise to the mature miRNA that is incorporated into the RNA-induced silencing complex (RISC), guiding it to the target mRNAs.16 It is now clear that miRNAs play an important role during development and disease and many of the genes encoding microRNAs are found at sites frequently deleted or mutated in cancers.17 While the role of miRNAs in various systems is starting to be elucidated, the mechanism by which their expression is regulated is still poorly understood.
We report here the regulation of a miRNA, mir-196b, by wild-type MLL as well as its dramatic overexpression by leukemic MLL fusion proteins. Mir-196b is located in a highly evolutionarily conserved region between HOXA9 and HOXA10 genes, at chromosome band 7p15.2 in human and 6qB3 in mouse. Our studies indicate that MLL regulates mir-196b expression in a pattern similar to that of the surrounding genes. Mir-196b is overexpressed specifically in primary leukemia samples from MLL patients, but not from other types of leukemia. Furthermore, expression of MLL fusion proteins in primary bone marrow cells causes overexpression of mir-196b. Treating these cells with specific antagomir targeting mir-196b significantly compromises the increased replating potential of MLL-AF9–expressing cells. Mir-196b expression increases proliferation and survival, and also partially blocks differentiation of normal bone marrow hematopoietic progenitor cells. Overexpression of mir-196b is an important component in leukemia development caused by MLL fusion proteins.
Methods
Cell lines
miRNA detection
RNA was isolated using TRI Reagent (Sigma-Aldrich, St Louis, MO) according to the manufacturer's protocol. Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Real-time PCR was performed in triplicate on an ABI 7300 machine using TaqMan analysis according to the manufacturer's instructions (miR196b probe set 000496, normalized to RNU6B probe set 001093; Applied Biosystems).
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay (ChIP) was performed using the EZ-ChIP Kit (Upstate/Millipore, Billerica, MA) according to the manufacturer's protocol. Chromatin was immunoprecipitated using anti–dimethyl H3 K79, anti–dimethyl H3 K4, anti–trimethyl H3 K4, and anti–dimethyl H3 K9 (Upstate/Millipore) and analyzed by qPCR in triplicate on an ABI 7300 machine using iTaq SYBR Green Supermix with Rox (Bio-Rad, Hercules, CA). Percentage enrichment was normalized to input DNA and calculated as previously reported.22 Amplification of the AB region was performed with primers previously described.21
Embryonic stem cell differentiation and isolation of CD41+ cells
Embryonic stem (ES) cells were maintained, differentiated, and day 10 embryoid bodies collected as described previously.23 Isolation of hematopoietic progenitors was performed using anti-CD41 antibody (BD Pharmingen, San Jose, CA). Cells not bound by the nanoparticles were collected as CD41− population.
Bone marrow cell sorting
Bone marrow (BM) was harvested from wild-type C57BL6/J mice using a Loyola University Institutional Animal Care and Use Committee–approved protocol. After red blood cell lysis, BM nucleated cells were dispersed into a single-cell suspension. The long-term hematopoietic stem cells (HSCs;Lin−Sca1+C-kit+Flk2−, LSKF−), short-term HSCs (Lin−Sca1+C-kit+Flk2+, LSKF+), and the committed progenitors (Lin−Sca1−C-kit+, LK) were enriched by lin+ cell depletion (EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit; StemCell Technologies, Vancouver, BC), and purified by FACSAria flow cytometer (BD Biosciences, San Jose, CA) sorting after fluorescein isothiocyanate–conjugated lineage (FITC-Lin) cocktail, phycoerythrin (PE)–Sca1, APC-c-kit and PE-Cy5.5-Flk2 staining. Gr1+ myeloid cells and B220+ B cells were sorted from BM cells after FITC-Gr1 and APC-B220 staining. All fluorescent antibodies used were purchased from eBioscience (San Diego, CA).
RT-PCR and qRT-PCR
RT-PCRs were performed in a Hybaid PCR Express machine. Each 50 μL reaction contained 1.5 mM MgCl2, 100 ng of each primer, 0.125 mM dNTP mix, 5 μL 10× PCR buffer (Fermentas, Glen Burnie, MD), and 2.5 units Taq DNA polymerase (GenScript, Piscataway, NJ). PCR products were analyzed with ethidium bromide by electrophoresis on a 2% agarose gel.
For quantitative RT-PCR, cDNA was analyzed using ABI Prism 7300. For SYBR Green, each 20 μL reaction contained 2 μL 10× iTaq SYBR Green Supermix with Rox (Bio-Rad) and 0.25 μM forward and reverse primers. All reactions were performed in triplicate and gene expression levels were normalized to hprt expression and fold difference calculated using the 2−ΔΔCt method. Primer sequences are available on request.
Cloning mir-196b expression vector
A 200-bp DNA fragment surrounding mir-196b was amplified using the primers: forward 5′ GAAGATCTTTCCTTGGCGGCGACA 3′ and reverse 5′CCCAAGCTTGATGGCCCGCCTA 3′. Gel-purified PCR product and vector were both digested with HindIII and BglII and ligated into pSuper.retro vector (OligoEngine, Seattle, WA).
Retrovirus production and colony assay
Retroviruses were produced as described previously.24 Bone marrow colony assays were performed as we have previously described with minor changes.24 Briefly, bone marrow cells were collected from 4- to 12-week-old C57Bl6 mice and c-kit–positive progenitors were selected using an Easy Sep Selection kit (StemCell Technologies). Lineage-depleted cells were separated using the Negative Selection Kit (StemCell Technologies). After the second infection, cells were plated in methylcellulose with cytokines interleukin-6 (IL-6), IL-3, stem cell factor (SCF), and (±) granulocyte macrophage-colony stimulating factor (GM-CSF), in the presence of puromycin (8.75 μg/mL). Cytospin slides of colony assay cells were stained with the Hema 3 Stain Set (Fisher Scientific, Kalamazoo, MI). Pictures were taken on an Olympus BH-2 microscope (Tokyo, Japan), under a 100×/1.25 NA oil-immersion lens, with a Sony 3CCD camera, model DXC-76OMD. Images were acquired with Adobe Premier software, version 4.2.1. Pictures of colonies in methylcellulose were taken through air on a Leica model DMIL microscope (Wetzlar, Germany), through a 4×/0.10 NA lens, with a Canon PowerShot S40 digital camera. Images were acquired with Canon ZoomBrowser EX, version 8. All images were processed using Adobe Illustrator CS3, version 13.0.0.
Antagomir treatment of MLL-AF9–transduced bone marrow cells
Antagomir oligoribonucleotides were synthesized (Dharmacon, Lafayette, CO; 2′-O-Me on each ribonucleotide, phosphorthioate on the first 2 and last 4 ribonucleotides, and a 3′ cholesterol modification linked through a hydroxyprolinol linkage; miR-196, 5′-CCCAACAACAGGAAACUACCUA-3′; control (mutant) miR-196, 5′-CCCAAGAACAGGUA AGUACGUA-3′). Underlined letters in the sequences represent mutated nucleotides. Wild-type Lin− bone marrow cells were treated with 100 nM antagomir. To confirm efficacy, RNA was extracted after 72 hours using TriZol (Sigma-Aldrich), then analyzed for steady-state mature miR-196b expression. The level of mature miR196b was not detectable by TaqMan assay after antagomir treatment. For colony assay, wild-type Lin− bone marrow cells were transduced with an MLL-AF9 expressing retroviral vector and treated with 100 nM control antagomiR-196 or antagomiR-196 before 15 000 cells were plated in duplicate in methylcellulose medium containing IL-3, IL-6, and SCF (M3534; StemCell Technologies). Colonies were enumerated after 8 days of culture and the average number plotted (± SD). For serial replating, colonies were pooled and 15 000 cells were treated with control antagomiR or antagomiR-196b and replated.
Bead-based miRNA expression profiling assay
A large-scale, genome-wide miRNA expression profiling analysis was performed using a bead-based flow cytometric method.25 A total of 55 acute myeloid leukemia (AML) primary patient samples, including 29 MLL-associated acute leukemias (10 acute lymphocytic leukemias [ALLs] and 19 AMLs) and 26 AMLs with other abnormalities, along with 3 normal bone marrow controls were included in the expression assay. To control for data quality, only samples with total miRNA signals greater than or equal to 15 000 were analyzed further. All patient samples were collected with approval of the Institutional Review Board at the University of Chicago and after informed consent was obtained in accordance with the Declaration of Helsinki. After normalization, only probes for human miRNAs with maximum expression in any sample being greater than or equal to 7.25 were retained for further analyses. TIGR Multiple Array Viewer software package (TMeV version 4.0; The Institute for Genetic Research [TIGR], Rockville, MD) was used to perform data analysis and to visualize the results.26 The details of this assay were reported elsewhere.27
Myeloid and B-cell differentiation assay
Bone marrow cells harvested after 1-week culture in the methylcellulose colony assay were used for the differentiation assays. Totals of 35 000 mir-196b or vector-infected cells were plated on 20 000 OP9 cells per well to a 24-well dish in RPMI, 20% fetal bovine serum (FBS), 1% Pen/Strep. Wells were supplemented either with 10 ng/mL GM-CSF (myeloid) or 10 ng/mL IL-7 and 10 ng/mL FLT3L (B cell). After 5 days, cells were stained with allophycocyanin (APC)–conjugated CD117 and PE-conjugated CD11b or PE-conjugated B220 antibodies. Percentage of cells expressing these surface markers was determined using flow cytometry.
Results
MLL-dependent expression of mir-196b
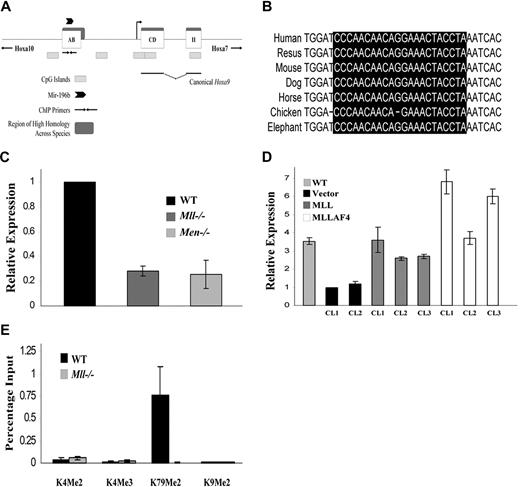
MLL is a known regulator of HOX gene expression. We have recently described expression of an Mll-dependent transcript from a region highly conserved across species located approximately 4.5 kb upstream of the canonical Hoxa9 promoter.21 Mll protects this DNA region from becoming methylated, thus allowing expression of the transcript. Mir-196b is located in the center of the conserved region and we hypothesized that the transcript encodes the precursor of mir-196b (Figure 1A,B). Our initial approach used ligation-mediated PCR to isolate the mature form of mir-196b (data not shown). While our method was successful, we were not able to determine the expression levels quantitatively. To overcome this problem, we used a microRNA assay specific for mir-196b that uses TaqMan probe chemistry, thus allowing for quantitative measurement. Analysis of mouse embryonic fibroblasts (MEFs), wild-type and null for Mll, showed a 3.5-fold higher level of expression of mir-196b in the presence of Mll (Figure 1C). As previously reported and confirmed in our laboratory (data not shown), canonical Hoxa9 transcript expression shows a similar Mll dependency.18 Dependence of mir-196b expression on MLL was further confirmed by exogenous expression of wild-type MLL in Mll−/− MEFs. MLL restored expression of mir-196b in these cells (Figure 1D).
Expression of mir-196b is dependent on Mll and Menin. (A) Schematic illustration of the murine Hoxa9 locus on chromosomal band 6qB3. Open boxes represent 3 exons and gray boxes indicate CpG islands. Exon II is the homeodomain (HD) containing exon. The canonical Hoxa9 is encoded by exons CD and II. Regions of high homology between species are noted as dark boxes and the region amplified by ChIP primers is indicated by arrows. The location of mir-196b is labeled above exon AB with an arrowhead. (B) Sequence alignment of the conserved mir-196b sequence among different species. The mature mir-196b sequence is highlighted in black. (C) Quantitative RT-PCR of mir-196b levels in WT, Mll−/−, and Men−/− MEFs. The experiment was run in triplicate and the results represent average expression plus or minus SD. (D) Quantitative RT-PCR of mir-196b expression in individual clones expressing an empty vector, MLL, or MLLAF4 in Mll−/− MEF background. Transfection of Mll−/− MEFs and clone selection was performed as previously described. Mir-196b expression was determined from total RNA. (E) ChIP assay performed in WT and Mll−/− MEFs. Chromatin was precipitated using indicated antibodies and qPCR performed with primers spanning mir-196b region. All samples were run in triplicate and were normalized to the input chromatin.
Expression of mir-196b is dependent on Mll and Menin. (A) Schematic illustration of the murine Hoxa9 locus on chromosomal band 6qB3. Open boxes represent 3 exons and gray boxes indicate CpG islands. Exon II is the homeodomain (HD) containing exon. The canonical Hoxa9 is encoded by exons CD and II. Regions of high homology between species are noted as dark boxes and the region amplified by ChIP primers is indicated by arrows. The location of mir-196b is labeled above exon AB with an arrowhead. (B) Sequence alignment of the conserved mir-196b sequence among different species. The mature mir-196b sequence is highlighted in black. (C) Quantitative RT-PCR of mir-196b levels in WT, Mll−/−, and Men−/− MEFs. The experiment was run in triplicate and the results represent average expression plus or minus SD. (D) Quantitative RT-PCR of mir-196b expression in individual clones expressing an empty vector, MLL, or MLLAF4 in Mll−/− MEF background. Transfection of Mll−/− MEFs and clone selection was performed as previously described. Mir-196b expression was determined from total RNA. (E) ChIP assay performed in WT and Mll−/− MEFs. Chromatin was precipitated using indicated antibodies and qPCR performed with primers spanning mir-196b region. All samples were run in triplicate and were normalized to the input chromatin.
The tumor suppressor protein menin was previously biochemically purified as part of an MLL protein complex and subsequently shown to be an essential cofactor in MLL-dependent gene expression.10,28 Binding of Mll to some of its target genes that have been studied requires menin; absence of menin leads to decreased expression of Mll-target genes, including Hoxa9.10,29 Thus, we hypothesized that if mir-196b is regulated by Mll, then the absence of menin should also diminish mir-196b levels. To test this hypothesis, Men−/− MEFs were assessed for mir-196b expression. As predicted, mir-196b expression in Men−/− MEFs was 4-fold lower than in the wild-type counterparts (Figure 1C).
Several studies have documented that Mll regulates expression of its target genes at least in part by altering chromatin structure.8-10,30 Previous studies have shown that the SET domain of MLL possesses histone methyltransferase activity specific for lysine 4 on histone H3.8,9 Using ChIP, we investigated histone modifications in the mir-196b region in wild-type and Mll null cells. We and others have previously reported binding of endogenous Mll to this region.21,31 Surprisingly, we did not observe any differences in H3K4 methylation between the 2 cell types (Figure 1E). However, we did notice dramatically higher levels of H3K79 dimethylation in the Mll wild-type cells compared with Mll−/− cells (Figure 1E). H3K79 methylation by the protein Dot1 has been linked to active transcriptional elongation suggesting that this mark may also represent transcriptional elongation in the conserved region only in the wild-type but not Mll−/− cells.32,33
Mll-dependent mir-196b expression during ES cell differentiation
Mll regulates expression of a subset of genes in the Hox cluster during embryonic development.19,34 Mice lacking Mll are embryonic lethal and display gross defects in the early stages of hematopoiesis.19,23 Absence of Mll leads to deficiencies in proliferation and survival of hematopoietic progenitors in the yolk sac.35 To investigate the dependency of mir-196b expression on Mll during development, we used wild-type and Mll−/− murine embryonic stem (ES) cells. On removal of leukemia inhibitory factor (LIF), ES cells form embryoid bodies (EBs) and the pattern of gene expression in the embryoid bodies generally follows that of early hematopoiesis in a developing embryo.36 Previous studies have shown that the expression of several Hox genes depends on the presence of Mll during ES cell differentiation.37 We wanted to determine whether Mll regulates the timing of mir-196b expression in a manner similar to that of the neighboring Hoxa9 and Hoxa10 genes. As previously reported, and confirmed in our laboratory, Mll−/− cells form embryoid bodies indistinguishable from those derived from the wild-type cells.37 Levels of Hoxa9 and Hoxa10 genes are similar in both cell types in undifferentiated ES cells and during the first few days of differentiation (Figure 2A,B). At the later stages of differentiation, transcription of both of these genes is up-regulated in the wild-type cells. Cells carrying homozygous Mll mutations show lower levels of Hoxa9 and Hoxa10 expression. Similarly, levels of mir-196b are decreased up to 14-fold in the absence of Mll. Mir-196b is expressed during ES cell differentiation in a pattern of expression similar to that of Hoxa9 and Hoxa10 (Figure 2C). Sharing of cis-regulatory elements has been previously described for genes in the HoxB and C clusters, however, it remains unclear whether mir-196b shares regulatory elements with the neighboring Hox genes or if its expression is controlled by Mll independently of Hoxa9 and Hoxa10.38,39
Mir-196b is regulated by Mll during ES cell differentiation, similar to Hoxa9 and Hoxa10. (A,B) Quantitative RT-PCR of Hoxa9 (A) and Hoxa10 (B) mRNA using SYBR green. All values are compared with WT day 0 and this value is set to 1. Experiments were performed in triplicate and the results represent average fold change plus or minus SD. (C) Quantitative RT-PCR of mir-196b expression using mir-196b TaqMan primers and probe.
Mir-196b is regulated by Mll during ES cell differentiation, similar to Hoxa9 and Hoxa10. (A,B) Quantitative RT-PCR of Hoxa9 (A) and Hoxa10 (B) mRNA using SYBR green. All values are compared with WT day 0 and this value is set to 1. Experiments were performed in triplicate and the results represent average fold change plus or minus SD. (C) Quantitative RT-PCR of mir-196b expression using mir-196b TaqMan primers and probe.
Embryoid bodies contain a highly heterogenous population of cells. To more specifically investigate hematopoietic cells we isolated CD41+ cells from day 10 embryoid bodies. This marker was shown to be present on definitive hematopoietic progenitor cells giving rise to all blood lineages.40 As reported by Ernst et al, wild-type and Mll−/− ES cells form a comparable number of CD41+ cells however Mll−/− CD41+ cells are less efficient at producing hematopoietic colonies.37 We found that both the canonical Hoxa9 as well as mir-196b are expressed at a lower level in Mll−/− CD41+ cells in comparison to the wild-type cells (Figure 3A,B). In addition, expression of mir-196b was also Mll dependent in the CD41 negative population, suggesting that Mll is also required for expression of mir-196b in other cell types and tissues (Figure 3B).
Mll regulates mir-196b expression in hematopoietic progenitors. (A) RT-PCR for canonical Hoxa9 in CD41+ ES cells. Day 10 EBs were disrupted and CD41+ cells isolated using a magnetic column. Total RNA was isolated from the CD41+ cells and used for cDNA synthesis. Hoxa9 primers used span the junction between exon CD and exon II of the gene. Gapdh is used as a loading control. Space between panels indicates repositioned gel lanes. (B) Quantitative RT-PCR of mir-196b expression in WT and Mll−/−CD41+ and CD41− murine embryonic stem cells from day 10 embryoid bodies. WT levels were set to 1 for comparison. Experiment was performed in triplicate and the results represent average expression plus or minus SD. (C) Quantitative RT-PCR of mir-196b expression in sorted mouse bone marrow cells. Mouse bone marrow cells were sorted in various populations based on the expression of cell surface markers. Expression of LT-HSCs is set to1 for comparison. The results represent average fold change plus or minus SD.
Mll regulates mir-196b expression in hematopoietic progenitors. (A) RT-PCR for canonical Hoxa9 in CD41+ ES cells. Day 10 EBs were disrupted and CD41+ cells isolated using a magnetic column. Total RNA was isolated from the CD41+ cells and used for cDNA synthesis. Hoxa9 primers used span the junction between exon CD and exon II of the gene. Gapdh is used as a loading control. Space between panels indicates repositioned gel lanes. (B) Quantitative RT-PCR of mir-196b expression in WT and Mll−/−CD41+ and CD41− murine embryonic stem cells from day 10 embryoid bodies. WT levels were set to 1 for comparison. Experiment was performed in triplicate and the results represent average expression plus or minus SD. (C) Quantitative RT-PCR of mir-196b expression in sorted mouse bone marrow cells. Mouse bone marrow cells were sorted in various populations based on the expression of cell surface markers. Expression of LT-HSCs is set to1 for comparison. The results represent average fold change plus or minus SD.
Mir-196b expression in hematopoietic progenitors
MiRNA expression patterns vary between cell types. We wanted to examine if mir-196b is generally expressed in hematopoietic progenitor cells or if it is limited to expression in specific stages of hematopoiesis. Mouse bone marrow cells were separated into more differentiated versus less differentiated populations. Expression of mir-196b was on average 2.5-fold higher in lineage negative cells (data not shown). Similarly, the c-Kit+ population of bone marrow cells expressed 6-fold higher levels of mir-196b than the c-Kit− cells (data not shown). These results suggest that mir-196b is indeed expressed in the hematopoietic progenitor population and that it may be playing a role at early stages of hematopoiesis.
To determine which population(s) of hematopoietic progenitors expresses mir-196b, mouse bone marrow cells were sorted into long-term hematopoietic stem cells (LT-HSC), short-term hematopoietic stem cells (ST-HSC), multipotent progenitors (MPP) and more mature myeloid (GR1+) and lymphoid (B220+) cells. Expression levels of mir-196b are the highest in ST-HSC and decrease as cells become more differentiated (Figure 3C). The tightly regulated pattern of mir-196b expression suggests that misregulation of mir-196b expression may lead to drastic defects in normal hematopoiesis.
MLL fusion proteins induce expression of mir-196b, which is required for increased proliferative capacity
Overexpression of certain HOX genes is a hallmark of MLL-associated leukemias, however, the exact mechanism by which MLL fusion proteins exert their effect in the leukemic cells is still poorly understood. Because MLL regulates expression of various HOX transcripts, we wanted to investigate whether an MLL fusion protein causes a change in mir-196b expression. Transfection of constructs expressing the MLL-AF4 fusion protein into Mll−/− MEFs reactivated expression of mir-196b to levels higher than transfection of constructs expressing wild-type MLL (Figure 1D). We also used a bone marrow progenitor colony assay to investigate the effect of MLL-AF9 on mir-196b expression. C-Kit+ bone marrow cells infected with MLL-AF9 retroviruses show on average more than a 100-fold increase in mir-196b expression in comparison to vector infected cells (data not shown). Expression of canonical Hoxa9 is also greatly increased (> 300-fold; data not shown).
Although MLL fusion proteins caused increased mir-196b expression, we next determined whether mir-196b expression was necessary to manifest the increased in vitro proliferative capacity observed for MLL fusion–transduced bone marrow progenitor cells. For this experiment, MLL-AF9–transduced progenitors were treated with antagomir specific to mir-196b or with control (mutant) antagomir and assessed for replating activity. We found that mir-196b function is required to maintain the increased proliferative capacity of MLL fusion-expressing bone marrow progenitors in vitro (Figure 4). The role of mir-196b in immortalization and enhanced proliferation of cells transduced by MLL fusion proteins was intriguing and suggested that further investigation of mir-196b in MLL leukemia was warranted.
MLL fusion proteins induce expression of mir-196b in primary bone marrow progenitors, which is required for their MLL fusion-dependent increased proliferative capacity in vitro. Wild-type Lin− bone marrow cells were transduced with MLL-AF9 retroviral vectors and treated with either control antagomiR-196 or antagomiR-196, then plated in duplicate. Colonies were enumerated after 8 days and the average number was plotted. Cells were then replated after additional antagomir treatment. The cycle was repeated twice. Data shown are representative of 2 independent experiments with similar results.
MLL fusion proteins induce expression of mir-196b in primary bone marrow progenitors, which is required for their MLL fusion-dependent increased proliferative capacity in vitro. Wild-type Lin− bone marrow cells were transduced with MLL-AF9 retroviral vectors and treated with either control antagomiR-196 or antagomiR-196, then plated in duplicate. Colonies were enumerated after 8 days and the average number was plotted. Cells were then replated after additional antagomir treatment. The cycle was repeated twice. Data shown are representative of 2 independent experiments with similar results.
Mir-196b is overexpressed in the majority of MLL-associated leukemias
To investigate mir-196b expression in primary leukemia patient samples, we used bead-based expression profiling technology.27 To this end, 55 different patient samples (10 ALL and 45 AML) and 3 normal bone marrow samples were analyzed for expression of mir-196b. The 29 MLL leukemia samples (10 ALL and 19 AML) expressed consistently higher levels of mir-196b than non-MLL leukemia samples (Figure 5). It is of interest that multiple cell lines derived from MLL leukemia patients also express high levels of mir-196b (data not shown). As a molecule that has the potential of regulating expression levels of many different proteins, high levels of mir-196b in MLL leukemias may play a crucial role in the disease process.
Mir-196b is overexpressed in the majority of MLL-associated leukemias, but not in non-MLL leukemias, irrespective of their phenotype. Heat map of relative mir-196b expression of 55 leukemia samples and 3 normal controls using bead-based technology.
Mir-196b is overexpressed in the majority of MLL-associated leukemias, but not in non-MLL leukemias, irrespective of their phenotype. Heat map of relative mir-196b expression of 55 leukemia samples and 3 normal controls using bead-based technology.
Mir-196b causes increased proliferative capacity and decreased differentiation
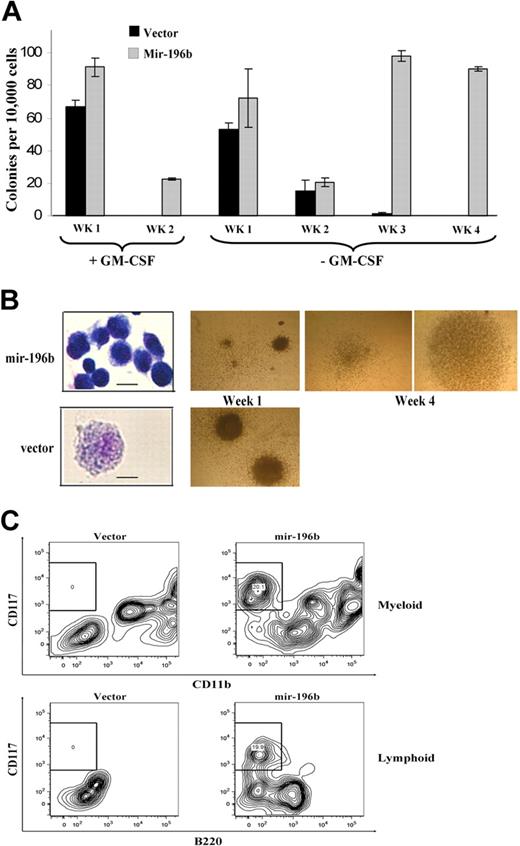
To examine possible roles of mir-196b overexpression in hematopoietic progenitor cells we cloned a 200 bp region surrounding the mature miRNA into a retroviral expression vector. Increased mir-196b expression was confirmed after transfection in MEFs (data not shown). In vitro serial replating colony assays have been used extensively to determine the leukemogenic potential of MLL fusion proteins. Because we found mir-196b overexpressed in primary MLL leukemia samples, we wished to determine whether overexpression of mir-196b could contribute to the immortalization process by providing increased proliferative capacity to progenitor cells. Bone marrow c-kit+ cells were transduced using retroviruses that express mir-196b precursor or with the empty vector alone and serially replated in methylcellulose containing either SCF, IL-6, IL-3 and GM-CSF (referred to as +GM-CSF) or SCF, IL-6 and IL-3 (referred to as −GM-CSF). Transduced cells formed colonies of various sizes after the first week in culture for both vector and mir-196b under both conditions, as expected (Figure 6B). Mir-196b was expressed approximately 2-fold higher in the mir-196b–transduced cells compared with controls (data not shown). In the presence of GM-CSF, mir-196b–expressing cells survived into the third week (after the second replating) and increased in cell number on average 5-fold, in contrast to control-infected cells that did not survive after the first replating (Figure 6A). Overexpression of mir-196b was not sufficient to completely immortalize cells under these conditions. However, using cytokine mix −GM-CSF, a more dramatic effect of mir-196b expression on increased colony-forming ability and proliferative capacity was observed. Under these conditions, mir-196b expressing cells formed tight colonies even after the fourth plating of cells, in contrast to the vector-transduced cells, which formed primarily loose colonies after the second plating and almost no colonies after the third plating (Figure 6A,B). Mir-196b–expressing cells after 4 platings in methylcellulose became a relatively homogenous population resembling blast cells and expressed intermediate levels of CD117 and Sca1, with about 50% also expressing Gr-1, whereas vector-transduced cells after the second and third plating (very few cells remaining after the third plating) became CD117 and Sca1 high, but negative for lineage markers and resembled mast cells (Figure 6B and data not shown). To rule out the possibility that simply activating the microRNA processing pathway was responsible for the observed increased proliferative capacity, we also transduced cells with retroviruses that express short hairpin RNAs that target 2 different molecules, CtBP and Bmi-1. Cells transduced with either of these retroviruses formed colonies in the first week, confirming effective infection of the cells; however, the colony-forming ability of both of these were similar to vector-transduced cells (data not shown). Our data demonstrate that mir-196b expression causes an increased survival and proliferative capacity of bone marrow progenitor cells.
Mir-196b expression enhances colony formation and partially blocks hematopoietic progenitor cell differentiation. (A,B) Serial replating myeloid colony assay using c-Kit+ bone marrow cells. Cells were transduced with retroviruses producing mir-196b construct or with an empty vector and plated in methylcellulose with 2 different cytokine mixes. The number of colonies (A) and colony and cell morphology (B) at 1 and 4 weeks are shown. Only mir-196b–expressing cells are able to form colonies at the fourth week. Scale bars on cytospin pictures represent 10 μm in top panel and 25 μm in bottom panel. (C) Representative FACS profiles of in vitro–differentiated bone marrow cells. After 1 week of growth and selection in methylcellulose, mir-196b or vector-expressing bone marrow cells were plated on the OP9 cell line in the presence of GM-CSF or IL-7/Flt3L. After 5 days, expression of CD117 and CD11b (top panels, cells on GM-CSF) or B220 (bottom panels, cells on IL-7/Flt3L) was determined. Under both conditions, overexpression of mir-196b causes a partial block in differentiation.
Mir-196b expression enhances colony formation and partially blocks hematopoietic progenitor cell differentiation. (A,B) Serial replating myeloid colony assay using c-Kit+ bone marrow cells. Cells were transduced with retroviruses producing mir-196b construct or with an empty vector and plated in methylcellulose with 2 different cytokine mixes. The number of colonies (A) and colony and cell morphology (B) at 1 and 4 weeks are shown. Only mir-196b–expressing cells are able to form colonies at the fourth week. Scale bars on cytospin pictures represent 10 μm in top panel and 25 μm in bottom panel. (C) Representative FACS profiles of in vitro–differentiated bone marrow cells. After 1 week of growth and selection in methylcellulose, mir-196b or vector-expressing bone marrow cells were plated on the OP9 cell line in the presence of GM-CSF or IL-7/Flt3L. After 5 days, expression of CD117 and CD11b (top panels, cells on GM-CSF) or B220 (bottom panels, cells on IL-7/Flt3L) was determined. Under both conditions, overexpression of mir-196b causes a partial block in differentiation.
The normal process of terminal differentiation is often abrogated during development of leukemia. We investigated whether mir-196b helps hematopoietic progenitor cells retain a more undifferentiated state. To this end, equal numbers of bone marrow progenitor cells cultured in methylcellulose with puromycin selection for 1 week after transduction with vector or mir-196b–expressing constructs were replated on OP9 feeder cells in the presence of GM-CSF or IL-7/FLT3L. The 2 conditions support myeloid and lymphoid differentiation, respectively. After 5 days, the cells were analyzed for the expression of CD117/CD11b or CD117/B220 markers. Under both conditions, the mir-196b–infected cells retained a significant percentage (∼ 20%) of CD117 positive cell population, whereas vector expressing cells did not (Figure 6C). This suggests that overexpression of mir-196b can partially block differentiation in hematopoietic progenitor cells.
Discussion
MLL, a large multidomain protein, is a master regulator of gene expression during development and hematopoiesis. MLL positively regulates expression of genes by binding to their promoters and recruiting various chromatin-associated proteins and through the H3K4 histone methyltransferase activity of its SET domain.8,9,28 Translocations in MLL give rise to in-frame fusions with various partner genes and the formation of chimeric proteins leading to misregulation of MLL target genes. While transcriptional activation and histone methyltransferase domains of MLL are both lost in the chimeras, the fusion proteins still cause overexpression of downstream MLL targets, including members of the HOX clusters.41,42 Dysregulation of certain HOX genes plays a central role in MLL-associated leukemogenesis, however, the mechanism is still very poorly understood.12,28 In our study, we demonstrate that MLL and MLL fusion proteins regulate expression of a microRNA located in the HOX cluster, mir-196b. Mir-196b is an attractive target of MLL regulation for various reasons: (1) mir-196b is situated between HOXA9 and HOXA10, 2 MLL targets that are frequently overexpressed in MLL leukemias; (2) some of the mir-196b targets are HOX genes themselves and by regulating mir-196b expression, MLL can simultaneously regulate boundaries of HOX expression43 ; and (3) as with any other miRNA, strict regulation of mir-196b expression is essential for its proper function and possible misregulation by MLL fusion proteins may lead to misregulated expression of mir-196b target mRNAs, and thus contribute to large-scale changes in gene expression often seen in leukemic cells.
In this study, we provide several lines of evidence to support mir-196b as a target of MLL regulation. Our previous findings demonstrated that the presence of Mll selectively prevents DNA methylation of a region adjacent to mir-196b that allows for expression of transcripts that are mir-196b precursors.21 Using a method that detects the mature form of miRNA, we demonstrate that Mll−/− MEFs express lower levels of mir-196b than wild-type cells. We and others have previously shown that the expression of other Mll targets, such as Hoxa7, a9, a10, are also decreased in these cells.18 One of the components of an MLL protein complex is menin, a tumor suppressor protein often deleted in multiple endocrine neoplasms.44 Genome-wide mapping of menin and MLL binding to chromatin shows that the 2 proteins are often bound to the same loci.31 Our data show that mir-196b expression is also dependent on menin and that in the absence of menin, mir-196b levels fall to similar low levels as measured in Mll−/− cells. Transfection of MLL or MLL-AF4 into the Ml−/− cells boosts expression of mir-196b to and above wild-type levels, respectively, which is accompanied by the reversal of DNA methylation.21
The SET domain of MLL contains histone methyltransferase activity specific for lysine 4 on histone H3 and this function contributes to activation of MLL target genes.8,9 Surprisingly, ChIP data do not show any changes in H3K4 methylation between wild-type and Mll−/− cells in the region surrounding the microRNA. It has been previously noted that trimethylation of H3K4 is a mark usually found close to the transcription start site and it is possible that the mir-196b precursor transcript starts further upstream. Interestingly, our ChIP data show that the mir-196b region is greatly enriched in H3K79 dimethylation only in the presence of Mll. H3K79 methylation is a mark associated with transcriptional elongation suggesting that Mll is necessary for proper transcriptional elongation in this region.32,33 Dot1 is the only H3K79 histone methyltransferase isolated to date. It is still unclear if any of these histone marks are altered in the presence of MLL fusion proteins, however, several MLL partners, including AF10, AF9, ENL, and AF4 interact with Dot1 in a multiprotein transcription elongation complex with pTEFb.45,46 It is possible that this interaction may also play a role in overexpression of MLL target genes in the MLL-associated leukemias.
Using in vitro differentiation of wild-type and Mll−/− murine embryonic stem cells we show that Mll regulates expression of mir-196b similarly to other 5′ Hox genes. These data suggest that Mll regulates mir-196b temporally and studies by Mansfield et al showed that the paralogous mir-196a is also regulated spatially in a Hox-like pattern during embryogenesis.47 Considering that a subset of mir-196 targets are also members of the HOX cluster, it is likely that the pattern of mir-196b expression plays a role in fine-tuning the levels of HOX gene expression. In a developing organism, this mechanism would help generate the boundaries of HOX expression without the need for turning off the genes completely.
Microarray profiles of miRNA expression patterns have shown that mir-196b is expressed at the highest levels in bone marrow and spleen, suggesting it has a role in hematopoiesis.48 Our analysis of c-Kit+ hematopoietic progenitors demonstrates that mir-196b is expressed at higher levels in this population than in the more differentiated c-Kit− population. In addition, separation of mouse bone marrow into lineage-negative and lineage-positive cells confirms that mir-196b is expressed at higher levels in less differentiated cells. Of the early hematopoietic progenitors, expression of mir-196b is the highest in ST-HSCs. Isolation of CD41+ definitive hematopoietic progenitors from differentiated embryoid bodies confirms mir-196b expression in hematopoietic progenitor cells only in the presence of Mll. Aberrant hematopoietic colony formation of Mll−/−CD41+ cells was rescued by the addition of certain Hox genes or the homeodomain containing transcription factor Cdx4.37 Cdx4 positively regulates expression of Hox genes and, similar to MLL, binding of Cdx4 to the Hoxa9 locus is dependent on menin.49 In a mouse model, Cdx4 overexpression generates acute myeloid leukemia while dysregulating Hox expression.50 It would be of interest to determine if Cdx4 also regulates mir-196b expression and if this regulation plays a role in Cdx4-induced leukemogenesis.
A hallmark of MLL-associated leukemia is overexpression of specific HOX genes.41,42 Likewise, our study shows that MLL fusion proteins also drastically increase levels of mir-196b. C-Kit+ bone marrow cells transduced with MLL-AF9 retroviruses express on average more than 100-fold increased levels of mir-196b than cells infected with control retrovirus. The increased proliferative capacity of the MLL fusion-expressing progenitors was dependent on mir-196b expression because antagomir specifically targeting mir-196b abrogated this effect. Importantly, expression analysis of primary patient leukemia samples confirms high levels of mir-196b expression specifically in MLL leukemias. This increase in expression is observed in both acute myeloid, as well as acute lymphoid, MLL leukemia samples. Significantly, non-MLL leukemias do not express high levels of mir-196b. Abnormal expression of mir-196b influences expression of many downstream targets that likely contribute to the development of leukemia. Targets of mir-196b include Hoxb8 and Hoxa7, along with an extensive list of additional putative mir-196b targets. To either rule in or rule out a direct contribution of mir-196b regulation of Hox target expression in the transformation process, we quantitatively assessed levels of multiple Hox genes after expression of mir-196b in bone marrow progenitor cells. These include Hoxa5, Hoxa7, Hoxa9, Hoxa10, Hoxb3, Hoxb4, Hoxb6, Hoxb8, Hoxc8, and Meis1. Hoxc8 and Hoxb8 were below the level of detection in both control- and mir-196b–expressing cells, suggesting that they do not play a role in the observed phenotypes. Of the other genes measured, Meis1 and Hoxc6 do show decreased expression when mir-196b is overexpressed, however, these are not predicted to be direct targets of mir-196b. Further studies are needed to determine which mir-196b targets are relevant in MLL leukemogenesis.
Using a serial replating hematopoietic colony assay, we show that increased levels of mir-196b stimulate the proliferative capacity of hematopoietic progenitor cells, but are not sufficient to completely immortalize cells. Furthermore, overexpression of mir-196b in hematopoietic cells also results in a partial differentiation block such that a significant percentage of progenitors retain expression of c-Kit under conditions that cause differentiation of cells not expressing this miRNA. Increased proliferation and block in differentiation are essential to leukemia development. Our results suggest that misregulation of mir-196b expression by MLL fusion proteins plays an important role in the disease. This evidence establishes a connection between misregulation of miRNA expression and increased proliferation, survival, and differentiation block in leukemia (Figure 7). In this model, expression of mir-196b is tightly controlled by MLL in hematopoietic precursor cells. The presence of MLL fusion proteins leads to aberrant mir-196b expression causing increased proliferation, abnormal hematopoietic differentiation and contributes to leukemogenesis. MiRNAs may be used as new molecular targets for the development of novel therapeutic strategies.
Model for the role of mir-196b in hematopoietic cells and MLL fusion-mediated leukemia. MLL regulates expression of mir-196b and MLL fusion proteins cause a block in differentiation of progenitor cells. This effect may be due partially to high levels of mir-196b expression caused by the MLL fusion protein. High levels of mir-196b may also provide a survival advantage to cells expressing the MLL fusion protein.
Model for the role of mir-196b in hematopoietic cells and MLL fusion-mediated leukemia. MLL regulates expression of mir-196b and MLL fusion proteins cause a block in differentiation of progenitor cells. This effect may be due partially to high levels of mir-196b expression caused by the MLL fusion protein. High levels of mir-196b may also provide a survival advantage to cells expressing the MLL fusion protein.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof.
During revision of this manuscript, Schotte and colleagues51 showed that mir-196b is specifically overexpressed in MLL-rearranged ALL but not in other B-ALL pediatric patient samples.
Acknowledgments
Wild-type and Mll null MEFs were kindly provided by Dr Patricia Ernst and Dr Stanley Korsmeyer. We thank Dr Xianxin Hua for providing wild-type and Men null MEFs and Dr Todd Golub for help with the bead-based miRNA profiling.
This work was funded by the G. Harold and Leila Y. Mathers Charitable Foundation (White Plains, NY; J.C.), National Institutes of Health (NIH)/National Cancer Institute (NCI, Bethesda, MD; CA105152, H.L.G.), Leukemia & Lymphoma Society (White Plains, NY) Translational Research Grant (J.D.R.), the Dr Ralph and Marian Faulk Medical Research Trust (Barrington, IL), and NIH/NCI grants CA105049 and HL087188 (N.J.Z.-L.).
National Institutes of Health
Authorship
Contribution: R.P. designed and performed research and wrote the manuscript; L.E.R., C.S.V., A.C., J.Z., F.E.E., and J.L. designed and performed research; N.J.A. and K.E. performed research; H.L.G. and J.C. designed research, analyzed data, and edited the manuscript; J.D.R. analyzed data and edited the manuscript; and N.J.Z.-L. designed research and wrote and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nancy J. Zeleznik-Le, 2160 South 1st Ave, Building 112, Room 337, Maywood, IL 60153; e-mail: nzelezn@lumc.edu.