Abstract
Granzyme H (GzmH) belongs to a family of 5 human serine proteases that are expressed by cytotoxic immune effector cells. Although GzmH is most closely related to the caspase-activating granzyme B (GzmB), neither a natural substrate nor a role in immune defense reactions has been demonstrated for this orphan granzyme. In rodents, multiple related genes exist, but none of these can be regarded as functional homologs. Here we show that host cells are efficiently killed by GzmH after perforin and streptolysin O–mediated delivery into the cytosol. Dying cells show typical hallmarks of programmed cell death, such as mitochondrial depolarization, reactive oxygen species (ROS) generation, DNA degradation, and chromatin condensation. Contrary to GzmB, cell death by GzmH does not involve the activation of executioner caspases, the cleavage of Bid or inhibitor of caspase-activated DNase (ICAD), or the release of cytochrome c. The high expression levels of GzmH in naive natural killer (NK) cells and its potent killing ability strongly support the role of the protease in triggering an alternative cell-death pathway in innate immunity.
Introduction
Granzyme H (GzmH) is regarded as an orphan granzyme with unknown biologic functions in immune defense cells.1 Recent reports show that this serine protease is predominantly expressed at high levels in natural killer (NK) cells and has chymotrypsin-like (chymase) activity.2 No functional studies have as yet been reported. Granzymes (granule enzymes) are of particular interest due to their different proteolytic specificities and potential abilities to trigger cell death in tumor and virally infected cells. Indeed GzmA, a “tryptase” that cleaves after basic residues, and GzmB, an “aspase” preferring acidic residues, have evolved distinct apoptotic pathways. The cytotoxic action of GzmB on target cells is largely caspase dependent,3 while that of GzmA is caspase independent.4 In particular, GzmA, transferred into target cells via perforin, leads to a very fast generation of reactive oxygen species (ROS) and induces the translocation of the previously described endoplasmic reticulum (ER)–associated suppressor of variegation, enhancer of zeste, and trithorax (SET) complex into the nucleus. There, GzmA relieves the active nuclease NM23-H1 from the complex by destroying its bound inhibitor SET.5 Recently, a caspase-independent cell-death mechanism was also observed for murine GzmC, which rapidly induced mitochondrial swelling and membrane depolarization.6 GzmM, which cleaves after residues with long, uncharged side chains such as methionine and leucine,7 was also shown to induce a very rapid form of caspase-independent cell death.8 More recently, GzmM-deficient mice were generated and shown to display increased susceptibility to murine cytomegalovirus (CMV) infections but displayed a normal NK and T-cell development with normal NK-mediated cytotoxicity.9
The 5 human granzyme genes (GZMA, GZMB, GZMH, GZMK, GZMM) are clustered on 3 different chromosomes.10 GZMA and GZMK are located on chromosome 5 and GZMM on chromosome 19.11 GZMB and GZMH share high structural homology (71% amino acid identity) and belong to a tightly linked gene cluster on chromosome 14, which also harbors cathepsin G and mast cell chymase. Despite their high sequence homology, both enzymes bear very distinct enzymatic activities. GzmB cleaves caspase 8–like specific sequences after acidic residues.12 In contrast, GzmH is shown to have chymotrypsin-like thioester activity with a preference for hydrophobic, aromatic amino acid residues (Phe or Tyr) at the P1 site.2 In a recent study, Sedelies and coworkers analyzed the expression of GzmH in human blood leukocytes.13 Using a new GzmH-specific antibody they showed the discordant regulation of GzmH and GzmB. GzmH was constitutively expressed in NK56+ CD3- NK cells irrespective of its activation status, and in contrast to GzmB was not present in activated CD8+ T cells. This finding thereby suggests a pivotal role for GzmH in NK cell–induced cell death.
In the rodent genomes, a highly variable number of paralogous genes were identified in the region bordered by cathepsin G and GZMB. No structure-activity relationships have so far been experimentally determined. The murine granzymes D to G and RNKP-7 from rats are predicted to cleave after phenylalanine similarly to GzmH in humans.1,14 The GZMH gene has been shown to be the result of a recent intergenic recombination event between a chymase-like ancestor and GZMB.15 Because this gene conversion has occurred in the primate lineage after mammalian radiation, a genuine ortholog with conserved substrate specificity is unlikely to exist in rats and mice.
Here, we demonstrate for the first time that GzmH is capable of inducing cell death. In a reconstituted system, pure recombinant GzmH was transferred into the target cell cytosol not only by the bacterial pore-forming protein, streptolysin O (SLO), but also by its natural pore-forming translocator, perforin (PFN). We show that GzmH achieves cell death by acting in a distinct fashion on mitochondrial and nuclear targets but not through the activation of the hallmark apoptotic substrates such as executioner caspase activation, bid cleavage, cytochrome c release, or ICAD cleavage. GzmH is thus a functional cytotoxic serine protease of NK cell granules, which expands the cell death–inducing repertoire of the innate immune system.
Materials and methods
Treatment and analyses of target cells
K562, U937, and HL60 cells were grown in cell culture as reported;16 the NK cell lines, YT, NK92, and NKL, were maintained as described for NK92 cells.17 Killing assays and target cell staining with annexin V (AV)–FITC and propidium iodide (PI) were performed, as described,17 in triplicates in 96-microwell plates (Nunc, Wiesbaden, Germany) using 5 × 104 cells in 50 μL per well. Sublytic concentrations of every new batch of perforin (a generous gift from C. J. Froelich, Evanston, IL), SLO (Sigma-Aldrich, St. Louis, MO), and SLOC530A (a kind gift from S. Bhakdi, Mainz, Germany) were determined separately for K562, U937, and HL60 cells and were defined as the concentration required to induce cell death in 10% to 20% of all cells. Sublytic concentrations of PFN or SLO varied depending on the cell line. PFN was used at final concentrations of 75 to 80 ng/mL with HL60 cells and 200 to 225 ng/mL with K562 cells. SLO was used at 75 ng/mL and 125 to 175 ng/mL, respectively. If not otherwise indicated, GzmB, GzmH, and GzmHS195A were added to cells at final concentrations of 5(10), 5(20), or 10(25) μg/mL, respectively; the concentrations in parentheses were used in combination with PFN. In some experiments the caspase inhibitors zVAD-fmk, zDEVD-fmk, and zVDVAD-fmk (Bachem, Bubendorf, Switzerland) were applied at a final concentration of 50 μM. Trypan blue staining was performed by mixing equal volumes of cell suspension with 0.1% trypan blue solution (T8154; Sigma-Aldrich). Trypan blue–positive cells were then scored using a hemocytometer.
Cellular morphology and nuclear fragmentation
To distinguish living cells from those with apoptotic nuclei, bright field pictures were taken with an inverted microscope (Axiovert 200M [Zeiss, Oberkochen, Germany]; 40×/0.6 numerical aperture objective lens) connected to a Coolsnap-HQ camera (Photometrics, Roper Scientific, Ottobrunn, Germany). Hoechst staining was performed as described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article.) For quantification one half of a visual field showing about 50 to 200 cells was evaluated. Sub-G1 staining with PI (Sigma-Aldrich) and fluorescence-activated cell sorter (FACS) measurements were performed as described.18 For analysis, the major cell population (more than 90%) was gated according to forward and side scatter criteria, thereby avoiding aggregated cells. Apoptotic DNA laddering was done as described.19
Transmembrane potential loss and ROS production
For measurement of mitochondrial depolarization, cells were stained with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl-carbocyanine iodide (JC-1; Merck Biosciences, Darmstadt, Germany) for 15 minutes at 10 μg/mL in phosphate-buffered saline (PBS) and directly analyzed with the FACS. Intracellular ROS production was monitored by adding 2 μM Dihydroethidium (DHE) (Molecular Probes, Eugene, OR) just before FACS analysis to cells treated for the indicated times at 37°C. For ROS measuring experiments, SLOC530A, a SLO variant mutated at the activating residue cysteine 530 was used to circumvent the need for prior activation under reducing conditions. In these cases mercaptoethanol was omitted from the medium. As a positive control, cells were exposed to 1% to 3% H2O2.
Caspase activity in living cells
Caspase activity in intact (nonlysed) target cells was measured with fluorogenic substrates. The cells were mixed with the substrates Ac-DEVD-AMC, Ac-LEHD-AMC, and zVDVAD-AFC (Merck Biosciences) at a 100 μM final concentration in RPMI 1640 medium (without phenol red) containing 0.5% BSA (Sigma-Aldrich), Hepes (25 mM), and 100 U/mL penicillin and streptomycin each and 2 mM l-glutamine (Invitrogen, Karlsruhe, Germany) and then plated on 96-well plates with flat bottoms for 1 hour at 37°C in a 5% CO2 atmosphere. In several experiments caspase inhibitors were included in the cell culture medium before treating the cells with granzymes and sublytic concentrations of PFN or SLO. Substrate hydrolysis was directly monitored using a Wallac Victor 2 multilabel counter, model 1420, (Perkin Elmer, Fremont, CA) at 37°C. 7-amino-4-methyl coumarin (AMC) was monitored by excitation at 380 nm and emission at 460 nm and 7-amino-4-trifluoromethyl coumarin (AFC) by excitation at 420 nm and emission at 520 nm.
Cytochrome c release from mitochondria
Cytochrome c release to the cytosol was assessed by immunoblotting after cell fractionation with 0.1% digitonin (Sigma-Aldrich) using a mouse monoclonal antibody against human cytochrome c as described.20 The anti-actin mouse monoclonal antibody (clone JLA20; Merck Biosciences) at a 1:5000 dilution was used to control loading. Specific mouse and rabbit antibodies were detected with goat anti-mouse (1:2000; Perbio Science, Bonn, Germany) or goat anti-rabbit antibodies (1:5000; Jackson Immunoresearch, Hamburg, Germany) conjugated to horseradish peroxidase (HRP), respectively. Bound HRP antibody conjugates were visualized with the SuperSignal West Pico substrate (Perbio Science).
Results
Recombinant GZMH is an active chymase of NK cells
Recombinant, nonglycosylated GzmH and the point-mutated inactive GzmHS195A were expressed in Escherichia coli and produced similarly as described.21 Figure S1A shows proform, active, and inactive GzmH on a silver-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Recombinant GzmH cleaved the thiobenzyl ester Suc-Phe-Leu-Phe-SBzl and is inhibited by the general protease inhibitor 3,4-dichloroisocoumarin (DCI) (Figure S1B-C). In addition, GzmH hydrolyzed the 2 peptide substrates, PTSY-AMC and ATSY-AMC (Figure S1D), which were predicted as potential substrates in a previously reported peptide library scan.22
We next analyzed the expression of GzmH in different human cell lines and purified cell populations using the specificity of a new monoclonal antibody (R&S Systems, Minneapolis, MN) that was first confirmed in Western blots and enzyme-linked immunosorbent assay (ELISA) using all known human granzymes produced in our laboratory (Figure S2A). Of several NK cell lines, only the YT cell line, which is a poor killer, was found to express low levels of GzmH. Using two different isolation methods (positive selection by magnetic cell separation (MACS) beads or negative selection by Dynal-beads) we confirmed the recent findings that NK cells are the major GzmH-expressing human lymphocyte subpopulation.13 Neither monocytes nor activated CD8+ CTLs showed any GzmH expression. In the residual cell population, after depletion of monocytes and NK cells, a weak GzmH signal was observed (Figure S2B), probably corresponding to weak expression of GzmH in CD4+ T cells.13
GzmH is a cytotoxic protease able to induce cell death
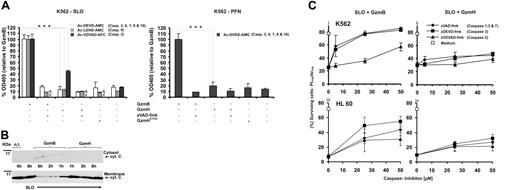
The cell death–inducing potential of GzmH, in comparison with that of GzmB, was assessed in an effector cell–independent system using recombinant GzmH and purified perforin (PFN) or streptolysin (SLO) as translocators. After target cell treatment with PFN or SLO, GzmH was nearly as potent as GzmB, reaching between 60% to 70% total cell death (AV+/PI− + AV+/PI+ cells). Importantly, PFN or SLO in combination with GzmHS195A revealed only a 5% to 10% increase compared with PFN/SLO background values (Figure 1Ai,ii). To ascertain the unique apoptosis-inducing specificity of GZMH, K562 cells were additionally treated with mast cell chymase, a closely related chymotrypsin-like serine protease and a member of the GZMH-GZMB gene cluster on chromosome 14q11.2. Although mast cell chymase efficiently cleaved our chymase substrates, the enzyme was not able to induce SLO-mediated target cell death (Figure 1Ai).
Cell death induced by recombinant GzmH. (A) K562 or HL60 cells exposed to GzmH and sublytic concentrations of SLO (i) or PFN (ii) resulted in near-complete cell death. Viability of K562 and HL60 cells was examined by annexin V–FITC (AV) and propidium iodide (PI) staining 10 to 12 (SLO) and 24 (PFN) hours after the indicated treatments. Depending on the translocator of choice, GzmB or GzmH was used at final concentrations of 5 or 10 μg/mL and 5 or 20 μg/mL, respectively, with the lower concentration used in combination with perforin. GzmHS195A and mast cell chymase were used at 25 μg/mL. Presented for both perforin- and SLO-mediated experiments is the sum of apoptotic (AV+ positive and PI- negative) and necrotic (AV+ and PI− cells from 3 pooled independent experiments (n = 3; ± standard deviation [SD]). Statistical significance is shown as *P > .005 (significant) and **P > .001 (very significant). In contrast to GzmH, mast cell chymase did not induce cell death. (B) Effective range of GzmH concentrations that induced cell death. K562 or HL60 cells were treated for 24 hours with sublytic SLO and GzmH or GzmB. The experiment shows the mean of triplicates ± SD and is representative of 2 independent experiments. (C) Time course of GzmH killing (shown is the mean of triplicates, representative of 2 independent experiments). GzmH induced cell death within 10 hours. Sublytic SLO induces 10% cell death on top of the natural background values. (D) Representative FACS data for K562 cells 12 hours after treatment with the indicated proteases, GzmB (7.5 μg/mL), GzmH and GzmHS195A (15 μg/mL each), and sublytic SLO. Apoptotic and dead cells were identified by annexin V and PI staining (ii). Dead cells are characterized by low FCS and high SSC signals (i). (E) Morphology of GzmH-treated K562 cells (magnification × 40). Ten hours after GzmH treatment, K562 cells displayed a characteristic morphology with increased granularity, condensation of nuclei, and membrane irregularities. Importantly, inactive GzmHS195A together with SLO did not trigger similar changes. (F) GzmH-induced cell death leads to the quick loss of membrane integrity. GzmH/SLO-treated K562 cells were stained with trypan blue at various time points (n = 3; ± SD). Contrary to GzmB, GzmH-induced cell death resulted in a much more pronounced trypan staining, with nearly 50% of cells positive after 4 hours. At this time point, GzmB-treated cells, in late-phase apoptosis, accounted for only 30%. Med. indicates medium; Chy., Chymase; n.d, not determined.
Cell death induced by recombinant GzmH. (A) K562 or HL60 cells exposed to GzmH and sublytic concentrations of SLO (i) or PFN (ii) resulted in near-complete cell death. Viability of K562 and HL60 cells was examined by annexin V–FITC (AV) and propidium iodide (PI) staining 10 to 12 (SLO) and 24 (PFN) hours after the indicated treatments. Depending on the translocator of choice, GzmB or GzmH was used at final concentrations of 5 or 10 μg/mL and 5 or 20 μg/mL, respectively, with the lower concentration used in combination with perforin. GzmHS195A and mast cell chymase were used at 25 μg/mL. Presented for both perforin- and SLO-mediated experiments is the sum of apoptotic (AV+ positive and PI- negative) and necrotic (AV+ and PI− cells from 3 pooled independent experiments (n = 3; ± standard deviation [SD]). Statistical significance is shown as *P > .005 (significant) and **P > .001 (very significant). In contrast to GzmH, mast cell chymase did not induce cell death. (B) Effective range of GzmH concentrations that induced cell death. K562 or HL60 cells were treated for 24 hours with sublytic SLO and GzmH or GzmB. The experiment shows the mean of triplicates ± SD and is representative of 2 independent experiments. (C) Time course of GzmH killing (shown is the mean of triplicates, representative of 2 independent experiments). GzmH induced cell death within 10 hours. Sublytic SLO induces 10% cell death on top of the natural background values. (D) Representative FACS data for K562 cells 12 hours after treatment with the indicated proteases, GzmB (7.5 μg/mL), GzmH and GzmHS195A (15 μg/mL each), and sublytic SLO. Apoptotic and dead cells were identified by annexin V and PI staining (ii). Dead cells are characterized by low FCS and high SSC signals (i). (E) Morphology of GzmH-treated K562 cells (magnification × 40). Ten hours after GzmH treatment, K562 cells displayed a characteristic morphology with increased granularity, condensation of nuclei, and membrane irregularities. Importantly, inactive GzmHS195A together with SLO did not trigger similar changes. (F) GzmH-induced cell death leads to the quick loss of membrane integrity. GzmH/SLO-treated K562 cells were stained with trypan blue at various time points (n = 3; ± SD). Contrary to GzmB, GzmH-induced cell death resulted in a much more pronounced trypan staining, with nearly 50% of cells positive after 4 hours. At this time point, GzmB-treated cells, in late-phase apoptosis, accounted for only 30%. Med. indicates medium; Chy., Chymase; n.d, not determined.
To determine the range of extracellular concentrations required for efficient killing, GzmH dilutions between 1 to 20 μg/mL were applied to SLO-treated K562 and HL60 cells (Figure 1B). After 24 hours, in a dose-dependent fashion, GzmH reached maximal killing efficiency at concentrations of 15 to 20 μg/mL (K562 cells) and 10 to 15 μg/mL (HL60 cells), whereas GzmB achieved comparable levels with both cell lines already at 10 μg/mL. Similarly, U937 cells were also efficiently killed by GzmH (data not shown).
To better define the time required for optimal GzmH-triggered killing, we performed a time course killing assay in which treated K562 cells were analyzed every 2 hours. The percentage of apoptotic and necrotic cells reached a plateau 10 hours after GzmH delivery, an effect GzmB achieved within only 2 hours (Figure 1C). The percentage of spontaneous cell death is also shown to indicate the minimal contribution of sublytic SLO (about 10%) to the total background values. Figure 1D shows representative FACS data of AV/PI-stained K562 cells 12 hours after treatment with the indicated proteases. The right column shows how active GzmH and GzmB strongly induced killing as deduced by the presence of high percentages of apoptotic and necrotic cells. The left column of plots, forward scatter (FSC) versus side scatter (SSC) signals, showed similar changes that occurred in both GZMH- and GZMB-treated cells, the dying apoptotic cells here characterized by low FSC and high SSC signals, typical of apoptotic cells (Figure 1D).
To assess the morphologic changes triggered by GzmH, we performed live cell imaging of GzmH-treated cells. Figure 1E reveals that GzmH, similar to GzmB, induces many of the typical morphologic changes that occur during apoptotic cell death, such as high granularity, nuclear condensation, apoptotic body formation, and irregularities of the cell surface membrane. Importantly, inactive GzmHS195A together with SLO did not elicit any of the above changes (Figure 1E).
Using trypan blue staining, the emergence of cell necrosis was assayed every 2 hours after GzmH and GzmB loading. Surprisingly, GzmH triggered the accumulation of trypan blue in target cells relatively quickly (45% of cells were positive after 4 hours), whereas the percentage of secondary cell necrosis was only 30% at the 4-hour time point (Figure 1F) in GzmB experiments. Membrane integrity was lost at a later time in response to GzmB, but GzmH required more time to induce noticeable cell death in most target cells. On the basis of these initial findings, the cell-death programs triggered by GzmH and GzmB appeared to be clearly distinguishable.
GzmH induces chromosomal condensation and nuclear fragmentation
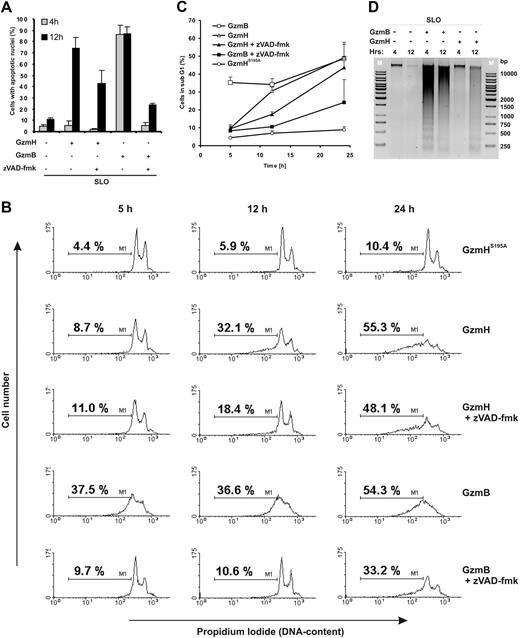
To further investigate the cellular changes after 12 hours of GzmH treatment, K562 cells were stained with the Hoechst dye 33342 and annexin V–FITC and analyzed by immunofluorescence microscopy after fixation. In contrast to cells treated with GzmHS195A, most cells stained positive for annexin V (dead and apoptotic cells) and displayed strong fragmented nuclei with condensed chromatin (Figure S3B). At the 4-hour time point, when maximum apoptotic nuclei were already visible after GzmB exposure, GzmH-treated cells still showed normal nuclei (Figure 2A). After 12 hours, however, most cells displayed apoptotic nuclei, fitting well to GzmH's delayed killing capacity (Figure 1C). GzmB-induced nuclear fragmentation was strongly inhibited by zVAD-fmk after 4 (94% inhibition) and 12 hours (70% inhibition) and was thus caspase dependent. In contrast, the effects of GzmH after 12 hours were only moderately inhibited by about 30%.
Nuclear condensation and fragmentation induced by GzmH. (A) Quantification of apoptotic nuclei after Hoechst 33342 staining 4 and 12 hours after the treatments indicated. The columns represent the mean with its standard deviations from triplicate measurements. (B) DNA content analysis of differentially treated K562 cells after 5, 12, and 24 hours. The pancaspase inhibitor zVAD-fmk was used at 100 μM. The 2 characteristic peaks (G1 and G2M, left and right peaks, respectively) of dividing cells were gradually lost, while cells with lower DNA content values in the so-called sub-G1 area appeared (indicated by M1). (C) Quantitative representation (n = 3; ± SD) of experiment B. Following the same trends as seen in the FACS data, GzmB-treated cells show early DNA degradation while GzmH works a slow but progressive diminution of DNA, with the percentage of sub-G1 cells matching that induced by GzmB at the 24-hour time point. (D) GzmH treatment does not cause apoptotic DNA laddering. K562 cells were treated for 4 and 12 hours as indicated, and cellular DNA was analyzed on a 1% agarose gel. The experiment was repeated 3 times.
Nuclear condensation and fragmentation induced by GzmH. (A) Quantification of apoptotic nuclei after Hoechst 33342 staining 4 and 12 hours after the treatments indicated. The columns represent the mean with its standard deviations from triplicate measurements. (B) DNA content analysis of differentially treated K562 cells after 5, 12, and 24 hours. The pancaspase inhibitor zVAD-fmk was used at 100 μM. The 2 characteristic peaks (G1 and G2M, left and right peaks, respectively) of dividing cells were gradually lost, while cells with lower DNA content values in the so-called sub-G1 area appeared (indicated by M1). (C) Quantitative representation (n = 3; ± SD) of experiment B. Following the same trends as seen in the FACS data, GzmB-treated cells show early DNA degradation while GzmH works a slow but progressive diminution of DNA, with the percentage of sub-G1 cells matching that induced by GzmB at the 24-hour time point. (D) GzmH treatment does not cause apoptotic DNA laddering. K562 cells were treated for 4 and 12 hours as indicated, and cellular DNA was analyzed on a 1% agarose gel. The experiment was repeated 3 times.
To explore the GzmH-induced damaging of cellular nuclei more explicitly, cell cycle experiments were performed. As expected, K562 cells treated with sublytic amounts of SLO and GzmHS195A revealed the characteristic peaks of proliferating cells in the G1, S, and G2 phases, with no sub-G1 peaks visible indicating the absence of fragmented apoptotic DNA. On the other hand, cells treated with either active GzmB or GzmH eventually lost the characteristic DNA peaks of living cells while the number of cells located in the sub-G1 area (M1) increased (Figure 2B). After GzmH treatment, the cell population in the sub-G1 area was evident only after 12 hours. Compared with GzmB, GzmH acted more slowly but induced similar levels of nuclear disintegration over a period of 24 hours. Consistent with the differential effects of zVAD-fmk on the nuclear morphology of GzmB- and GzmH-treated cells, the pancaspase inhibitor also moderately prevented GzmH-triggered DNA degradation, whereas inhibition by zVAD-fmk was highly efficient in protecting GzmB-treated target cells. Figure 2C is a quantitative presentation of our observations in Figure 2B specifying the percentage of sub-G1 cells (marker M1) after treatment (see figure legend).
Nuclear disintegration was then analyzed by agarose gel electrophoresis (Figure 2D). After 4 hours no laddering was detected in postnuclear lysates of GzmH-treated cells. A slight background smear became apparent at the 12-hour time point. On the other hand, GzmB, which efficiently activates procaspase 3, revealed a clear-cut laddering pattern after 4 hours. Moreover, levels of ICAD, the inhibiter of caspase-activated DNase (CAD), remained untouched during the time course experiment, with minimal decline only detectable at the 12-hour time point (Figure S4A). These observations strongly indicate that CAD activation does not take place during GzmH-induced cell death and suggest the late decline in ICAD is the result of secondary, indirect events.
Degradation and release of radioactively labeled nuclear DNA into the cytosol was, moreover, monitored by the 3H-thymidine release assay (Figure S4B). In line with the above experiments, GzmH induced slower 3H-thymidine release than GzmB, reaching maximum levels over 24 hours after cell-death induction. By contrast, the catalytically inactive GzmHS195A mutant did not induce any significant effects above background levels. Again, treatment with zVAD-fmk totally abolished the initial effects of GzmB after 4 hours. After 12 and 24 hours, however, cells treated with zVAD-fmk and either GzmB or GzmH displayed consistent 3H-thymidine release at similar levels. In summary, GzmH induced a slow form of DNA degradation, distinct from that of GzmB, which was only partially inhibited by zVAD-fmk.
GzmH induces mitochondrial damage and ROS production
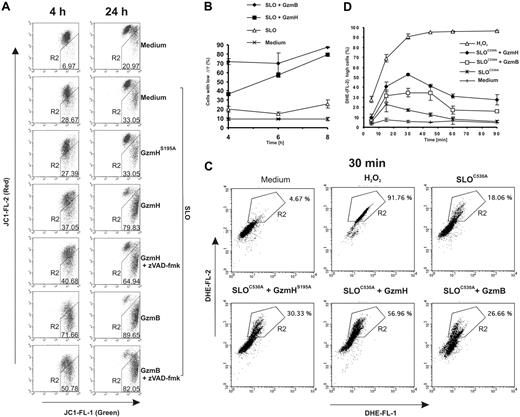
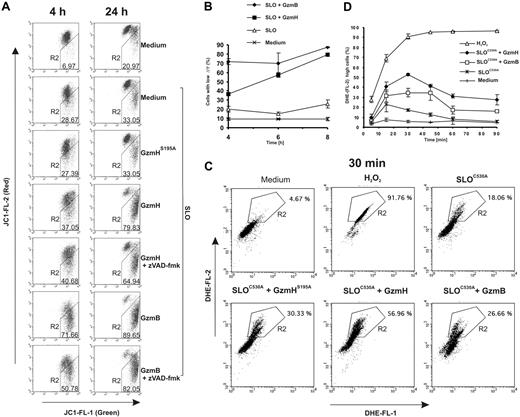
Using the green fluorescent probe JC-1, which accumulates and aggregates in the inner membrane space and appears red in intact mitochondria, we monitored the depolarization and potential loss (ΔΨm) of inner mitochondrial membranes in K562 cells. As expected, GzmB exerted its strong depolarizing effect on mitochondria already after 4 hours (Figure 3A, left column). A near-complete loss of red fluorescence was visible by FACS analyses after 24 hours (Figure 3A, right column). GzmH-treated cells displayed a similar loss of the membrane potential within 24 hours, but this effect occurred with some delay (Figure 3A-B). Treatment with GzmHS195A clearly confirmed that the decline was due to the proteolytic activity of GzmH (Figure 3A). Treatment with zVAD-fmk revealed that mitochondrial depolarization was in both cases not dependent on caspase activation (Figure 3A). Additionally, we performed a time course experiment and analyzed ΔΨm for up to 8 hours (Figure 3B). Compared with GzmB, mitochondrial depolarization induced by GzmH is less efficient but nevertheless is closely correlated, in terms of time, with the extent of cell death (Figure 1C).
GzmH triggers mitochondrial depolarization and ROS. (A) GzmH induces a loss of the mitochondrial membrane potential ΔΨm. K562 cells were treated with GzmH or GzmB in the presence or absence of the pancaspase inhibitor for 4 or 24 hours and analyzed by flow cytometry after JC-1 staining. The loss of ΔΨm is detected by a decrease of red fluorescence (FL2) and has been quantified by the percentage of cells appearing in the R2 region. (B) Accumulation of cells with loss of ΔΨm over time. The experiment shows the mean of triplicates (± SD) and is representative of 2 experiments. Depolarization in most cells was already observed at the 4-hour time point after GzmB treatment; comparable effects were visible after 8 hours in GzmH-treated cells. (C) ROS levels measured by DHE staining in GzmH- and GzmB-treated K562 cells after 30 minutes and over the first 90 minutes. K562 cells were treated with the indicated granzymes and SLOC530A, a mutated SLO variant that needs no prior reductive activation. The dye is oxidized by ROS to the strongly red fluorescent ethidium (R2 in the dot blots). H2O2 (1%) was used as positive control. (D) GzmH induced a rapid but transient ROS increase that peaked after 30 minutes. The data represent the average percentage of cells in area R2 of the raw FACS data. Shown is the mean of triplicates ± SD of 1 experiment representative of 2 experiments.
GzmH triggers mitochondrial depolarization and ROS. (A) GzmH induces a loss of the mitochondrial membrane potential ΔΨm. K562 cells were treated with GzmH or GzmB in the presence or absence of the pancaspase inhibitor for 4 or 24 hours and analyzed by flow cytometry after JC-1 staining. The loss of ΔΨm is detected by a decrease of red fluorescence (FL2) and has been quantified by the percentage of cells appearing in the R2 region. (B) Accumulation of cells with loss of ΔΨm over time. The experiment shows the mean of triplicates (± SD) and is representative of 2 experiments. Depolarization in most cells was already observed at the 4-hour time point after GzmB treatment; comparable effects were visible after 8 hours in GzmH-treated cells. (C) ROS levels measured by DHE staining in GzmH- and GzmB-treated K562 cells after 30 minutes and over the first 90 minutes. K562 cells were treated with the indicated granzymes and SLOC530A, a mutated SLO variant that needs no prior reductive activation. The dye is oxidized by ROS to the strongly red fluorescent ethidium (R2 in the dot blots). H2O2 (1%) was used as positive control. (D) GzmH induced a rapid but transient ROS increase that peaked after 30 minutes. The data represent the average percentage of cells in area R2 of the raw FACS data. Shown is the mean of triplicates ± SD of 1 experiment representative of 2 experiments.
To test whether mitochondrial dysfunction, as indicated by ΔΨm loss, was associated with increased ROS formation, we stained K562 cells with the oxidation-sensitive fluorogenic probe DHE, which is oxidized by ROS to products with strong red fluorescence.23 We found that GzmH induced an early but partially transient ROS increase (indicated by region 2 in Figure 3C) that peaks at about 30 minutes after SLO addition (Figure 3D). Thereafter, ROS declined to slightly elevated levels but gradually increased from 4 hours onward as cell death became more and more apparent (data not shown).
GzmH does not activate main apoptosis executioner molecules
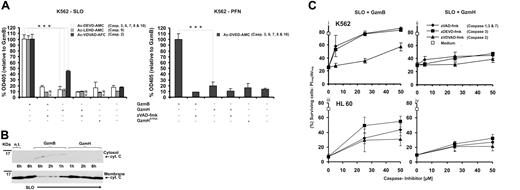
Because loss of the inner mitochondrial membrane potential (ΔΨm) and accumulation of ROS can be induced by caspase activation, we continuously monitored the activity of caspases 3, 6, and 7 in living cells for up to 10 hours using the cell-penetrating fluorogenic substrate Ac-DEVD-AMC. Figure 4A shows the efficient cleavage of Ac-DEVD-AMC in response to GzmB after K562 cells were exposed to sublytic PFN or SLO. Conversely, GzmH or GzmHS195A resulted only in baseline readings. To rule out any late surge in caspase activity, prolonged measurements of up to 15 hours were performed but indeed revealed no increase in activity (data not shown). The addition of zVAD-fmk completely inhibited GzmB-induced executioner caspase activity. In line with these experiments, catalytically inactive variants of main executioner caspases 3 and 7 (C285A mutants) and also purified Bid, which damages the mitochondrial outer membrane, were not directly cleaved by GzmH (Figure S5B-C).
GzmH kills independent of caspases. (A) Using cell-penetrating caspase substrates we analyzed caspase activities in living cells 10 hours after granzyme treatment. Data were normalized to GzmB and pooled from 3 (left) and 2 (right) independent experiments ± SD. In contrast to GzmB cell death mediated by sublytic concentrations of perforin (right) or SLO (left), GzmH did not induce cleavage of Ac-DEVD-AMC, which monitors the activities of caspases 3, 6, 7, 8, and 10. Addition of zVAD-fmk completely blocked the activation of caspases by GzmB. The same observations were seen with the caspase 9 substrate, Ac-LEHD-AMC, here only monitored using SLO as a translocator. Cleavage of the caspase 2 substrate, Ac-VDVAD-AFC, was observed in both GzmB- and GzmH-SLO treated K562 cells. In comparison with GzmB, the effect of GzmH was weaker but clearly above the background levels caused by sublytic SLO alone. (B) Western blot analysis (representative of 2 independent experiments) reveals that cytochrome c release is not a hallmark of GzmH-induced cell death. K562 cells were exposed to GzmH or GzmB and sublytic SLO for 6 hours. After cytosolic and membrane supernatant preparation, cytochrome c, in the case of GzmH treatment, could only be detected in the membrane fraction. The membrane fractions prepared 1, 2, and 6 hours after exposure to GzmB show a gradual decline of cytochrome c and gradually reappear in the cytosolic fractions. (C) GzmH kills independently of caspases in K562 and HL60 cells. Shown is the percentage of surviving cells (AV-/PI-) that had been treated with SLO and GzmB (i, iii) or GzmH (ii, iv) for 12 hours. Values represent the average of 3 experiments ± SD. The caspase inhibitors zVAD-fmk and zDEVD-fmk rescued cells from GzmB-induced apoptosis in a dose-dependent fashion, whereas killing by GzmH was minimally affected. Notably, the caspase 2 inhibitor zVDVAD-fmk did not protect the cells against the actions of GzmH. n.d indicates not determined.
GzmH kills independent of caspases. (A) Using cell-penetrating caspase substrates we analyzed caspase activities in living cells 10 hours after granzyme treatment. Data were normalized to GzmB and pooled from 3 (left) and 2 (right) independent experiments ± SD. In contrast to GzmB cell death mediated by sublytic concentrations of perforin (right) or SLO (left), GzmH did not induce cleavage of Ac-DEVD-AMC, which monitors the activities of caspases 3, 6, 7, 8, and 10. Addition of zVAD-fmk completely blocked the activation of caspases by GzmB. The same observations were seen with the caspase 9 substrate, Ac-LEHD-AMC, here only monitored using SLO as a translocator. Cleavage of the caspase 2 substrate, Ac-VDVAD-AFC, was observed in both GzmB- and GzmH-SLO treated K562 cells. In comparison with GzmB, the effect of GzmH was weaker but clearly above the background levels caused by sublytic SLO alone. (B) Western blot analysis (representative of 2 independent experiments) reveals that cytochrome c release is not a hallmark of GzmH-induced cell death. K562 cells were exposed to GzmH or GzmB and sublytic SLO for 6 hours. After cytosolic and membrane supernatant preparation, cytochrome c, in the case of GzmH treatment, could only be detected in the membrane fraction. The membrane fractions prepared 1, 2, and 6 hours after exposure to GzmB show a gradual decline of cytochrome c and gradually reappear in the cytosolic fractions. (C) GzmH kills independently of caspases in K562 and HL60 cells. Shown is the percentage of surviving cells (AV-/PI-) that had been treated with SLO and GzmB (i, iii) or GzmH (ii, iv) for 12 hours. Values represent the average of 3 experiments ± SD. The caspase inhibitors zVAD-fmk and zDEVD-fmk rescued cells from GzmB-induced apoptosis in a dose-dependent fashion, whereas killing by GzmH was minimally affected. Notably, the caspase 2 inhibitor zVDVAD-fmk did not protect the cells against the actions of GzmH. n.d indicates not determined.
We also measured caspase 9 activity using the fluorogenic substrate Ac-LEHD-AMC (Figures 4A and S5Aii). Whereas caspase 9 activity was easily detected in GzmB-treated K562 cells over time, no activity toward Ac-LEHD-AMC was found after SLO-mediated delivery of GzmH or GzmHS195A. Despite the lack of caspase 9 activity after GzmH treatment, we nevertheless chose to verify cytochrome c release, the smallest of the proapoptotic mitochondrial intermembrane space proteins. If the outer mitochondrial membrane were damaged by GzmH, cytochrome c should then be released into the cytosol, thereby triggering the formation of the apoptosome complex and procaspase 9 activation.24 To ascertain the lack of cytochrome c release, cytosolic fractions of GzmH-treated K562 cells were analyzed at different time points following cell-death induction. GzmH-treated target cells did not show any detectable cytochrome c loss from the inner membrane space of mitochondria (Figure 4B). As expected, cytosolic levels of cytochrome c increased over time in GzmB-treated K562 cells, while levels of cytochrome c declined in the respective membrane fractions (Figure 4B).
Activation of caspase 2 in response to DNA damage and genotoxic stress has recently been shown to trigger an alternative cell-death pathway.25 Because we demonstrated the lack of caspase 3 activity in response to GzmH-induced cell death, cleavage of the fluorogenic Ac-VDVAD-AFC substrate should uniquely monitor caspase 2 activity in GzmH-treated cells. GzmH, delivered to K562 cells with SLO, generated some Ac-VDVAD-AFC cleaving activity, whereas GzmB again showed a much greater effect at every recorded time point (Figures 4A left and S5Aiii). In experiments repeatedly performed with GzmHS195A, only background signals were recorded, whereas the caspase 2 inhibitor zVDVAD-fmk, as well as zVAD-fmk, completely prevented GzmH-dependent caspase 2–like activity (data not shown). GzmH apparently does not directly activate caspase 2, because, in contrast to GzmB, activity against Ac-VDVAD-AFC was not induced in a cytosolic K562-S100 lysate after GzmH addition (data not shown).
To investigate whether caspase activation, especially that of caspase 2 (Figures 4A and S5Aiii), was essential for GzmH-induced cell death, we pretreated K562 or HL60 cells with various concentrations (5, 25, 50 μM) of different inhibitors for caspases zVAD-fmk (a pancaspase inhibitor), zVDVAD-fmk (inhibiting caspase 2), and zDEVD-fmk (inhibiting the executioner caspases 3, 6, and 7) and monitored cell survival by AV/PI staining after 10 hours of incubation (Figure 4C). In GzmB-treated cells, zVAD-fmk and zDEVD-fmk restored the percentage of surviving cells to the level of untreated cells (Figure 4Ci,iii), while none of these inhibitors, even at the highest concentrations (50 μM), were able to offer protection against GzmH after 10 hours. By contrast, progression of GzmH-induced cell death, as monitored by the reduction of surviving cells after 10 hours, was poorly affected by all 3 inhibitors, implying that caspase activation, and especially caspase 2 activity, was not crucial for GzmH-mediated cell death (Figure 4Cii,iv).
Discussion
Because primary CD56+ NK cells express the strongly proapoptotic GzmA and GzmB along with perforin,26-28 other granzymes of NK cells, like GzmH and GzmM, have so far received little attention as genuine cell death–inducing effector proteases. Several older and more recent studies inferred a number of extracellular activities for granzymes, even in the case of GzmA29 and GzmB30 with regard to matrix remodeling and killer cell migration at the site of target cell attack. Similar extracellular functions are easily envisaged for the cathepsin G–related GzmH, and hence it was important to explore the death-inducing potential of human GzmH by a well-controllable delivery system.31 When we analyzed the distribution and expression levels of GzmH in lymphocyte populations of peripheral blood, we indeed confirmed CD3-, CD56+ NK cells (Figure S2B) as the only naive lymphocyte subset that contained relatively large amounts of GzmH.
To obtain milligram amounts of highly active and pure GzmH for various functional tests, we expressed the human GzmH with a cathepsin C–cleavable prosequence in E coli and developed an optimized refolding and purification protocol by modifying previously established procedures for GzmA,32 GzmB,16 and GzmK21 refolding. Compared with preparations of GzmH produced by baculovirus-infected cells,2 the bacterially expressed GzmH was not modified by heterologous and heterogeneous carbohydrates or a His tag at the C-terminus, which may alter cell surface interactions and its internalization by target cells. Modifications of GzmH at 2 putative N-linked glycosylation sites may occur (Figure S2B), but the structure and functions of potential carbohydrate modifications have so far not been clarified.
In this study the cell death–inducing activity of recombinant GzmH was investigated and compared with that of GzmB in an effector cell–independent system using two completely different translocators, purified PFN and SLO. Sublytic SLO is known to trigger a plasma membrane repair response like PFN, which is characterized by the exocytosis and fusion of lysosomes with the plasma membrane.33 GzmB-dependent events obtained with both SLO and PFN as delivering agents were highly reproducible and mirrored those previously observed in response to GzmB and PFN.34 Hence our findings obtained with SLO were not distorted by the mode of GzmH delivery. Target cells exposed to external GzmH and a translocator underwent biologic and morphologic changes indicative of nuclear damage and membrane perturbation. These effects were triggered neither by GzmHS195A nor by GzmH in the absence of translocators, implying that GzmH-induced cell death required the limited proteolysis of intracellular substrates that are only accessible after membrane translocation.
The induction of cell death by GzmH, as opposed to that by GzmB, was not mediated by caspase activation, cytochrome c release, Bid degradation, or the inactivation of the inhibitor of the CAD nuclease, which are the major hallmarks of GzmB. Although some caspase 2 activity was observed following GzmH delivery, the zVDVAD-fmk inhibitor did not suppress target cell death, as was the case for other major caspase-targeting fluoromethylketone-based inhibitors. By contrast, GzmB-triggered cell death was efficiently blocked by all inhibitors except for zVDVAD-fmk.
GzmH-induced cell death displayed several features that were reminiscent of murine GzmC-induced cytotoxic effects.6 After 12 hours of GzmH exposure but not before 4 hours, nuclei appeared condensed and fragmented. The morphologic changes of cellular nuclei were paralleled by the loss of mitochondrial membrane potential that was already noticeable at the 4-hour time point but affected almost all cells after 8 hours. GzmH cell death was also marked by an increase in intracellular ROS levels, which initially surged within 30 minutes of exposure to the protease. This late ROS accumulation was slower than in GzmB-treated cells and occurred with kinetics similar to the mitochondrial membrane depolarization of target cells. The zVAD-fmk caspase inhibitor did not prevent the loss of membrane potential during the first 4 hours and had only a moderate protecting effect during the later stages.
To further evaluate the causative role of ROS in cell-death induction, we applied several antioxidants, including Tiron, D1417, and MnTBAP (Mn(III)tetrakis(4-benzoic acid) porphyrin chloride), a cell-permeable superoxide dismutase (SOD) mimetic, to target cells. Under these experimental conditions we encountered a principal obstacle in delivering GzmH and GzmB with the same efficacy (data to be reported elsewhere). The usage of the redox-insensitive translocator protein, C530A variant of SLO, did not alleviate the inhibitory effects of antioxidants on granzyme delivery.
Although the chain of cause and effect has not yet been reconstructed for any of the known granzymes, cytotoxic activities of GzmA, GzmB, GzmH, and murine GzmC appear to converge and culminate in irreversible mitochondrial damage. Mitochondrial damage was observed with all of these granzymes in the absence of caspase activation despite the broad diversity in cleavage specificities with chymase-like, tryptase-like, and caspase-like activities. Thus, the initial chain of proteolytic events in the cytosol of target cells is most likely distinct for each granzyme and may hit on different cell-death pathways.
The biologic importance of the human GzmH gene is underscored by the fact that structural equivalents exist in all placental mammals. Primates like chimps and macaques, but also dogs and rabbits, possess a single copy GzmH gene on the 5′ side of the GzmB gene. The structural hallmarks of all GzmH homologs are Thr(Ser, Ala)189, Lys(Arg)192, and Gly226, which determine the S1 subsite, and Phe-99, which affects S2 (Figure S6). GzmB homologs of all these species differ from GzmH by Arg226 but otherwise are identical with GzmH at positions 192 and 99 (Figure S6). Indeed, replacement of Arg226 by Gly226 converted human GzmB into a GzmH-like variant that cleaved Suc-Ala-Ala-Pro-Phe-SBzl and Boc-Ala-Ala-Phe-SBzl like the wild-type GzmH.35
GzmH-like equivalents have not yet been identified unambiguously in the genomes of rodents (rats, mice) and artiodactyls (cattle). Local gene duplications and conversions generated a highly diverse and much greater repertoire of paralogous genes in these species.15,36 Due to a very recent sequence transfer from the adjacent GZMB locus to the ancestral \E GZMH locus affecting residues 63 to 142, similarity between human GZMH and human GZMB is very high. Likewise, the mouse GZMC ancestor, which had already branched off from the granzyme D, E, F, G precursor, was converted by adjacent GZMB sequences but differently and independently in the rodent lineage.15 The relatively high similarity between murine GZMC and GZMH (61% amino acid identity) and their immediate vicinity to the GZMB gene in the human and murine genome strongly suggest that GZMH is the human counterpart of murine GZMC. Predictive structure-function comparisons, however, indicate that, in addition to GzmC, murine GzmD and GzmE and the rat granzyme RNKP-7 should be considered as functional homologues of human GzmH. In GzmD and GzmE the key residues at positions 99 (His in RNKP-7), Lys192, and Gly226 are conserved but not in mouse and rat GzmC (Figure S6). Suc-Phe-Leu-Phe-SBzl–specific activity has indeed been reported to be involved in rat natural killer cell–mediated killing,37 but this peptidolytic activity has not been experimentally linked to the activity of purified rat GzmC. Because substrate specificities and cleavage activities of purified GzmC, GzmD, and GzmE have not yet been determined, it remains to be seen if any of the rodent granzymes C to E fulfill the same molecular functions as GzmH in humans.
In summary, our findings clearly indicate the importance of GzmH as an alternative cytotoxic effector protease. The biologic value and niche of this weapon, which was added to the arsenal of natural killer cells late during mammalian evolution, have yet to be deciphered. Once identified, however, they will contribute to a broader understanding of NK cell function, not only in tumor elimination and viral clearance but also as a regulator of inflammation and tissue repair.38 Notably, Andrade and colleagues have recently demonstrated an additional antiadenoviral activity of both GzmB and GzmH.39 Direct cleavage of adenoviral components by these granzymes reduced virus replication and resistance of infected host cells to killer cells.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisment” in accordance with 18 U.S.C. section 1734.
Acknowledgments
The authors thank E. Stegmann and W. Essbauer for excellent technical assistance and H. Wekerle for his continuous interest in the project. We thank R&D Systems for supplying us with pro-GzmH– and GzmH-specific antibodies; Dr C. J. Froelich for generous quantities of recombinant perforin as well as his helpful advice; G. Salvesen and S. Riedl for recombinant caspases; S. Bhakdi for recombinant SLOC530; and Novabiochem (Laufelfingen, Switzerland) for the provision of tailormade GzmH-AMC substrates.
This work was supported by the German Research Council (SFB571 and Je194/2–2).
Authorship
Contribution: E.F. designed and performed research, analyzed data, and wrote the paper; S.G.-P. contributed reagents and analytical tools; D.E.J. designed research and wrote the paper; and F.C.K. designed and performed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Florian C. Kurschus, Department of Neuroimmunology, Max-Planck-Institute of Neurobiology, Am Klopferspitz 18, 82152 Martinsried, Germany; e-mail: kurschus@neuro.mpg.de.

![Figure 1. Cell death induced by recombinant GzmH. (A) K562 or HL60 cells exposed to GzmH and sublytic concentrations of SLO (i) or PFN (ii) resulted in near-complete cell death. Viability of K562 and HL60 cells was examined by annexin V–FITC (AV) and propidium iodide (PI) staining 10 to 12 (SLO) and 24 (PFN) hours after the indicated treatments. Depending on the translocator of choice, GzmB or GzmH was used at final concentrations of 5 or 10 μg/mL and 5 or 20 μg/mL, respectively, with the lower concentration used in combination with perforin. GzmHS195A and mast cell chymase were used at 25 μg/mL. Presented for both perforin- and SLO-mediated experiments is the sum of apoptotic (AV+ positive and PI- negative) and necrotic (AV+ and PI− cells from 3 pooled independent experiments (n = 3; ± standard deviation [SD]). Statistical significance is shown as *P > .005 (significant) and **P > .001 (very significant). In contrast to GzmH, mast cell chymase did not induce cell death. (B) Effective range of GzmH concentrations that induced cell death. K562 or HL60 cells were treated for 24 hours with sublytic SLO and GzmH or GzmB. The experiment shows the mean of triplicates ± SD and is representative of 2 independent experiments. (C) Time course of GzmH killing (shown is the mean of triplicates, representative of 2 independent experiments). GzmH induced cell death within 10 hours. Sublytic SLO induces 10% cell death on top of the natural background values. (D) Representative FACS data for K562 cells 12 hours after treatment with the indicated proteases, GzmB (7.5 μg/mL), GzmH and GzmHS195A (15 μg/mL each), and sublytic SLO. Apoptotic and dead cells were identified by annexin V and PI staining (ii). Dead cells are characterized by low FCS and high SSC signals (i). (E) Morphology of GzmH-treated K562 cells (magnification × 40). Ten hours after GzmH treatment, K562 cells displayed a characteristic morphology with increased granularity, condensation of nuclei, and membrane irregularities. Importantly, inactive GzmHS195A together with SLO did not trigger similar changes. (F) GzmH-induced cell death leads to the quick loss of membrane integrity. GzmH/SLO-treated K562 cells were stained with trypan blue at various time points (n = 3; ± SD). Contrary to GzmB, GzmH-induced cell death resulted in a much more pronounced trypan staining, with nearly 50% of cells positive after 4 hours. At this time point, GzmB-treated cells, in late-phase apoptosis, accounted for only 30%. Med. indicates medium; Chy., Chymase; n.d, not determined.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/2/10.1182_blood-2006-10-051649/4/m_zh80130704560001.jpeg?Expires=1768228086&Signature=ycjF~Ztn6-aW7ErTIlwsMm6bOXHHKvxRxwlRq4tjO44YrfNf8vZ7G6ksM7kItKc1qNCuCUxMm2q7tN5qjFVTPVtVVVNHWTMbGrbEY8oj0B3asCAM4RK8MJgIdDRtL2tF93OKbv5duiSd2FjVRVnq1RGVqDcfrBzcRenaSQDRtkD91bG3gFGCCNh12A7LbrJyauySh21YDyX3V4xtrVsy4CmWeSzm81WtVRzvhyMGZ7kdiCZiK~ak2c-KE891LZqZuhFeGji-mxbphlR0TJn0oZ8Q5Ftw10uIa2e00Suwnmx-Gdmmz96Zrsohwq25GNQl6-QR0F1FtRkQGX7H9DiwLA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 1. Cell death induced by recombinant GzmH. (A) K562 or HL60 cells exposed to GzmH and sublytic concentrations of SLO (i) or PFN (ii) resulted in near-complete cell death. Viability of K562 and HL60 cells was examined by annexin V–FITC (AV) and propidium iodide (PI) staining 10 to 12 (SLO) and 24 (PFN) hours after the indicated treatments. Depending on the translocator of choice, GzmB or GzmH was used at final concentrations of 5 or 10 μg/mL and 5 or 20 μg/mL, respectively, with the lower concentration used in combination with perforin. GzmHS195A and mast cell chymase were used at 25 μg/mL. Presented for both perforin- and SLO-mediated experiments is the sum of apoptotic (AV+ positive and PI- negative) and necrotic (AV+ and PI− cells from 3 pooled independent experiments (n = 3; ± standard deviation [SD]). Statistical significance is shown as *P > .005 (significant) and **P > .001 (very significant). In contrast to GzmH, mast cell chymase did not induce cell death. (B) Effective range of GzmH concentrations that induced cell death. K562 or HL60 cells were treated for 24 hours with sublytic SLO and GzmH or GzmB. The experiment shows the mean of triplicates ± SD and is representative of 2 independent experiments. (C) Time course of GzmH killing (shown is the mean of triplicates, representative of 2 independent experiments). GzmH induced cell death within 10 hours. Sublytic SLO induces 10% cell death on top of the natural background values. (D) Representative FACS data for K562 cells 12 hours after treatment with the indicated proteases, GzmB (7.5 μg/mL), GzmH and GzmHS195A (15 μg/mL each), and sublytic SLO. Apoptotic and dead cells were identified by annexin V and PI staining (ii). Dead cells are characterized by low FCS and high SSC signals (i). (E) Morphology of GzmH-treated K562 cells (magnification × 40). Ten hours after GzmH treatment, K562 cells displayed a characteristic morphology with increased granularity, condensation of nuclei, and membrane irregularities. Importantly, inactive GzmHS195A together with SLO did not trigger similar changes. (F) GzmH-induced cell death leads to the quick loss of membrane integrity. GzmH/SLO-treated K562 cells were stained with trypan blue at various time points (n = 3; ± SD). Contrary to GzmB, GzmH-induced cell death resulted in a much more pronounced trypan staining, with nearly 50% of cells positive after 4 hours. At this time point, GzmB-treated cells, in late-phase apoptosis, accounted for only 30%. Med. indicates medium; Chy., Chymase; n.d, not determined.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/2/10.1182_blood-2006-10-051649/4/m_zh80130704560001.jpeg?Expires=1768632997&Signature=nQQF631nzpF0dn0TLBTw1trpZIzjcwcdSbvML4DKNeUtKm7oo58Qi3PlDKLHP4QfDDlD7ADUjcbsNN43~hatxfrcsCmt~JCq8NOta4Y5ljqXEdzsMt70-4oojS2PoPIsaEE~Uj3kkToLvNy-36MuYEs6VTiYvvVX2i8nXcSStjJmPSuQAl0rJpTW5SD4CgJV9W4TGO2ZGYEFCHrRszTXDV1Cq6csB4uv8wj1vNWpBdbP8wNI3j5ciYv0xSzTeytzqex1lckNPDIyAo6Vtz1ZPjdaNXSY0oMhMFpmndGwcedH7fCoUZb~EH3Oigzw8OoLRNh8iox5p~Le1cPgIrWHHA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)