Abstract
Expression of the K1 gene of human herpesvirus 8 activates nuclear factor-κB and induces lymph node hyperplasia and lymphomas in transgenic mice. To further delineate its role in cell survival, we determined whether K1 altered apoptosis of lymphoma cells. K1 protein is expressed in Kaposi sarcoma and primary effusion lymphoma. We retrovirally transfected BJAB lymphoma, THP-1, U937, and Kaposi sarcoma SLK cells to express K1 and a K1 mutant with the deleted immunoreceptor tyrosine-based activation motif (K1m). We challenged cells with an agonistic anti-Fas antibody, Fas ligand, irradiation, and tumor necrosis factor–related apoptosis-inducing ligand. K1 transfectants but not K1m transfectants exhibited reduced levels of apoptosis induced by the anti-Fas antibody but not apoptosis induced by the tumor necrosis factor–related apoptosis-inducing ligand or irradiation. K1 expression resulted in reduced apoptosis rates as shown in several assays. K1 induced a modest reduction in levels of Fas-associated death domain protein, and procaspase 8 recruited to the death-inducing signaling complex. Finally, K1 transfectants cleaved procaspase 8 at significantly lower rates than did K1m transfectants. K1-transfected mice, compared with vector-transfected mice, showed lower death rates after challenge with anti-Fas antibody. K1 may contribute to lymphoma development by stimulating cell survival by selectively blocking Fas-mediated apoptosis.
Introduction
Human herpesvirus 8 (HHV-8) has a pathogenic role in primary effusion lymphoma (PEL), Kaposi sarcoma (KS), and perhaps some cases of Castleman disease.1,2 One HHV-8 gene with transforming properties is K1. This gene is positionally homologous with transforming genes of Epstein-Barr virus. Indeed, K1 has transforming activity in rodent cells and in marmosets after K1 functionally replaces the STP gene in the herpesvirus saimiri genome.3 Ubiquitous expression of K1 in transgenic mice induces lymphoproliferation, splenomegaly, and lymphomas.4 Furthermore, K1 expression in mice induces signaling of nuclear factor-κB activation, which is associated with enhanced vascular endothelial growth factor expression and down-regulated interleukin-12 expression. Lymphocytes in K1 transgenic mice exhibit abnormal proliferation in response to antigens. Additionally, K1 expression induces activation-associated cytokine dysregulation, and the immunoreceptor tyrosine-based activation motif (ITAM) of K1 is constitutively phosphorylated, resulting in constitutive signaling.3–6 Finally, K1 stimulates Lyn tyrosine kinase activity in lymphocytes and lymphoma cells of K1 transgenic mice in an ITAM-dependent manner. In other models, ITAM signaling activates Lyn and ZAP70 by binding these kinases and activating the ITAM.7–9
Investigators have observed K1 expression in cases of Castleman disease and PEL. K1 RNA is present in PEL tissues and in PEL cell lines that can be up-regulated after treatment of cells with phorbol esters.6,10 Some KS cells express K1 RNA in the absence of lytic gene Orf26 expression.11 K1 is expressed in chronically infected cells and is up-regulated when cells enter the lytic phase of the virus life cycle.10,12,13 Further characterization of K1 protein in tissues has been limited because of its highly variable amino-acid sequence and propensity to complex with itself and other membrane proteins.12
Despite extensive studies, levels of K1 protein and the role of K1 expression in lymphomas and in mouse hyperplasias are not known. We previously hypothesized that because K1 activates nuclear factor-κB, it would also promote cell survival.14–16 Others have shown that K1 immortalizes cells.11 To determine whether K1 regulates apoptosis, we focused on the Fas (Apo-1/CD95) pathway, the most important apoptosis pathway that determines the fate of lymphocytes.17,18 To delineate the role of K1 in cell survival, in our present study we tested the ability of this protein to interfere with apoptosis in K1-transfected mice and in BJAB lymphoma cells and hematopoietic and endothelium-related cell lines (THP-1 macrophages/monocytes, U937 monocytes, and KS SLK cells) expressing K1 or a K1 mutant with an ITAM (K1m) with the Fas ligand, an anti-Fas antibody, and other agents that induce apoptosis.
Materials and methods
Cells
BJAB, THP-1, U937, BC3, BCBL1, KS1, HBL6, and KS SLK cells (American Type Culture Collection, Manassas, VA; E. Cesarman) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 50 U/mL penicillin, 50 μg/mL streptomycin, and 2 mM l-glutamine (all from Gibco Invitrogen, Grand Island, NY). Cultures of human embryonic kidney (HEK) 293 cells and PT67 NIH3T3 packaging cells (RetroPack PT67; Clontech, Mountain View, CA) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum.
Plasmids
The K1 gene was cloned by polymerase chain reaction (PCR) with DNA from HHV-8–infected BC-3 lymphoma cells.19 The gene was tagged at the carboxyl terminus with the DNA sequence encoding the myc tag and subcloned into the pSG5 (Stratagene, La Jolla, CA). The DNA sequence of K1 was confirmed by DNA sequencing. HEK 293 cells were transfected with pSG5 plasmids containing K1myc (pSG5-K1myc) or vector alone using the FuGENE 6 transfection reagent (Roche, Basel, Switzerland) and subjected to immunoprecipitation and immunoblotting with anti-myc antibody, which confirmed expression of K1 (Figure 1A). The cDNA of K1 and K1m were subcloned into the packaging plasmid pLXSN (Clontech). A reporter gene encoding alkaline phosphatase was also expressed in pLXSN. Mice were transfected by tail-vein inoculation of 100 μg/g mouse weight pCDEFK1Flag or vector alone.
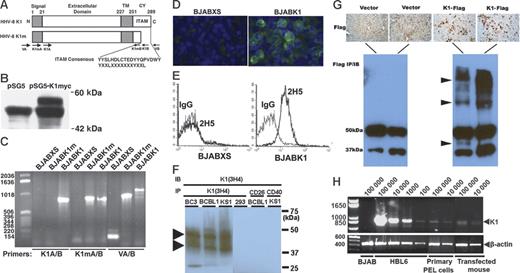
K1 encodes a transmembrane protein with an immunoreceptor tyrosine-based activation motif (ITAM). (A) Schematic of K1 protein with the ITAM consensus amino acid sequence. The K1 gene encodes for a single transmembrane (TM) protein with a short cytoplasmic (CY) sequence that contains an ITAM. The mutant K1 gene (K1m) has a truncated ITAM. Above the bars are the amino acid numbers. The arrows below the bars indicate the locations and names of the oligonucleotide primers used for PCR. (B) K1myc expression in transfected HEK 293 cell extracts subjected to immunoprecipitation and immunoblotting with an anti-myc antibody. A unique band with an estimated size of approximately 50 kDa was present in lysates of HEK 293 cells transfected with pSG5K1myc. (C) PCR analysis of K1 expression in the BJAB cells stably transfected with pLXSN vectors expressing K1 (BJABK1) or K1m (BJABK1m) or with pLXSN alone (BJABXS) after selection of cells with G-418 antibiotic. The sizes of the bands are consistent with the predicted DNA sizes. (D) Expression of K1-encoded protein in BJAB cells. Cells were stained with the primary anti-K1 antibody 2H5 and a secondary anti–mouse IgG–FITC antibody. Images were visualized using an Olympus BX51 TF fluorescence microscope (Olympus Optical, Tokyo, Japan) equipped with a 40×/0.75 numerical aperture (NA) objective. Images were captured using an Olympus DP21 camera system. (E) Flow-cytometry analysis of the expression of K1 on the surface of BJAB transfectants. BJAB transfectants were stained with 2H5 and the anti–mouse IgG–FITC antibody. As a control, each experimental histogram is overlaid with mouse isotype IgG-stained BJAB transfectants. (F) HHV-8–infected cells show the presence of K1 in monomer and aggregate forms by immunoprecipitation-immunoblotting analysis. (G) Mice transfected with K1-Flag show the presence of K1 (arrowheads). Images were visualized using a Zeiss Axioskop 2 plus (Zeiss Microimaging, Thornwood, NY) equipped with a 20×/0.50 NA objective. Images were captured using a Zeiss MC 80 DX camera, and were scanned using an HP Scanjet 8200 (Hewlett-Packard, Palo Alto, CA). (H) Total RNA was isolated from transfected mouse liver and cells. RNA from equal numbers of BJAB cells, cells from the HBL6 cell line, primary human PEL cells, and mouse liver cells was subjected to RT-PCR. β-actin mRNA was amplified as an internal control for input mRNA.
K1 encodes a transmembrane protein with an immunoreceptor tyrosine-based activation motif (ITAM). (A) Schematic of K1 protein with the ITAM consensus amino acid sequence. The K1 gene encodes for a single transmembrane (TM) protein with a short cytoplasmic (CY) sequence that contains an ITAM. The mutant K1 gene (K1m) has a truncated ITAM. Above the bars are the amino acid numbers. The arrows below the bars indicate the locations and names of the oligonucleotide primers used for PCR. (B) K1myc expression in transfected HEK 293 cell extracts subjected to immunoprecipitation and immunoblotting with an anti-myc antibody. A unique band with an estimated size of approximately 50 kDa was present in lysates of HEK 293 cells transfected with pSG5K1myc. (C) PCR analysis of K1 expression in the BJAB cells stably transfected with pLXSN vectors expressing K1 (BJABK1) or K1m (BJABK1m) or with pLXSN alone (BJABXS) after selection of cells with G-418 antibiotic. The sizes of the bands are consistent with the predicted DNA sizes. (D) Expression of K1-encoded protein in BJAB cells. Cells were stained with the primary anti-K1 antibody 2H5 and a secondary anti–mouse IgG–FITC antibody. Images were visualized using an Olympus BX51 TF fluorescence microscope (Olympus Optical, Tokyo, Japan) equipped with a 40×/0.75 numerical aperture (NA) objective. Images were captured using an Olympus DP21 camera system. (E) Flow-cytometry analysis of the expression of K1 on the surface of BJAB transfectants. BJAB transfectants were stained with 2H5 and the anti–mouse IgG–FITC antibody. As a control, each experimental histogram is overlaid with mouse isotype IgG-stained BJAB transfectants. (F) HHV-8–infected cells show the presence of K1 in monomer and aggregate forms by immunoprecipitation-immunoblotting analysis. (G) Mice transfected with K1-Flag show the presence of K1 (arrowheads). Images were visualized using a Zeiss Axioskop 2 plus (Zeiss Microimaging, Thornwood, NY) equipped with a 20×/0.50 NA objective. Images were captured using a Zeiss MC 80 DX camera, and were scanned using an HP Scanjet 8200 (Hewlett-Packard, Palo Alto, CA). (H) Total RNA was isolated from transfected mouse liver and cells. RNA from equal numbers of BJAB cells, cells from the HBL6 cell line, primary human PEL cells, and mouse liver cells was subjected to RT-PCR. β-actin mRNA was amplified as an internal control for input mRNA.
Retroviral infection
Recombinant virions were produced using the pLXSN retroviral vector containing K1 and K1m (Clontech) and added to BJAB, THP-1, U937, and KS SLK cells. Stably expressing cell lines were selected with 1000 μg/mL G-418 (Gibco Invitrogen). One microgram of genomic DNA was used as a template for amplification. The primers used were as follows: K1A, 5′-GTAGTTGTTTGCAGTCTGG-3′; K1B, 5′-TGGTTGCGTATAGTCTTCCG-3′; K1mA, 5′-CGCGAATTCATGTTCCTGTATATGTTGTTTGCATGCTGG-3′; K1mB, 5′-GATGGATCCCTAATCCTGCAATTGTTGTGGCA-3′; VA, 5′-CCCTTGAACCTCCTCGTTCGACC-3′; and VB, 5′-GAGACCTGGGACTTTCCACACCC-3′. BJAB cells containing the K1 gene, the K1m gene, and the pLXSN vector alone were assigned the names BJABK1, BJABK1m, and BJABXS, respectively. Names for transfected THP-1, U937, and KS SLK cells were assigned similarly.
Immunofluorescence staining and fluorescence-activated cell sorter (FACS) analysis
Cells were layered onto coated slides using the cytospin method, fixed with 1% paraformaldehyde at 4°C, and washed with phosphate-buffered saline (PBS). BJAB transfectants were incubated with the primary anti-K1 antibody 2H5 (a gift from Dr Jae U. Jung) 1:5 in dilution buffer, at ambient temperature for 30 minutes as reported previously.20 Slides were washed 3 times with PBS and incubated at ambient temperature for 15 minutes with a secondary antibody at a concentration of 20 μg/mL in dilution buffer. Slides were washed again and coverslips were mounted onto them using Vectashield mounting medium (Vector Laboratories, Burlingame, CA), followed by 4′, 6-diamidino-2-phenylindole (DAPI) staining. Representative images were recorded at an original magnification of × 400 with an Olympus BX51 TF fluorescence microscope. BJAB transfectants were also stained with 2H5 (1:5 in dilution buffer) and incubated at 37°C for 30 minutes. Cells were then washed 3 times with PBS and incubated with anti–mouse IgG fluorescein isothiocyanate (FITC; 1:100 dilution) at 37°C for 30 minutes and analyzed using the FACSCalibur system (BD Biosciences, San Jose, CA). Sections of paraffin-embedded tissue were incubated with anti-Flag antibody (1 μg/mL) followed by anti–mouse avidin-biotin (Vector Laboratories).
Immunoprecipitation and immunoblotting assays
BJAB transfectants were harvested by centrifugation and washed with cold PBS, and cell extracts were prepared in lysis buffer (1% Nonidet P-40, 50 mM HEPES [pH 7.5], 100 mM sodium chloride, 2 mM ethylenediaminetetraacetic acid, 1 mM pyrophosphate, 10 mM sodium orthovanadate, 3 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium fluoride, 4 μg/mL aprotinin, 1 μg/mL leupeptin; all from Sigma Chemical, St Louis, MO) for 30 minutes on ice. The lysate solution was spun at 13 000g for 10 minutes at 4°C, and supernatants were collected. Fifty micrograms of protein extract (Bio-Rad Laboratories, Hercules, CA) was separated on a sodium dodecyl sulfate polyacrylamide gel and transferred to a nitrocellulose membrane (Schleicher & Schuell, Keene, NH). For detection of the death-inducing signaling complex (DISC), 300 μg of cell extract was incubated at 4°C with 5 μg of the primary anti-Fas antibody for 1 hour; protein A/G sepharose (Pierce Biotechnology, Rockford, IL) was then added to the extract and the mixture was incubated for 4 hours. Pellets were washed 5 times in lysis buffer and boiled in sodium dodecyl sulfate–loading buffer, and supernatants were analyzed by an immunoblot assay with antibodies against Fas (B-10; sc-8009), Fas-associated death domain (FADD) protein (H-181; sc-5559), FLICE-inhibitory protein (cFLIP; G-11; sc-5276), myc (9E10; sc-40; Santa Cruz Biotechnology, Santa Cruz, CA), caspase 8 (1C12; Cell Signaling Technology, Beverly, MA), and hepatocyte growth factor receptor (c-Met). To identify K1, cell extracts were immunoprecipitated and immunoblotted with the 3H4 antibody (from Dr Jae U. Jung). Anti-CD26 and anti-CD40 antibodies (National Institutes of Health [NIH] AIDS Research and Reference Reagent Program) were used as controls. Bands were scanned using the Universal Hood II (Bio-Rad Laboratories) with Quantity One 4.5.0 software.
Apoptosis analysis
Apoptosis in transfected cells was induced by treating aliquots of cells with 0 to 50 ng/mL anti-Fas antibody CH-11 (Upstate, Lake Placid, NY), 100 ng/mL Fas ligand (Alexis Biochemical, San Diego, CA), 50 ng/mL tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), and 20 μg/mL cycloheximide (R&D Systems, Minneapolis, MN), or 6 or 8 Gy radiation.21 BJAB cells expressed TRAIL receptor–1 and TRAIL receptor–2.22,23 Radiation was delivered using a Nasatron 137Cs irradiator (US Nuclear, Burbank, CA).24 Typically, 5 Gy radiation induces cell death in 50% to 60% of HeLa cells.25,26 Cells with morphologic pattern of apoptosis were counted from 3 wells, and results were reported as means ± standard deviation.
Genomic DNA visualization and apoptosis analysis
Genomic DNA was isolated and visualized on a 1% agarose gel stained with ethidium bromide. Aliquots of cells were fixed with 4% formaldehyde and stained with Hoechst 33342 dye (Sigma Chemical) or DAPI. Cells were visualized at an original magnification of × 200 or × 400, and cells with morphologic features of apoptosis were counted. Also, 2 × 105 cells were fixed in 70% ethanol, treated with RNase A for 30 minutes, and stained with 25 μg/mL propidium iodide (Sigma Chemical) for 30 minutes. Also, living cells (2 × 105) were stained with 25 μg/mL propidium iodide and FITC-labeled annexin V (BD Biosciences). Cells were analyzed with a Coulter XL fluorescence-activated cell sorter (Beckman Coulter, Miami, FL). Aliquots of 104 cells were read using 525- and 630-nm filters.27
RT-PCR analysis
Total RNA was isolated from BJAB, HBL6, primary effusion lymphoma primary cells, and transfected mouse liver cells using TRI Reagent (Sigma Chemical). RNA was treated with DNase I, and cDNA was synthesized and amplified in 35 cycles using the One-Step reverse transcriptase–PCR (RT-PCR) System (Invitrogen) with K1 forward 5′-GTATGTTGTTTGCAGTCTGG-3′ and K1 reverse 5′-TGGTGCGTATAGTCTTCCG-3′ oligonucleotides. Dilutions of HBL6 RNA were used to generate standard curves. To control for relative RNA input in all the lanes, we used human β-actin with forward 5′-ACC TTC AAC CCA GCC ATG T-3′ and reverse 5′-CTG ATC CAC ATC TGC TGG AAG-3′ oligonucleotides for human cells, and mouse β-actin as control with forward 5′-TTC TTT GCA GCT CCT TCG TTG CCG-3′ and reverse 5′-TGG ATG GCT ACG TAC ATG GCT GGG-3′ oligonucleotides for transfected mouse tissue.28 The oligonucleotides selected are in different exons so that genomic DNA would produce an amplified signal of large size. To exclude the possibility of DNA contamination, RNA was treated with DNase I prior to amplification.
Caspase-8 activity assay
To show whether K1 interfered with caspase-8–related apoptosis, BJABK1, BJABK1m, and BJABXS cells were treated with 50 ng/mL CH-11. At periodic intervals, 106 cells were harvested and cell extracts were incubated with a caspase-8 substrate (7-amino-4-trifluoromethyl coumarin [IETD-AFC]; Calbiochem, San Diego, CA) at 37°C for 2 hours in the dark. Recombinant caspase 8 was used as a positive control, and a caspase-8 inhibitor (z-IETD-fmk) was used to check for specificity. Fluorescence was monitored via a multi-detection microplate reader (FLx800; Bio-Tek Instrument, Winooski, VT).
Induction of apoptosis in mice transfected with plasmid-expressing K1
Mice were transfected by intravenous inoculation of 100 μg pCDEFK1Flag. Mice were injected intravenously with 0.4 μg/g mouse weight Jo2 antibody and monitored for survival.
Statistical analysis
Results are expressed as mean values ± standard deviations. The 2-tailed Student t test was used to estimate statistical significance of the differences in results from 3 wells of cells. The level of significance was set to P values less than or equal to .05.29
Results
Expression of K1 in transfected BJAB cells
We confirmed the level of expression of K1myc in transiently transfected HEK 293 cells by performing immunoprecipitation and immunoblotting with an anti-myc antibody (Figure 1B). We subcloned the K1 and K1m cDNA into the retroviral vector pLXSN and packaged the cDNA in the PT67 NIH3T3 cell line, using supernatants from transfected packaging cells to transfect BJAB, U937, THP-1, and KS SLK cells with vectors expressing K1 and K1m. After selection of cells with G-418 antibiotic, we designated stable transfectants expressing K1 and K1m as BJABK1 and BJABK1m, respectively, and designated other cell lines accordingly.
PCR analysis indicated that transgenes of expected sizes were present in BJABK1 and BJABK1m cells with the use of different primers (Figure 1C). Furthermore, immunofluorescence analysis with the anti-K1 antibody 2H5 showed expression of K1 on the cell membrane and organelles (Figure 1D). We confirmed that K1 is expressed on the surface of BJAB transfectants by using flow-cytometry analysis after staining cells with the anti-K1 monoclonal antibody 2H5 (Figure 1E). To determine the level of K1 expression in HHV-8–infected cells, extracts of BC3, BCBL1, and KS1 (HHV-8 infected) and 293 (noninfected) cells were analyzed by immunoprecipitation-immunoblotting, which showed the presence of K1 as a monomer and as aggregated protein (Figure 1F, arrowheads). K1-transfected mice showed the presence of K1-Flag in liver and extract as a monomer and as a complexed protein (Figure 1G, arrowheads).
To determine the level of K1 expression in human tumors and K1-transfected mice, total RNA was isolated and dilutions of RNA were subjected to RT-PCR. We found that cells from transfected mice liver and primary effusion lymphoma contained comparable levels of K1 RNA, chronically HHV-8–infected cells (HBL6) contained higher levels of K1 RNA, and β-actin RNA levels showed the relative input RNA (Figure 1H). We amplified β-actin, which produced no PCR products if genomic DNA was present.
K1 suppresses apoptosis induced by Fas in BJAB transfectants
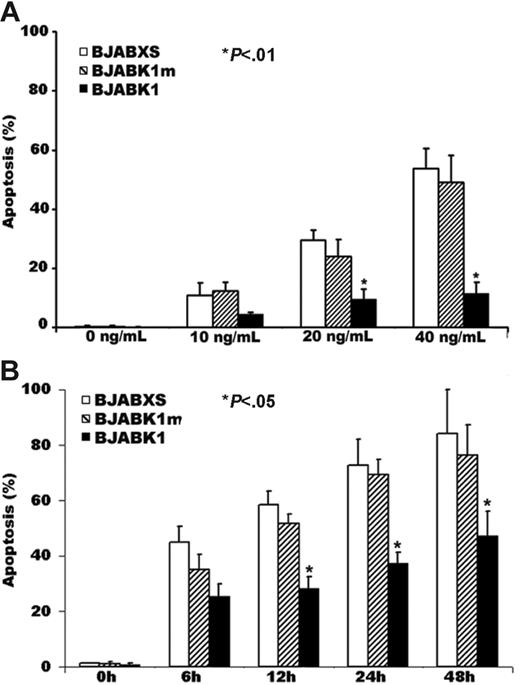
We treated BJAB transfectants with the anti-Fas antibody CH-11 for 24 hours, stained them with Hoechst 33342, and observed microscopically to detect morphologic features of apoptosis. BJABK1 cells but not BJABK1m or BJABXS cells showed lower rates of apoptosis (P < .05 for all comparisons; Figure 2A). These results suggest that the presence of the ITAM of K1 is required to suppress apoptosis.
Suppression of anti-Fas antibody–induced apoptosis in BJAB K1 transfectants. (A) Aliquots of cells were incubated for 24 hours with the anti-Fas antibody CH-11, stained with Hoechst 33342, and examined microscopically for morphologic features of apoptosis. Apoptosis rates in 4 wells were significantly lower in BJABK1 cells than in BJABK1m and BJABXS cells treated with 20 or 40 ng/mL CH-11 (*P < .01). The error bars indicate standard deviation. Similar results were obtained in 2 independent experiments. (B) Time-dependent killing effect in transfected BJAB cells. Cells were treated with 50 ng/mL CH-11, stained with Hoechst 33342, and examined microscopically for morphologic features of apoptosis. Apoptosis rates were significantly lower in BJABK1 cells than in BJABK1m and BJABXS cells at 12, 24, and 48 hours (*P < .05 for all comparisons). Similar results were obtained in 2 independent experiments.
Suppression of anti-Fas antibody–induced apoptosis in BJAB K1 transfectants. (A) Aliquots of cells were incubated for 24 hours with the anti-Fas antibody CH-11, stained with Hoechst 33342, and examined microscopically for morphologic features of apoptosis. Apoptosis rates in 4 wells were significantly lower in BJABK1 cells than in BJABK1m and BJABXS cells treated with 20 or 40 ng/mL CH-11 (*P < .01). The error bars indicate standard deviation. Similar results were obtained in 2 independent experiments. (B) Time-dependent killing effect in transfected BJAB cells. Cells were treated with 50 ng/mL CH-11, stained with Hoechst 33342, and examined microscopically for morphologic features of apoptosis. Apoptosis rates were significantly lower in BJABK1 cells than in BJABK1m and BJABXS cells at 12, 24, and 48 hours (*P < .05 for all comparisons). Similar results were obtained in 2 independent experiments.
We also treated BJAB transfectants with 50 ng/mL CH-11, stained them with DAPI, and observed microscopically to detect morphologic features of apoptosis. The percentage of transfectants undergoing apoptosis at 12, 24, and 48 hours was significantly lower in BJABK1 cells than in BJABK1m and BJABXS cells (P < .05 for all comparisons; Figure 2B).
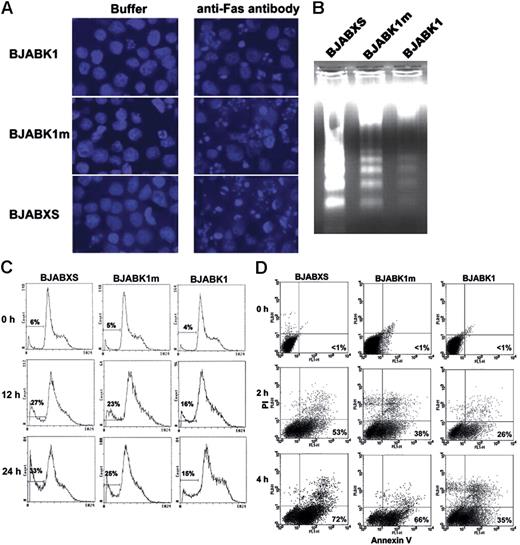
In another experiment, representative photomicrographs show BJAB transfectants 24 hours after treatment with an antibody buffer or 50 ng/mL CH-11 and staining with DAPI (Figure 3A). The apoptosis rates in buffer-treated BJABKI, BJABK1m, and BJABXS cells were 0.8%, 0.9%, and 2.0%, respectively, whereas the rates in CH-11–treated cells were 37% ± 4%, 72% ± 10%, and 72% ± 9%, respectively.
Protection of BJAB cells against anti-Fas antibody–induced apoptosis by K1 expression. (A) Transfected BJAB cells after treatment with a buffer or 50 ng/mL anti-Fas antibody CH-11 for 24 hours and staining with DAPI. Morphologic features of apoptosis were less frequent in BJABK1 cells than in BJABK1m and BJABXS cells. Representative photomicrographs are shown. Original magnification, × 400. Images are visualized and captured as described in Figure 1D. (B) BJABK1 cells showed less-fragmented DNA than did BJABK1m and BJABXS cells after induction of apoptosis with CH-11. Cells were incubated with 50 ng/mL CH-11 for 24 hours. Genomic DNA was then isolated and analyzed by electrophoresis on an agarose gel stained with ethidium bromide. Similar results were obtained in 2 independent experiments. (C) Transfected BJAB cells were treated with 50 ng/mL for 12 or 24 hours, stained with propidium iodide, and analyzed by flow cytometry. The number of cells containing subdiploid DNA is indicated by the curve under the horizontal bar at the left side of each panel. (D) Transfected BJAB cells were treated with an antibody buffer or 50 ng/mL CH-11 for 2 or 4 hours; cells were labeled with propidium iodide (PI) plus annexin V–FITC and analyzed by flow cytometry. Antibody-treated BJABK1 cells had lower apoptosis rates than did antibody-treated BJABK1m and BJABXS cells. Similar results were obtained in 2 independent experiments.
Protection of BJAB cells against anti-Fas antibody–induced apoptosis by K1 expression. (A) Transfected BJAB cells after treatment with a buffer or 50 ng/mL anti-Fas antibody CH-11 for 24 hours and staining with DAPI. Morphologic features of apoptosis were less frequent in BJABK1 cells than in BJABK1m and BJABXS cells. Representative photomicrographs are shown. Original magnification, × 400. Images are visualized and captured as described in Figure 1D. (B) BJABK1 cells showed less-fragmented DNA than did BJABK1m and BJABXS cells after induction of apoptosis with CH-11. Cells were incubated with 50 ng/mL CH-11 for 24 hours. Genomic DNA was then isolated and analyzed by electrophoresis on an agarose gel stained with ethidium bromide. Similar results were obtained in 2 independent experiments. (C) Transfected BJAB cells were treated with 50 ng/mL for 12 or 24 hours, stained with propidium iodide, and analyzed by flow cytometry. The number of cells containing subdiploid DNA is indicated by the curve under the horizontal bar at the left side of each panel. (D) Transfected BJAB cells were treated with an antibody buffer or 50 ng/mL CH-11 for 2 or 4 hours; cells were labeled with propidium iodide (PI) plus annexin V–FITC and analyzed by flow cytometry. Antibody-treated BJABK1 cells had lower apoptosis rates than did antibody-treated BJABK1m and BJABXS cells. Similar results were obtained in 2 independent experiments.
We monitored apoptosis and cell-cycle progression in BJAB transfectants by measuring DNA content using flow cytometry after treating cells with 50 ng/mL CH-11 and staining with propidium iodide. Fewer BJABK1 cells than BJABK1m cells and BJABXS cells contained subdiploid DNA at 12 and 24 hours (Figure 3C). Consistently, there appeared to be more BJABK1 cells than BJABK1m or BJABXS cells in S phase after 24 hours of treatment. Repeated experiments had similar results. Thus, K1-transfected cells were protected from CH-11–triggered apoptosis, and the ITAM of K1 was required for this protection.
Given these novel findings, we tested for K1 suppression against apoptosis using combined staining with propidium iodide and annexin V–FITC. BJABK1, BJABK1m, and BJABXS cells were treated with 50 ng/mL CH-11 for 0, 2, and 4 hours. We then doubly stained cells and analyzed them using flow cytometry. At 4 hours, BJABK1 cells had a lower apoptosis rate (35% ± 7%) than did BJABK1m cells (66% ± 11%) and BJABXS cells (72% ± 11%; Figure 3D). These results confirmed occurrence of CH-11–induced apoptosis and partial suppression of apoptosis by K1.
We induced apoptosis of BJAB transfectants by treating them with 100 ng/mL cross-linked Fas ligand. After 16 hours, we stained cells with Hoechst 33342 and assessed them microscopically for morphologic features of apoptosis. The apoptosis rate in BJABK1, BJABK1m, and BJABXS cells was 20% ± 6%, 45% ± 6%, and 50% ± 7%, respectively (data not shown). Thus, K1 partially suppressed the apoptotic effect of the Fas ligand as observed with anti-Fas antibody.
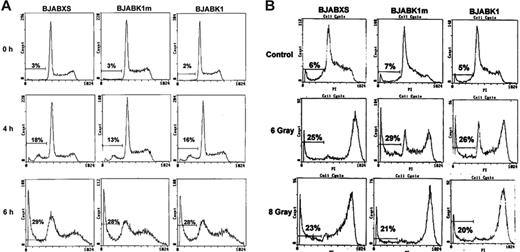
Apoptosis can be induced by various stimuli. Ligands other than Fas ligand can bind cognate receptors and induce apoptosis. We treated BJAB transfectants with TRAIL, stained them with propidium iodide, and analyzed them using flow cytometry. The subdiploid DNA content of the cells was higher after 6 hours of treatment than after 4 hours. The number of BJABK1 cells with subdiploid DNA was not significantly different from that of BJABK1m or BJABXS cells after 4 and 6 hours (Figure 4A). Thus, K1 did not selectively suppress apoptosis induced by TRAIL.
Expression of K1 in BJAB cells does not inhibit TRAIL-induced or irradiation-induced apoptosis. (A) BJAB transfectants were treated with 50 ng/mL TRAIL for 4 and 6 hours, fixed, stained with propidium iodide for DNA content, and analyzed by flow cytometry. The number of cells containing subdiploid DNA is indicated by the curve under the horizontal bar at the left of each panel. The percentage of apoptosis in BJABK1, BJABK1m, and BJABXS cells was similar at these intervals. Similar results were obtained in 2 independent experiments. (B) BJAB transfectants were irradiated at 6 or 8 Gy and then incubated for 24 hours, fixed, stained with propidium iodide (PI), and analyzed using flow cytometry. There were no differences in apoptosis rates of BJABK1 cells compared with those of BJABK1m and BJABXS cells. Similar results were obtained in 2 independent experiments.
Expression of K1 in BJAB cells does not inhibit TRAIL-induced or irradiation-induced apoptosis. (A) BJAB transfectants were treated with 50 ng/mL TRAIL for 4 and 6 hours, fixed, stained with propidium iodide for DNA content, and analyzed by flow cytometry. The number of cells containing subdiploid DNA is indicated by the curve under the horizontal bar at the left of each panel. The percentage of apoptosis in BJABK1, BJABK1m, and BJABXS cells was similar at these intervals. Similar results were obtained in 2 independent experiments. (B) BJAB transfectants were irradiated at 6 or 8 Gy and then incubated for 24 hours, fixed, stained with propidium iodide (PI), and analyzed using flow cytometry. There were no differences in apoptosis rates of BJABK1 cells compared with those of BJABK1m and BJABXS cells. Similar results were obtained in 2 independent experiments.
When we irradiated BJAB transfectants at 6 Gy to induce apoptosis, we found that irradiation killed BJABXS, BJABK1m, and BJABK1 cells at similar rates (23%, 21%, and 20%, respectively). Irradiation at 8 Gy also resulted in no difference in the apoptosis rates (Figure 4B). We also found that the irradiation induced polyploidy in BJAB transfectants. Irradiated transfectants had a high level of p53 protein expression as determined by immunoblot analysis (data not shown). Results indicated that K1 neither preferentially suppressed the up-regulation of p53 protein nor protected cells from radiation-induced apoptosis.30
Suppression of apoptosis by K1 in other transfected cells
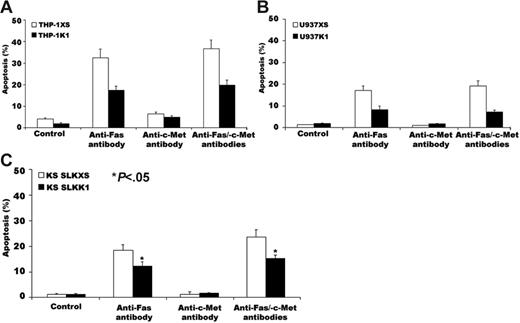
For 24 hours, we stimulated transfected THP-1, U937, and KS SLK cells (selected because they have an intact Fas-signaling system) with 50 ng/mL anti-Fas antibody CH-11; we then stained cells with DAPI and assessed microscopically for morphologic features of apoptosis.27 The apoptosis rate was 17% ± 2% in THP-1 cells containing the K1 gene; 33% ± 4% in THP-1 cells with the pLXSN vector alone (Figure 5A); 8% ± 2% in U937 cells containing the K1 gene; 17% ± 2% in U937 cells with the pLXSN vector alone (Figure 5B); 13% ± 3% in KS SLK cells containing the K1 gene; and 20% ± 4% in KS SLK cells with the pLXSN vector alone (Figure 5C). Treatment with control anti-HGFR antibody alone or in combination with CH-11 did not alter apoptosis rates in the transfectants.
Expression of K1 in THP-1, U937, and KS SLK cells blocks anti-Fas antibody–induced apoptosis. THP-1, U937, and KS SLK transfectants were incubated with 50 ng/mL CH-11, 1 μg/mL anti-HGFR antibody, or both for 24 hours. Cells were then fixed and analyzed for apoptosis by DAPI staining. (A) Apoptosis of THP-1 transfectants induced by CH-11, anti-HGFR antibody, or both. (B) Apoptosis of U937 transfectants induced by CH-11, anti-HGFR antibody, or both. (C) Apoptosis of KS SLK transfectants induced by CH-11, anti-HGFR antibody, or both. The error bars indicate standard deviation (*P < .05 for all comparisons). Similar results were obtained in 2 independent experiments.
Expression of K1 in THP-1, U937, and KS SLK cells blocks anti-Fas antibody–induced apoptosis. THP-1, U937, and KS SLK transfectants were incubated with 50 ng/mL CH-11, 1 μg/mL anti-HGFR antibody, or both for 24 hours. Cells were then fixed and analyzed for apoptosis by DAPI staining. (A) Apoptosis of THP-1 transfectants induced by CH-11, anti-HGFR antibody, or both. (B) Apoptosis of U937 transfectants induced by CH-11, anti-HGFR antibody, or both. (C) Apoptosis of KS SLK transfectants induced by CH-11, anti-HGFR antibody, or both. The error bars indicate standard deviation (*P < .05 for all comparisons). Similar results were obtained in 2 independent experiments.
K1 and formation of the DISC
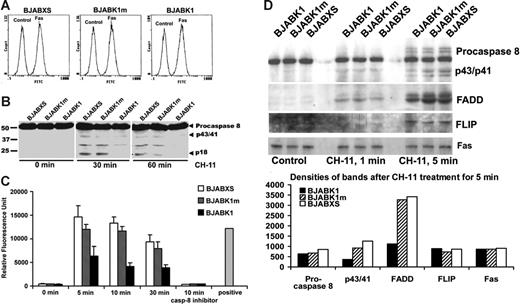
We found similar levels of expression of Fas receptors in BJABK1, BJABK1m, and BJABXS cells (Figure 6A). Thus, altered expression of Fas receptor did not explain the suppression of apoptosis in K1-expressing cells. DISC formation culminates in the binding and cleavage of procaspase 8.23,31,32 We monitored proteolytic caspase 8 activities in BJAB transfectants treated with 50 ng/mL anti-Fas antibody CH-11 for 30 or 60 minutes. We probed cell extracts by immunoblotting with an anti–caspase 8 antibody that detects cleavage peptides. BJABK1 cells had lower levels of caspase 8 cleavage peptides than did BJABK1m and BJABXS cells at both 30 and 60 minutes (Figure 6B).33 BJABK1 cell extracts did not have detectable levels of caspase 8 subunit p18 products, whereas the same extracts had equal levels of β-actin. To assess caspase activity, we subjected cell extracts to fluorescently labeled peptide substrates. At 5, 10, and 30 minutes after treatment with CH-11, BJABK1 cells had lower levels of caspase 8 activities than did BJABK1m and BJABXS cells (Figure 6C). This assay monitored caspase 8 activity because addition of the caspase 8 inhibitor z-IETD-fmk completely abolished activity.
BJAB transfectants have similar levels of expression of Fas receptor, and BJABK1 cells recruit fewer subunits of Fas, FADD, and procaspase 8 to the DISC after stimulation with an anti-Fas antibody. (A) Expression of Fas on the surface of BJAB transfectants. Stably transfected BJAB cells were stained with the anti–mouse IgG–FITC antibody, and expression of Fas was detected by flow cytometry. The levels of Fas expression were the same in BJABK1, BJABK1m, and BJABXS cells. (B) Suppression of the cleavage of procaspase 8 by expression of K1. BJAB transfectants were evaluated for their level of procaspase 8 cleavage with the anti-Fas antibody CH-11. After 30 or 60 minutes of incubation with CH-11, BJABK1 cells contained low levels of procaspase 8 cleavage products (indicated by arrowheads). Recombinant caspase 8 was used as a positive control. Similar results were obtained in 3 independent experiments. (C) Caspase 8 activity in BJAB transfectants. BJAB transfectants were incubated with CH-11; at periodic intervals, extracts were removed and incubated with the caspase 8 substrate IETD-AFC at 37°C for 2 hours, and caspase 8 activity was analyzed by using a fluorescence reader. BJABK1 cells had a significantly lower level of caspase 8 activity than BJABK1m and BJABXS cells (P < .005). Caspase 8 activity was completely inhibited by treatment with the caspase 8 inhibitor z-IETD-fmk. Error bars represent SD. Similar results were obtained in 2 independent experiments. (D) BJABK1, BJABK1m, and BJABXS cells were treated with 1 μg/mL CH-11 for the indicated times. Cell extracts were subjected to immunoprecipitation (IP) with CH-11 and immunoblotting (IB) with antibodies targeting procaspase 8, FADD, and FLIP. All of the transfectants recruited these DISC subunits as indicated by the bands shown. The intensity of the p43/p41 and FADD bands was lower in K1 compared with vector-transfected cells. Fas and cFLIP levels remained unchanged. Similar results were obtained in 3 independent experiments.
BJAB transfectants have similar levels of expression of Fas receptor, and BJABK1 cells recruit fewer subunits of Fas, FADD, and procaspase 8 to the DISC after stimulation with an anti-Fas antibody. (A) Expression of Fas on the surface of BJAB transfectants. Stably transfected BJAB cells were stained with the anti–mouse IgG–FITC antibody, and expression of Fas was detected by flow cytometry. The levels of Fas expression were the same in BJABK1, BJABK1m, and BJABXS cells. (B) Suppression of the cleavage of procaspase 8 by expression of K1. BJAB transfectants were evaluated for their level of procaspase 8 cleavage with the anti-Fas antibody CH-11. After 30 or 60 minutes of incubation with CH-11, BJABK1 cells contained low levels of procaspase 8 cleavage products (indicated by arrowheads). Recombinant caspase 8 was used as a positive control. Similar results were obtained in 3 independent experiments. (C) Caspase 8 activity in BJAB transfectants. BJAB transfectants were incubated with CH-11; at periodic intervals, extracts were removed and incubated with the caspase 8 substrate IETD-AFC at 37°C for 2 hours, and caspase 8 activity was analyzed by using a fluorescence reader. BJABK1 cells had a significantly lower level of caspase 8 activity than BJABK1m and BJABXS cells (P < .005). Caspase 8 activity was completely inhibited by treatment with the caspase 8 inhibitor z-IETD-fmk. Error bars represent SD. Similar results were obtained in 2 independent experiments. (D) BJABK1, BJABK1m, and BJABXS cells were treated with 1 μg/mL CH-11 for the indicated times. Cell extracts were subjected to immunoprecipitation (IP) with CH-11 and immunoblotting (IB) with antibodies targeting procaspase 8, FADD, and FLIP. All of the transfectants recruited these DISC subunits as indicated by the bands shown. The intensity of the p43/p41 and FADD bands was lower in K1 compared with vector-transfected cells. Fas and cFLIP levels remained unchanged. Similar results were obtained in 3 independent experiments.
To assess structural formation of the DISC, we incubated BJAB transfectants with agonistic CH-11 and analyzed them using immunoprecipitation and immunoblotting. Recruitment of p43/p41 and FADD to the DISC was consistently and modestly lower in BJABK1 cells compared with BJABK1m and BJABXS cells (Figure 6D). We repeated this analysis 3 times with similar results. Therefore, suppression of Fas-mediated apoptosis may be partially explained by the lower level of DISC formation by K1. The decreased levels in DISC formation were associated with significantly reduced caspase 8 activity (Figure 6C).
Survival of mice transfected with plasmid-expressing K1 after challenge with anti-Fas antibody
Activating anti-Fas (Jo2) antibody induces death of mice primarily by inducing apoptosis of hepatocytes.34–36 After 6 hours, 10 of 10 pCDEFK1Flag-transfected mice and 1 of 14 vector-transfected mice survived (P < .001; Table 1). K1Flag-transfected mice at necropsy showed livers with pink color and some hemorrhaging, whereas vector-alone–transfected mice showed consistently black-colored livers and extensive hemorrhaging. In K1Flag-transfected mice, 5% of liver cells expressed K1Flag (Figure 1G). This observation is consistent with prior studies indicating that Jo2 induces apoptosis of hepatocytes as the primarily insult causing mouse death, which in our series is protected by K1.34–36
Discussion
In this study, we showed that expression of the HHV-8 protein K1 suppresses apoptosis induced by the anti-Fas antibody CH-11 or Fas ligand in several cell types, including cells of hematopoietic or endothelial origin. Expression of K1 in transfected mice also protected mice against lethal effects of activating anti-Fas antibody. This suppression of apoptosis appears to be selective for Fas because K1 did not suppress apoptosis induced by TRAIL or irradiation. Furthermore, K1 appears to interfere with apoptosis early in the signaling pathway of Fas-mediated apoptosis, as we found that K1 partially suppressed formation of DISC and significantly blocked caspase 8 activity.
Suppression of apoptosis by K1 does not appear to be a late-stage or global cellular effect. TRAIL uses receptors (TRAIL receptor-4 and TRAIL receptor-5) that are different from those of Fas ligand. TRAIL and Fas ligand signaling share the subunits of FADD and procaspase 8 in apoptosis signaling.37 Therefore, K1 must directly interfere with Fas or another DISC subunit selectively present only in the Fas pathway. Irradiation induces apoptosis through expression of p53 and can also secondarily involve Fas-mediated signaling.38 Despite not having defined a mechanism for blockage of Fas, these studies are important and significant enough to protect mice from Fas-mediated death.
Fas is a target of numerous agents that regulate apoptosis. For example, receptors can regulate apoptosis by binding and sequestering Fas, thus preventing DISC formation and apoptosis.39,40 Viral proteins that regulate Fas-mediated apoptosis mimic cFLIP by blocking caspase 8 by competition in binding to the DISC. We observed recruitment of lower levels of caspase 8 subunits with K1 and significantly lower caspase 8 activity and apoptosis rates with K1. These alterations in DISC function are consistent with K1-related suppression of apoptosis.
K1 requires its ITAM to suppress apoptosis. ITAMs are found in the subunits of immune receptor complexes of lymphocytes, including B-cell receptors (BCRs) and T-cell receptors.41–43 K1 without the ITAM (K1m) lost most of its ability to suppress apoptosis, indicating that ITAM-dependent signaling is required. Furthermore, ITAM-binding signaling kinases such as Lyn kinase bind to the DISC and are capable of modulating apoptosis. There may be a connection between K1 and apoptosis because K1 activates Lyn and related kinases.9 K1 also may mediate regulation of apoptosis through protein kinase B and suppress Fas-mediated apoptosis in a PI3 kinase–dependent manner.44 However in our system, blockage appears to occur at the DISC.
K1 and other viral proteins with ITAMs disrupt the function of pathways of cellular proteins containing ITAMs. For example, K1 and Epstein-Barr virus LMP2 both modulate calcium mobilization and block BCR signaling.45 However, the roles in apoptosis of other viral proteins with ITAMs, such as hantavirus glycoprotein G1, bovine leukemia virus gp30, simian immunodeficiency virus PBj14 nef, and rhesus monkey rhadinovirus R1, are not clear.45–49 The apparent underlying primary effect of the expression of viral proteins with ITAMs is binding and inhibition of the function of host ITAM receptors in a dominant-negative manner. The DISC does not contain subunits with ITAMs, so DISC subunits may be an indirect target of K1.
Clues regarding how K1 regulates apoptosis may lie in its potential ability to bind other proteins. K1 binds with itself and other receptors such as IgGλ by intermolecular disulfide binding in its extracellular domain.12 Multiple cysteine residues of the extracellular domain of K1 are essential for the aggregation properties and may mediate regulation of apoptosis through binding and disruption of receptors with ITAMs. Even K1 with a deleted ITAM has some antiapoptosis activity (Figures 2–3), suggesting a dominant-negative interaction by binding and interference with receptors. Hepatocyte growth factor receptor expressed in cells and mice binds to Fas and blocks apoptosis.39 K1 and hepatocyte growth factor receptor may participate in a similar fashion to prevent apoptosis. Given its apoptosis-suppressive effects, we hypothesized that K1 promotes the survival of infected cells and the dissemination of HHV-8. Constitutive expression of K1 in transgenic mice produces lymph node hyperplasia and lymphomas with a non–B-cell, non–T-cell, and CD138+ marker phenotype resembling that of HHV-8–associated lymphomas, suggesting that K1 may be sufficient for expansion of the lymphocyte pool and phenotype of human lymphomas.9
Authorship
Contribution: Suizhao Wang performed apoptosis caspase 8 activity studies; Shu Wang performed experiments with the animal model and death-inducing signaling complexes; H.M. performed apoptosis studies; D.P.Y. performed transfections and cell-culture analysis; O.P. performed K1 animal studies; L.E.F. was responsible for human lymphoma cell characterization, K1 expression, and writing the paper; A.Y. was responsible for human lymphoma cell characterization, K1 expression, and writing the paper; and F.S. was responsible for the design of the research studies and for writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Suizhao Wang and Shu Wang contributed equally to this study.
Correspondence: Felipe Samaniego, Department of Lymphoma and Myeloma, Unit 429, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Boulevard, Houston, TX 77030; e-mail: fsamaniego@mdanderson.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute (grants CA106173, CA16672, and CA098412), The Leukemia & Lymphoma Society Translational Research Program, The Concern Foundation, NIH AIDS Research & Reagent Program, and The University of Texas M. D. Anderson Cancer Center Multidisciplinary Research Program and Institutional Research Grant.
We thank Jae V. Jung for the 3H4 antibody. We are grateful to the George Diamantis family for support of this research and thank Cindy Cabrera, Sharon Turner, and Alison Woo for assistance in the preparation of the manuscript.