Abstract
The polymerization of fibrin occurs primarily through interactions between N-terminal A- and B-knobs, which are exposed by the cleavage of fibrinopeptides A and B, respectively, and between corresponding a- and b-holes in the γ- and β-modules. Of the potential knob-hole interactions—A:a, B:b, A:b, and B:a—the first has been shown to be critical for fibrin formation, but the roles of the others have remained elusive. Using laser tweezers–based force spectroscopy, we observed and quantified individual B:b and A:b interactions. Both desA-fibrin with exposed A-knobs and desB-fibrin bearing B-knobs interacted with fragment D from the γD364H fibrinogen containing b-holes but no functional a-holes. The strength of single B:b interactions was found to be 15 to 20 pN, approximately 6-fold weaker than A:a interactions. B:b binding was abrogated by B-knob mimetic peptide, the (β15-66)2 fragment containing 2 B-knobs, and a monoclonal antibody against the β15-21 sequence. The interaction of desB-fibrin with fragment D containing a- and b-holes produced the same forces that were insensitive to A-knob mimetic peptide, suggesting that B:a interactions were absent. These results directly demonstrate for the first time B:b binding mediated by natural B-knobs exposed in a fibrin monomer.
Introduction
The fibrin clot is a branched protein polymer that provides the three-dimensional (3-D) scaffold for a thrombus in vertebrates. Fibrin is formed from a soluble precursor, fibrinogen, upon the action of thrombin, a blood enzyme. Thrombin normally cleaves 4 relatively small peptides from fibrinogen, producing fibrin monomers capable of spontaneous self-assembly into 2-stranded oligomeric structures called protofibrils. Once the protofibrils reach a critical size (approximately10 nm thick and 600-800 nm long), they aggregate laterally to form fibers, often 80 to 120 nm in diameter, organized into the branched network called a fibrin clot.1-3
Fibrinogen is a plasma protein composed of 3 pairs of polypeptides designated Aα-, Bβ-, and γ-chains, folded into 1 central and 2 pairs of distal globules connected by 2 coiled-coil strands (Figure 1A). Proteolytic cleavage of the fibrinogen molecule in the middle of the coiled-coil can yield 1 fragment E and 2 fragments D, corresponding to the E and D regions, respectively. Each D region contains 2 modules formed by the C-termini of the γ-chain (γ-module) and the Bβ-chain (β-module). The γ-module contains a cavity,4 the γ-chain hole or a-hole, which plays an important role in fibrin polymerization. The β-module also has a cavity,5 the β-chain hole or b-hole, which may also participate in fibrin polymerization.3,6
Schematic representation of fibrin(ogen) molecules and their fragments used in this study as sources of knobs and holes. The drawing is based on crystallographic data and represents approximately the relative positions and dimensions of the molecular parts. A- and B-knobs are highly flexible and, hence, have not been visualized in the crystal structure to date. Gray and black circles represent fibrinopeptides A (FpA) and B (FpB), respectively. (A) Fibrinogen is 45 nm long and consists of 3 parts, namely 2 D regions and one E region. The D regions contain the distal portions of the coiled-coil and the C-terminal β- and γ-modules. The E region contains the central N-terminal part of the molecule and the proximal portions of both sets of coiled-coils. The E region has 2 pairs of binding sites, A- and B-knobs, that are masked and become exposed after cleavage of FpA and FpB. The D regions have constitutively open a- and b-holes located in the γ- and β-modules, respectively. (B) DesA-fibrin monomer with only A-knobs exposed, obtained from normal fibrinogen by selective cleavage of FpA using batroxobin. (C) DesB-fibrin monomer with only B-knobs exposed, obtained from fibrinogen variant AαR16C by selective cleavage of FpB by thrombin. (D) Fibrinogen variant γD364H with impaired nonfunctional a-holes. (E) Fragment D from normal fibrinogen has a- and b-holes. (F) Fragment D from fibrinogen γD364H with only b-holes. (G) D-dimer from fibrin representing 2 D regions of the adjacent monomer molecules covalently cross-linked through the C-termini of the γ-chains. (H) Recombinant fragment (β15-66)2 representing dimeric N-terminal portions of the β-chain bearing B-knobs.
Schematic representation of fibrin(ogen) molecules and their fragments used in this study as sources of knobs and holes. The drawing is based on crystallographic data and represents approximately the relative positions and dimensions of the molecular parts. A- and B-knobs are highly flexible and, hence, have not been visualized in the crystal structure to date. Gray and black circles represent fibrinopeptides A (FpA) and B (FpB), respectively. (A) Fibrinogen is 45 nm long and consists of 3 parts, namely 2 D regions and one E region. The D regions contain the distal portions of the coiled-coil and the C-terminal β- and γ-modules. The E region contains the central N-terminal part of the molecule and the proximal portions of both sets of coiled-coils. The E region has 2 pairs of binding sites, A- and B-knobs, that are masked and become exposed after cleavage of FpA and FpB. The D regions have constitutively open a- and b-holes located in the γ- and β-modules, respectively. (B) DesA-fibrin monomer with only A-knobs exposed, obtained from normal fibrinogen by selective cleavage of FpA using batroxobin. (C) DesB-fibrin monomer with only B-knobs exposed, obtained from fibrinogen variant AαR16C by selective cleavage of FpB by thrombin. (D) Fibrinogen variant γD364H with impaired nonfunctional a-holes. (E) Fragment D from normal fibrinogen has a- and b-holes. (F) Fragment D from fibrinogen γD364H with only b-holes. (G) D-dimer from fibrin representing 2 D regions of the adjacent monomer molecules covalently cross-linked through the C-termini of the γ-chains. (H) Recombinant fragment (β15-66)2 representing dimeric N-terminal portions of the β-chain bearing B-knobs.
The self-assembly of fibrin monomers into a fibrin clot requires the exposure by thrombin action of new polymerization sites that are masked in fibrinogen. Thrombin initially cleaves the Arg16-Gly17 bond in the Aα-chains of human fibrinogen, releasing fibrinopeptide A (FpA). The cleavage leads to exposure of the N-terminal sequence Gly17-Pro18-Arg19 in the fibrin α-chains (ie, fibrinogen Aα-chains without the FpA), which is called A-knob (Figure 1B).7 A-Knobs are complementary to a-holes located in the γ-modules of another fibrin molecule, and their interaction is termed A:a binding. Cleavage of FpA and exposure of A-knobs, making desA-fibrin, are necessary and sufficient to form fibrin clots. Fibrin a-holes are also necessary for clot formation; if they are blocked by A-knob mimetic Gly-Pro-Arg-Pro (GPRP) peptide8 or impaired by a point mutation of residue γAsp364,9 thrombin-induced fibrin polymerization is prevented. Together, these data suggest that A:a interactions are the driving force of fibrin polymerization and clot formation. Recently, using laser tweezers–based force spectroscopy, we observed the interactions between A-knobs and a-holes at the single-molecule level and found the A:a bonds to be strong and stable.10 The laser tweezers technique that enables quantification of individual knob-hole interactions is based on the ability of the optical system to measure the binding strength of 2 surface-bound protein molecules.11,12
B-knob becomes exposed after thrombin cleaves the Arg14-Gly15 bond in the Bβ-chain; this is followed by the release of fibrinopeptide B (FpB). The sequence of the newly exposed N-terminal motif in the remaining β-chain is Gly15-His16-Arg17-Pro18, which is considered the working part of B-knob. Importantly, thrombin cleavage of FpB is much slower than that of FpA so that most, if not all, the FpB removal and exposure of B-knobs occur after fibrin polymers have already formed.13,14 Consistent with this, the Gly-His-Arg-Pro (GHRP) peptide, a B-knob mimetic, does not prevent fibrin clot formation,15 suggesting that B-knob–mediated interactions are not necessary for protofibril formation or for their lateral aggregation. On the other hand, the surmise about the existence of B:b interactions and their importance for lateral aggregation is based on a great deal of indirect, but consistent, data that can be roughly segregated into 3 groups: (1) differences in clot formation dynamics and structure after selective release of FpA or FpB16-23 ; (2) binding of GHRP or its derivatives to fibrin(ogen) and their fragments and effects of the peptide(s) on fibrin formation8,15,24-29 ; (3) consequences of naturally occurring or recombinant mutations of B-knobs or b-holes for fibrinogen conversion to fibrin.30-32 Perhaps the most significant, though not absolute, argument in favor of the occurrence of B:b and other B-knob-mediated interactions during fibrin polymerization is the formation of clots, at 15°C or lower, from desB-fibrin.33,34 The desB-fibrin, with exclusively or predominantly exposed B-knobs (Figure 1C), is produced upon selective cleavage of FpB over FpA and can be formed using variant fibrinogens with impaired release of FpA, such as fibrinogen Metz,34-36 Schwarzach,37 or Frankfurt XIII,38 or specific snake venom enzymes.20,33,39
Notwithstanding the evidence that the interactions mediated by B-knob and b-hole contribute to fibrin polymerization, no experimental approach has directly shown the existence of B:b knob-hole interactions using protein molecules rather than mimetic peptides. In our preceding work, we were also unable to detect B:b, B:a, or A:b binding in the presence of strong dominant A:a interactions.10 In this study, for the first time, direct evidence is provided for binding between b-holes and natural A- or B-knobs exposed in a fibrin monomer. The B:b and A:b bonds were found to be much weaker than the A:a bonds, suggesting that b-holes are involved in subsidiary knob-hole binding reactions that reinforce fibrin polymerization.
Materials and methods
Expression and purification of fibrinogens
Two recombinant fibrinogens, normal and variant γD364H, were used in this study as the precursors of D fragments. Fibrinogen γD364H cannot form fibrin upon thrombin action because of an impaired a-site that prevents A:a interactions (Figure 1D)9 Both fibrinogens were expressed and purified as previously described.9,40,41 Fibrinogen Frankfurt XIII (homodimeric AαR16C), whose FpA is not releasable by thrombin, was isolated from citrated patient plasma and was characterized as described.38
Preparation of D fragments from normal and γD364H fibrinogens
Fragment D from normal fibrinogen was purified as previously described.42 Trypsin digestion of γD364H was carried out similarly to that of normal fibrinogen. Digestion was stopped by filtration through a 0.22-μm filter (Costar, Corning, NY), and the fibrinogen fragments were purified on a 2-mL NH2-GPRPAA affinity column (15-mm diameter). Briefly, D-γD364H digest was diluted 2 times with water and loaded at 0.2 mL/min onto the affinity column equilibrated with 200 mM Tris/HCl buffer, pH 7.4, 20 mM CaCl2. D-γD364H was collected in the flow-through and was further purified from low-molecular–weight products in a centrifugal filter device (50-kDa cutoff; Millipore, Bedford, MA), by 3 washes of 20 mM HEPES buffer, pH 7.4, 150 mM NaCl. The polypeptide chain composition and the purity of D fragments were assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Expression and preparation of the recombinant fibrin (β15-66)2 fragment
Recombinant (Bβ1-66)2 fragment mimicking the dimeric arrangement of the Bβ-chain in fibrinogen, which forms 2 BβN-domains, was produced in Escherichia coli and purified, as has been described elsewhere.43 To produce the activated (β15-66)2 fragment corresponding to the fibrin βN-domain with 2 exposed B-knobs (Figure 1H), (Bβ1-66)2 was treated with thrombin and purified as described earlier.43
Purification of fibrin DD fragment
The DD fragment (Figure 1G) was purified from a trypsin digest of fully cross-linked fibrin based on a modification of a published procedure.5 Briefly, human fibrinogen (ERL, South Bend, IN) was clotted in the presence of CaCl2 and l-cysteine-HCl by the addition of α-thrombin and factor XIII (ERL). A trypsin solution (Sigma, St Louis, MO) was overlaid on the top of fibrin clot, and digestion proceeded until it was stopped by the addition of a soybean trypsin inhibitor (Sigma). The DD fragment was purified with the use of Superdex 200 (Amersham-Pharmacia, Piscataway, NJ).
Coating surfaces with desB- or desA-fibrin or recombinant (β15-66)2
Surfaces coated with the interacting proteins were prepared as described previously,10,44 with some important modifications. desB-Fibrin (Figure 1C) was formed from the naturally occurring homodimer fibrinogen variant Frankfurt XIII with impaired cleavage of FpA because of the AαR16C substitution, which makes the resultant AαCys16-Gly17 bond, unlike normal AαArg16-Gly17, insusceptible to thrombin.38 First, purified fibrinogen AαR16C was bound covalently to 5-μm spherical silica pedestals anchored to the bottom of a chamber. Pedestals coated with a thin layer of polyacrylamide were activated with 10% glutaraldehyde (30 minutes, 37°C), after which 1 mg/mL fibrinogen in 20 mM HEPES (pH 7.4)/150 mM NaCl/3 mM CaCl2 was immobilized for 2 hours at 4°C. After washing the chamber with 20 vol of the same buffer to remove noncovalently adsorbed protein, 2 mg/mL bovine serum albumin (BSA) in 0.055 M borate buffer (pH 8.5)/150 mM NaCl/3 mM CaCl2 was added as a blocker (30 minutes, 4°C). To form desB-fibrin–coated pedestals, the immobilized fibrinogen AαR16C was treated with human thrombin (1 U/mL, 37°C, 1 hour), and then chambers were washed with 20 vol cold (4°C) 100 mM HEPES (pH 7.4) containing 150 mM NaCl, 3 mM CaCl2, 2 mg/mL BSA, and 0.1% (vol/vol) Triton X-100 for approximately 30 minutes before the measurements (working buffer). To form desA-fibrin–coated pedestals (Figure 1B), normal recombinant fibrinogen was first immobilized as described for fibrinogen Frankfurt XIII and then were treated with batroxobin (Batroxobin moojeni) (1 U batroxobin activity [BU]/mL, 37°C, 1 hour), after which the chamber was washed with 20 vol working buffer. It is noteworthy that 1 BU was defined as the enzyme concentration yielding the same FpA release rate as that obtained with 1 U thrombin activity (U). The (β15-66)2 fragment with 2 B-knobs was immobilized on the activated pedestals similarly to fibrinogen from 1 mg/mL solution, as described.
Coating surfaces with the fragment D
Fragment D from normal fibrinogen or fibrinogen γD364H was bound covalently to carboxylate-modified 1.87-μm latex beads using N-(3-dimethylaminopropyl)-N'-ethyl carbodiimide hydrochloride (Sigma) as a cross-linking agent.10 BSA was used as a blocker. The immobilization step lasted 15 minutes at 4°C in 0.055 M borate buffer (pH 8.5)/150 mM NaCl/3 mM CaCl2. Based on the preliminary binding experiments with I125-labeled fibrinogen, the surface density of fragments D and D-γD364H and all other reacting proteins was at the point of surface saturation; nonetheless, the fraction of reactive molecules that have a conformation and orientation compatible with binding was indeterminate.
Model system to study knob-hole interactions
We used a laser tweezers–based model system to study interactions between 2 surface-bound proteins.10,12,44 A laser tweezers or gradient optical trap is formed by focusing a laser beam with a microscope objective to a spot in the specimen plane.45 This system permits the measurement of discrete rupture forces produced by surface-bound molecular pairs during repeated intermittent contact.12,44 To study particular pairs of knob-hole interactions, combinations of fibrin(ogen) molecules and their fragments, bearing either knobs or holes, were bound to pedestals and beads. For these studies, desB-fibrin or the (β15-66)2 fragment (both containing B-knobs) or desA-fibrin (containing A-knobs) were covalently bound to stationary pedestals anchored to the inner surface of a flow chamber. Suspension of latex beads (107/mL) coated covalently with fragments D (containing a- and b-holes) or D-γD364H (containing only b-holes) was then flowed into the chamber. One of the latex beads was trapped by a focused laser beam and moved in an oscillatory manner so that the bead was intermittently in contact with a stationary pedestal. The tension produced when fragment D or D-γD364H on the latex bead interacted with a complementary molecule on the anchored pedestal was sensed and displayed as a force signal that was proportional to the strength of protein–protein binding.10 Rupture forces from many interactions were collected and displayed as normalized force spectra histograms for each experimental condition.
Measurement of binding strength, data processing, and data analysis
The position of the optical trap and, hence, a fragment D–coated latex bead was oscillated in a triangular waveform at 0.5 Hz with a pulling velocity of 1.8 μm/s, which corresponded to a loading rate of 400 pN/s. All experiments were conducted at a trap stiffness of 0.22 ± 0.01 pN/nm, as computed from the bandwidth of Brownian motion. Contact duration between interacting surfaces varied from 10 to 200 milliseconds. Rupture forces were collected at 2000 scans/s (0.5-ms resolution). Results of many experiments under similar conditions were averaged so that each rupture force histogram represented 103 to 104 repeated contacts of more than 10 different bead–pedestal pairs. Individual forces measured during each contact–detachment cycle were collected into 5 pN–wide bins. The number of events in each bin was plotted against the average force for that bin after normalizing for the total number of interaction cycles. The percentage of events in a particular force range (bin) represented the probability of rupture events at that tension. Optical artifacts observed with or without trapped latex beads produced signals that appeared as forces smaller than 10 pN. Accordingly, rupture forces in this range were not considered when the data were analyzed. Rupture force histograms were fit with multimodal Gaussian curves (Origin 7.5; OriginLab, Northampton, MA) to determine the position of a peak that corresponded to the most probable rupture force and the area of each peak that reflected cumulative binding probability for the underlying interactions.12
Results
Comparison of A:a and B:b interactions
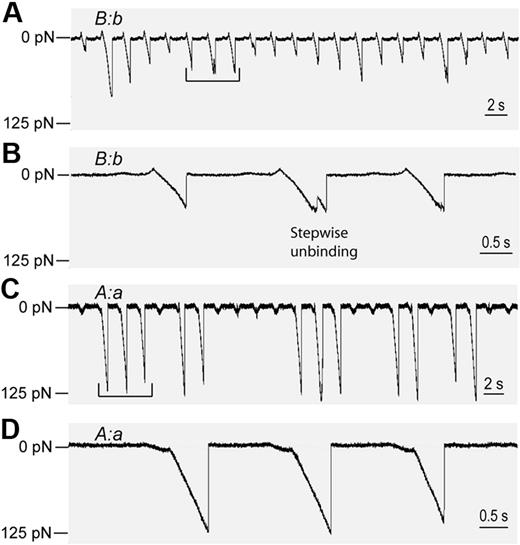
The basic model system to study the B:b interactions consisted of 2 surfaces, one coated with desB-fibrin from homodimeric dysfibrinogen Frankfurt XIII with the mutation AαR16C that precluded release of FpA38 and the other with fragment D from fibrinogen γD364H.9 DesB-fibrin had only B-knobs exposed (Figure 1C), and fragment D-γD364H had only functional b-holes (Figure 1F). Therefore, for this system, B:b was the only possible knob-hole interaction. Surfaces coated with thrombin-treated fibrinogen AαR16C (desB-fibrin monomer) reacted readily with the surfaces coated with fragment D-γD364H. The forces required to dissociate desB-fibrin from D-γD364H–coated surfaces (Figure 2A-B) were different from those required to dissociate desA-fibrin from fibrinogen-coated surfaces (Figure 2C-D), associations attributed primarily to A:a interactions.10 These differences were readily quantified in histograms compiled of the rupture forces or force spectra (Figure 3). First, the forces required to rupture the bonds between desB-fibrin and D-γD364H were mostly small; in fact, most were smaller than 30 pN. Second, the forces were variable and ranged from 10 to 80 pN (Figures 2A, 3A). Third, a substantial fraction (up to 20%) of the detachments appeared to occur as stepwise events (Figure 2B), indicative of the dissociation of multiple intermolecular bonds.12 In comparison, as previously found, the rupture forces of desA-fibrin:fibrinogen bonds were larger (approximately 125 pN) and relatively homogeneous in magnitude (Figure 2C), and the detachments occurred primarily in single steps (Figure 2D).10 Although the obvious stepwise detachments were excluded from the compiled force spectra, many of the multiple molecule-binding/unbinding events could not be resolved and, therefore contributed to the force spectra. To distinguish between single and multiple interactions, we fitted the force histograms with 3 empirically determined Gaussian curves (Figure 3, dashed lines) with the use of Origin 7.5 software (OriginLab). As expected for multiple, identical interactions, fitting analysis revealed that the centroids of the peaks were roughly quantized at 17 ± 4 pN, 33 ± 8 pN, and 55 ± 14 pN. The area under each of the 3 curves corresponded to the probability that a surface contact led to 1, 2, or 3 pairs of bound molecules, respectively (Table 1) Cumulative probability (all events with rupture forces greater than 10 pN) and relative probabilities for forming single or multiple bonds computed from this fit are given in Table 1 for each experimental condition measured. One can see that the rupture force spectrum of B:b interaction is different from that of A:a interaction,10 which contained a sharp peak at 125 to 130 pN and nonspecific forces smaller than 40 pN with no binding of intermediate strength.
Rupture forces of B:b and A:a knob-hole interactions detected as voltage signals calibrated in force units. Portions of digitized raw data obtained using different surface-bound molecular pairs showing the forces exerted by the trap on the latex bead during a series of contacts (upward deflections) and separations (downward deflections). (A) Extended sequence of the B:b interactions appearing as weaker and variable rupture force signals. (B) Extraction from the data in panel A, showing a stepwise unbinding event indicative of multiple knob-hole interactions. (C) Extended data trace and (D) extraction of the A:a interactions appearing as strong homogeneous signals determined using laser tweezers and described in detail elsewhere.10
Rupture forces of B:b and A:a knob-hole interactions detected as voltage signals calibrated in force units. Portions of digitized raw data obtained using different surface-bound molecular pairs showing the forces exerted by the trap on the latex bead during a series of contacts (upward deflections) and separations (downward deflections). (A) Extended sequence of the B:b interactions appearing as weaker and variable rupture force signals. (B) Extraction from the data in panel A, showing a stepwise unbinding event indicative of multiple knob-hole interactions. (C) Extended data trace and (D) extraction of the A:a interactions appearing as strong homogeneous signals determined using laser tweezers and described in detail elsewhere.10
Rupture force spectra demonstrating the interactions of desB-fibrin and fragment D-γD364H in the absence and presence of the GHRPam and GPRPam peptides. (A) Interactions between desB-fibrin and fragment D-γD364H. (B-D) The same interactions in the presence of 2 mM, 5 mM, and 10 mM GHRPam, respectively. (E) Plot derived from panels A to D reflecting the differential inhibitory effect of GHRPam on the weak, intermediate, and strong B:b interactions (normalized by the areas of each of the 3 peaks in the absence of the inhibitor). (F) Restoration of the baseline force profile after removal of the 10 mM GHRPam. (G) The same interactions in the presence of 10 mM GPRPam, showing that the inhibitory effect of the GHRPam observed in panel D was specific.
Rupture force spectra demonstrating the interactions of desB-fibrin and fragment D-γD364H in the absence and presence of the GHRPam and GPRPam peptides. (A) Interactions between desB-fibrin and fragment D-γD364H. (B-D) The same interactions in the presence of 2 mM, 5 mM, and 10 mM GHRPam, respectively. (E) Plot derived from panels A to D reflecting the differential inhibitory effect of GHRPam on the weak, intermediate, and strong B:b interactions (normalized by the areas of each of the 3 peaks in the absence of the inhibitor). (F) Restoration of the baseline force profile after removal of the 10 mM GHRPam. (G) The same interactions in the presence of 10 mM GPRPam, showing that the inhibitory effect of the GHRPam observed in panel D was specific.
Interactions between B-knobs and b-holes
To prove the specificity of observed B:b interactions, the rupture force measurement was repeated in the presence of increasing amounts of the GHRPam peptide, a B-knob mimetic (Figure 3B-D), or GPRPam peptide, a A-knob mimetic.8,15,46,47 Cumulative probability of interactions with rupture forces greater than 10 pN decreased monotonically with increasing GHRPam (Table 1). At 5 mM GHRPam, probability dropped to 34.5% ± 5.5%, and at 10 mM, cumulative probability decreased approximately10-fold to 7.2% ± 1.3% (Table 1). The inhibitory effect of the GHRPam peptide was reversible inasmuch as the rupture force profile (Figure 3F) was restored to the original profile after the peptide was removed, as was the cumulative binding probability (Table 1). For comparison, 10 mM GPRPam peptide had a much smaller effect than the same molar amount of GHRPam on force distribution (Figure 3G) and cumulative binding probability (Table 1), suggesting that GPRPam does not compete well for B:b interactions.
Although the inhibitory effect of GHRPam on the overall probability of interactions between desB-fibrin and D-γD364H was dose dependent, different components of the force spectrum had different susceptibility to the inhibitor. As inferred from the slopes in Figure 3E, the stronger interactions were more sensitive to the effect of GHRPam and disappeared earlier than the weaker ones, suggesting that the strong forces reflected multiple-molecule rather than single-molecule binding events.
Interactions between B-knobs and a-holes
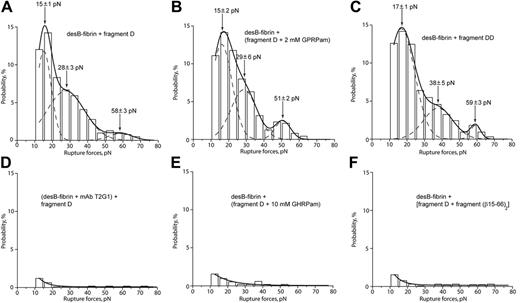
To further probe B-knob interactions, the force spectrum for desB-fibrin and fragment D from normal fibrinogen (Figure 1E) or D-dimer obtained from cross-linked normal fibrin (Figure 1G) was measured. Rupture force profiles of the interaction of desB-fibrin with fragment D (Figure 4A) and with D-dimer (Figure 4C) were nearly the same as the interaction of desB-fibrin with D-γD364H. Given that fragment D and D-dimer have intact a- and b-holes, B-knobs of desB-fibrin can potentially participate in B:b and B:a interactions. The similarity of the force spectra in the presence and absence of the functional a-holes, however, suggested that B:a contacts did not contribute in addition to the B:b interactions shown in Figure 3.
Rupture force spectra demonstrating the interactions of desB-fibrin with fragment D from normal fibrinogen and fragment DD from cross-linked fibrin in the absence and presence of specific inhibitors of B:b and B:a interactions. (A) Interactions between desB-fibrin and fragment D. (B) The same interactions in the presence of 2 mM GPRPam, the specific inhibitor of potential B:a binding. (C) Interactions between desB-fibrin with fragment DD. (D-F) Complete inhibition of the interactions between desB-fibrin and fragment D by the mAb T2G1 against the β15-21 portion of fibrin, by 10 mM GHRPam, and by the free soluble recombinant fragment (β15-66)2, respectively.
Rupture force spectra demonstrating the interactions of desB-fibrin with fragment D from normal fibrinogen and fragment DD from cross-linked fibrin in the absence and presence of specific inhibitors of B:b and B:a interactions. (A) Interactions between desB-fibrin and fragment D. (B) The same interactions in the presence of 2 mM GPRPam, the specific inhibitor of potential B:a binding. (C) Interactions between desB-fibrin with fragment DD. (D-F) Complete inhibition of the interactions between desB-fibrin and fragment D by the mAb T2G1 against the β15-21 portion of fibrin, by 10 mM GHRPam, and by the free soluble recombinant fragment (β15-66)2, respectively.
To test this hypothesis further, several competitive inhibition experiments were performed. To inhibit B:b interactions of desB-fibrin with fragment D, B-knob was blocked with 100 μg/mL mAb T2G1,48,49 an antibody against the β15-21 portion of fibrin comprising the B-knob (Figure 4D), and b-hole was blocked with either 10 mM GHRPam (Figure 4E) or 37 μM (400 μg/mL) recombinant (β15-66)2 fragment (Figure 1H) comprising the fibrin βN-domain with 2 B-knobs (Figure 4F). Force spectra and cumulative probability for interactions (Table 1) in all 3 cases were nearly fully suppressed. To block B:a interactions specifically, the 2-mM GPRPam peptide was added, but it caused no changes in the force distribution (Figure 4B) or the cumulative probability (Table 1). These data together suggest that B:a interactions do not contribute to fibrin binding.
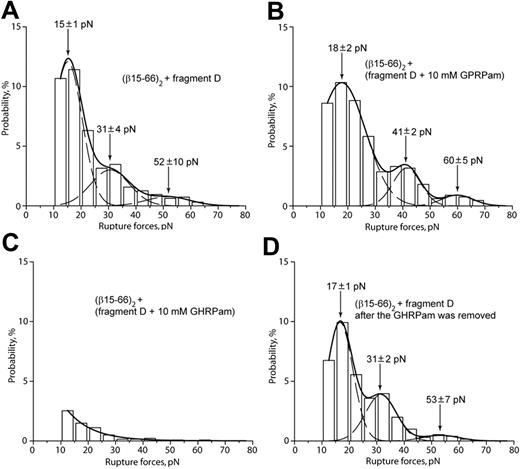
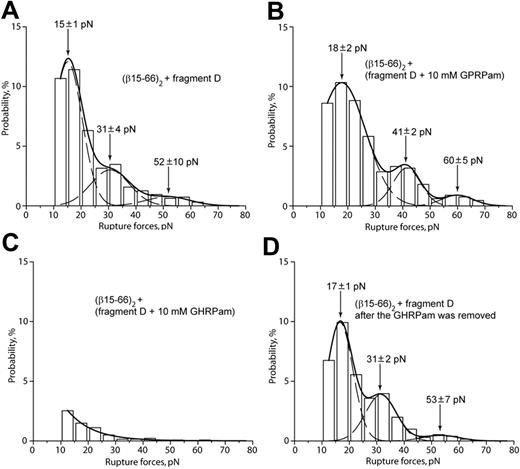
To reduce the possibility that desB-fibrin–fragment D binding is caused by portions of the fibrin molecule other than B-knob, the interactions of (β15-66)2 (Figure 1H) with fragment D (Figure 1E) were measured. Based on the observed ability of the (β15-66)2 fragment to compete with desB-fibrin for b-holes (Figure 4F), (β15-66)2 was expected to bind like a functional fibrin βN-domain containing B-knobs. Accordingly, the rupture force spectrum for (β15-66)2 and fragment D had 3 peaks at 15 ± 1 pN, 31 ± 4 pN, and 52 ± 10 pN (Figure 5A), similar to the force profile obtained for desB-fibrin and fragment D (Figure 4A). The interaction of (β15-66)2 with fragment D was insensitive to GPRPam (Figure 5B) and highly sensitive to GHRPam (Figure 5C), and the latter effect was reversible (Figure 5D). Thus, B:b interactions had the same characteristics irrespective of whether B-knob was a part of a fibrin molecule or recombinant isolated βN-domain.
Rupture force spectra demonstrating the interactions of recombinant fragment (β15-66)2 with fragment D obtained from normal fibrinogen. (A) Interactions of (β15-66)2 with fragment D. (B) The same interactions in the presence of 10 mM GPRPam. (C) The same interactions in the presence of 10 mM GHRPam. (D) Restoration of the baseline force profile after removal of 10 mM GHRPam.
Rupture force spectra demonstrating the interactions of recombinant fragment (β15-66)2 with fragment D obtained from normal fibrinogen. (A) Interactions of (β15-66)2 with fragment D. (B) The same interactions in the presence of 10 mM GPRPam. (C) The same interactions in the presence of 10 mM GHRPam. (D) Restoration of the baseline force profile after removal of 10 mM GHRPam.
Interactions between A-knobs and b-holes
The strength of A:b interactions was measured using desA-fibrin as the source of A-knobs and fragment D-γD364H as the source of b-holes. DesA-fibrin readily interacted with D-γD364H, producing a broad rupture force spectrum up to 75 pN, which could be fit with 3 partially overlapping Gaussian curves peaking at 11 ± 6 pN, 35 ± 8 pN, and 57 ± 5 pN (Figure 6A), similar to the force profile obtained for the B:b interactions. The interactions were almost completely abrogated by GHRPam (Figure 6B), suggesting that they were mediated by b-holes and, therefore, represented the A:b binding pair.
Rupture force spectra demonstrating the interactions of desA-fibrin and fragment D-γD364H in the absence and presence of the GHRPam peptide. (A) Pure interactions between desA-fibrin and fragment D-γD364H. (B) The same interactions in the presence of 10 mM GHRPam.
Rupture force spectra demonstrating the interactions of desA-fibrin and fragment D-γD364H in the absence and presence of the GHRPam peptide. (A) Pure interactions between desA-fibrin and fragment D-γD364H. (B) The same interactions in the presence of 10 mM GHRPam.
Discussion
It has been proposed that fibrin polymerization is mediated by the interactions of A- and B-knobs in central regions, with complementary a- and b-holes in distal regions, of fibrin monomers. Using an optical trap–based system to measure knob-hole interactions,44 we previously uncovered only strong A:a bonds. Neither B:b nor hypothetical A:b and B:a bonds were detected.10 The current experiments were designed so that the strong A:a bonds, which might have obscured weaker or less probable knob-hole interactions, were eliminated. The key experimental variation was to use only B-knob–bearing molecules represented by surface-bound thrombin-treated fibrinogen variant AαR16C with no release of FpA (desB-fibrin monomer) or the recombinant (β15-66)2 fragment so that A-knobs of fibrin were unavailable for interaction. To reduce the likelihood of interactions other than those mediated by knobs and holes, the other surface was coated with D fragments obtained from normal fibrinogen or fibrinogen variant γD364H, in which a-holes were not active.9 The lack of A:a interactions at these interfaces was confirmed by the absence of rupture forces stronger than 100 pN, which were typical for the A:a pairs (Figure 2),10 and the incapacity of GPRPam, the a-hole blocker, to abrogate interactions between desB-fibrin and fragment D (Figure 4B). With the A:a bonding suppressed, we found that fibrin fragments could interact through B:b and A:b, but not B:a, bonds.
The results indicated that a-holes, when they are exposed, do not bind to B-knobs. First, no difference was observed in rupture force profiles when desB-fibrin interacted with either D-γD364H or fragment D (Figures 3A, 4A) in the absence or presence of a-holes. Second, the interactions between desB-fibrin (Figure 4B) and (β15-66)2 (Figure 5B) with fragment D were insensitive to the presence of GPRPam peptide, the A-knob mimetic. The lack of measurable B:a binding conforms to general considerations regarding unavailability of a-holes for B-knobs50 and does not support the hypothesized role of B:a binding in the formation of β-fibrin.51
In contrast to the absence of measurable B:a binding, A-knobs in desA-fibrin displayed specific reactivity with b-holes in D-γD364H similar to that of B-knobs (Figure 6), indicating the existence of the A:b interactions. A similar conclusion was recently drawn from crystallographic data on the GPRVVEam peptide, mimicking the N-terminus of the α-chain, which was shown to reside in a- and b-holes in fragment D of recombinant human fibrinogen.29 The physiologic relevance of the A:b binding remains unclear because fibrinogen γD364H with nonfunctional a-holes does not form a clot when treated by thrombin.9
The surface-bound desB-fibrin monomer readily reacted with D-γD364H or fragment D, producing a similar spectrum of rupture forces. Therefore, the data on B:b interactions obtained with fragments D and D-γD364H could be combined and analyzed en masse. The susceptibility of the interactions of desB-fibrin with fragments D-γD364H and D to the inhibitory effect of the GHRPam peptide (Figures 3D, 4E) indicates that the binding is specifically mediated by b-holes, which are known to be selectively occupied by the peptide.25,26 The participation of B-knobs in the observed interactions is substantiated, first, by the inhibitory effect of the mAb against the B-knob-containing portion of the β-chain 15-21 (Figure 4D) and, second, by competitive inhibition in the presence of free (β15-66)2 (Figure 4F) containing 2 B-knobs. In addition, the rupture force spectrum of surface-bound (β15-66)2 with fragment D (Figure 5A) was the same as that of the desB-fibrin (Figure 4A) and was equally sensitive to the inhibitory action of the GHRPam peptide (Figure 5C). These findings convincingly show that the measured rupture forces reflect the B:b interactions. For the first time, B:b binding was shown to be mediated by a natural B-knob exposed in a full-length fibrin monomer, not by a B-knob mimetic peptide.
Several indirect arguments support the idea that the 3 decreasing peaks of rupture force histograms in Figures 3 to 5 are indicative of single, double, and triple B:b binding, respectively. First, the maximum values of the weak (15-20 pN), intermediate (30-40 pN), and strong (50-60 pN) force peaks are roughly quantized, as would be predicted if they represented multiples of the bimolecular interactions.52 Second, when the single-molecule binding probability was approximately 50%, the double-molecule interactions were statistically expected to occur in 25% (0.52) of the binding events, and the probability of triple interactions was 12.5% (0.53), which approximately corresponded to most of the experimentally observed peak areas (Table 1). Third, the stronger forces were more susceptible to the inhibitory effect of GHRPam (Figure 3E), which is consistent with the assumption that the stronger forces reflected multiple interactions and, therefore, disappeared first. Fourth, the high incidence of multiple intermolecular interactions was confirmed by the relatively common occurrence (up to 20%) of stepwise detachment (Figure 2D) of the interacting surfaces. Based on these considerations, the binding strength of the individual B:b interactions represented by the weakest peaks in the force spectra was estimated to be approximately 15 to 20 pN. Given that the experimental conditions were the same as those used to study the A:a interactions (125-130 pN),10 the B:b bonds seemed to be approximately 6- to 8-fold weaker than the A:a bonds, which is remarkably consistent with the estimation that α-fibrin binding to other fibrin molecules is 6.25-fold tighter than that of β-fibrin.53
We found that the inhibitory effect of the GHRPam peptide on the B:b interactions was fully pronounced only at 10 mM, whereas the GPRPam peptide effectively inhibited the A:a interactions at 1 mM.10 This finding is in good agreement with the approximately 1 order of magnitude difference in the affinities of the GPRP and GHRP peptides toward human fibrinogen, which were equal to 4 × 104 M-1 and 7 × 103 M-1, respectively.15 Importantly, in our experiments, the inhibitory effect of the GHRPam peptide resulted from specific and selective binding to b-hole and could not be attributable to nonspecific electrostatic shielding because 10 mM GPRPam, unlike the GHRPam peptide, induced only moderate suppression of the B:b interactions (Table 1), perhaps because of its ability to bind not only to a-holes25 but also to b-holes46 (O.V.G., unpublished data, April 1, 2006).
Surprisingly, in the previous work, we were unable to reveal the B:b interactions when desAB-fibrin was exposed to fibrinogen or fragment D despite the presence of B-knobs and b-holes on the touching surfaces.10 One likely explanation is that the weak B:b interaction could be masked by nonspecific forces up to 40 pN that were excluded from data analysis. Even if the A:a and B:b interactions occurred simultaneously, the probability of the B:b binding seemed to be substantially smaller, which was in line with the absolute prevalence of the A:a binding during earlier stages of fibrin polymerization. The alternative possibility is that the B:b binding could not occur concurrently with the much stronger (and perhaps faster) A:a interactions. If the latter assumption was true, it might have explained why the cleavage of FpB and the subsequent B:b interactions occurred only in the later stages of fibrin formation, when the A:a bonding was completed. The presumption about mutual exclusiveness of A:a and B:b interactions is consistent with a recently proposed mechanism for delayed FpB cleavage.54
At least 2 conceivable mechanisms explain the late B:b bonding by formation and/or activation of b-holes upon fibrin polymerization. The first implies assembly of a new B-knob-binding site by alignment of the D regions on 2 fibrin monomers on polymerization. Proposed more than 2 decades ago,55 this mechanism was disproved in the subsequent work56 and seems inconsistent with the crystallographic data showing 2 structurally and functionally independent b-holes in the D-dimer.25 The second mechanism presumes conformational rearrangement of the β-module, similar to the changes induced by GHRPam binding,26 followed by increased affinity of b-hole to B-knob. To retest these possibilities, desB-fibrin was exposed to the D-dimer (Figure 4C). However, neither the strength nor the binding probability of the interactions between desB-fibrin and fragment DD were different compared with the monomeric fragment D. Therefore, our result suggests that b-holes act independently and that polymerization by itself does not change the strength of B:b binding. This result agrees with the recent hypothesis that each B-knob first binds a single b-hole, and then locks the β-module in a different conformation that changes the properties of an entire clot.28
In conclusion, our data for the first time directly demonstrate the existence of B:b knob-hole interactions between fibrin molecules. We also observed A:b interactions but did not detect B:a binding. Although formation of the B:b bonds is in agreement with current notions of fibrin polymerization, particularly lateral aggregation of protofibrils, physiologic relevance of the A:b binding remains unclear. Two possibilities may explain how B:b interactions can promote lateral aggregation. The first is that B:b binding occurs between the protofibrils because of long and highly flexible B-knobs that can bind b-holes of the neighboring protofibrils without substantial conformational rearrangements (Figure 7A-B). Alternatively, B:b binding may occur within protofibrils (Figure 7C) promoting their lateral aggregation indirectly by a number of secondary mechanisms. First, the FpB cleavage has been shown to cause exposure of αC-domains so that they can enhance lateral aggregation through αC–αC bridges.57-59 Second, B:b binding may induce the protrusion of β-modules and the formation of contacts between the β-modules belonging to different protofibrils.7 Third, lateral aggregation may be accelerated kinetically through stabilization of protofibrils by the formation of B:b bonds in addition to A:a bonds.19,60 Whatever the mechanism, it is important to know that intermolecular B:b interactions really can exist, though their physiologic significance and the mechanisms during fibrin polymerization remain to be established.
Schematic representation of the hypothetical role of B:b interactions in the lateral aggregation of protofibrils during fibrin polymerization. Designations are the same as in Figure 1. (A) Portion of a protofibril assembled only through A:a interactions with B-knobs exposed on the outer surface of the structure. (B) Portion of a fibrin fiber built of 2 protofibrils aggregating laterally and connected through B:b interactions. (C) Portion of a protofibril assembled through A:a interactions and stabilized by B:b bonds within the protofibril.
Schematic representation of the hypothetical role of B:b interactions in the lateral aggregation of protofibrils during fibrin polymerization. Designations are the same as in Figure 1. (A) Portion of a protofibril assembled only through A:a interactions with B-knobs exposed on the outer surface of the structure. (B) Portion of a fibrin fiber built of 2 protofibrils aggregating laterally and connected through B:b interactions. (C) Portion of a protofibril assembled through A:a interactions and stabilized by B:b bonds within the protofibril.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: the authors declare no competing financial interests.
Contribution: R.I.L. and J.W.W. designed the research. R.I.L. conducted the research. O.V.G., D.K.G., S.Y., and L.M. contributed vital new reagents. R.I.L., H.S., and J.W.W. analyzed the data and wrote the paper.
Acknowledgments
This work was supported by National Institutes of Health grants HL-30954 (J.W.W.), HL-31048 and HL-56051 (L.M.) and by American Heart Association grant 0365340U (O.V.G.). O.V.G. was supported by the grant HL-31048 awarded to Susan T. Lord.
We thank Dr Susan T. Lord (Department of Pathology and Laboratory Medicine, University of North Carolina, Chapel Hill, NC) for her enthusiastic support and help with this research and Dr Inge Scharrer (Zentrum der inneren Medizin, J.W. Goethe Universität, Frankfurt, Germany) for identifying the patient and plasma samples of fibrinogen Frankfurt XIII.