Abstract
The endothelial cell protein C receptor (EPCR) is expressed by endothelial cells of large blood vessels and by hematopoietic stem cells. DNaseI hypersensitive (DH) site mapping across 38 kb of the human EPCR gene (hEPCR) locus identified 3 potential regulatory elements. By itself, the DH region spanning the proximal promoter (PP) was unable to direct cell-specific transcription in transgenic mice. A second DH element, located upstream of PP and termed –5.5HS was hypersensitive only in endothelial cells (ECs) and immature hematopoietic cell lines. Transgenes expressing LacZ under the control of –5.5HS coupled to either PP or the SV40 promoter were able to direct β-galactosidase activity to the endothelium of large vessels during embryogenesis and adulthood. The –5.5HS exhibited enhancer activity that was conferred by the interplay of transcription factors interacting with conserved Ets and composite GATA/Tal1 motifs. The third DH element, located in intron 2, was primarily hypersensitive in EPCR-negative cells, and capable of initiating antisense transcription, suggesting a role in hEPCR silencing. This study identifies critical elements required for the tissue specificity of hEPCR and suggests a mechanism for endothelial and hematopoietic stem cell–specific transcriptional regulation that reflects the common origin of these cell types.
Introduction
The protein C (PC) anticoagulant pathway plays a crucial role in the regulation of blood coagulation and inflammation.1 Activated PC (APC) generated in this pathway serves to confine the hemostatic plug to the site of vascular injury by inhibiting the cofactor function of clotting factors Va and VIIIa on intact endothelium. In addition, APC may also exert antiapoptotic and neuroprotective functions that influence inflammatory responses.2,3 The important regulatory roles of APC in coagulation and inflammation are illustrated by the increased risk of thrombosis incurred by individuals with deficiencies in components of the PC pathway4 and by the improved outcome of patients with severe sepsis treated with APC.5
PC is activated by thrombin, but only when thrombin is bound to its receptor, thrombomodulin, present on endothelial cells (ECs). Activation is further enhanced by another EC receptor, the endothelial cell protein C receptor (EPCR).6 By binding PC, EPCR helps present PC to the thrombin:thrombomodulin complex and so reduces the Km for PC activation.7 In this way, EPCR enhances APC generation by at least 5-fold in vitro.7,8 In vivo, blocking protein C-EPCR interactions results in an 88% decrease in circulating APC levels generated in response to thrombin infusion.9
EPCR function is critical for embryo development because EPCR knockout mice die in midgestation.10 The normal distribution of EPCR is highly tissue specific. During embryogenesis, EPCR is expressed by the embryonic giant trophoblast cells from approximately E7.5, and by certain embryonic EC from approximately E11.5.11 In adults, EPCR is expressed almost exclusively by ECs, particularly those of larger blood vessels.12 Its expression in microvascular ECs is either very low or absent. EPCR is, however, also expressed by primitive hematopoietic stem cells (HSCs).13 Initial reports on the transcriptional regulation of the murine EPCR (Procr, here termed mEPCR)14 and human EPCR (PROCR, here termed hEPCR)15 genes have failed to define the molecular mechanisms responsible for the rather unique in vivo cellular distribution of EPCR.
hEPCR consists of 4 exons that span approximately 5.4 kb (kilobase) of genomic DNA on chromosome 20, at position q11.2.16 mEPCR is structurally homologous to its human counterpart.17 Transcription of hEPCR is initiated from 2 major sites located at –79 and –82 bp (base pair) relative to the translation initiation point, and from an additional minor site at –162 bp.16,18 Previous investigations of the hEPCR 5′-flanking region characterized the 2.3-kb region upstream of the translation initiation site,15 referred to as the proximal promoter (PP). In transfection studies, PP (and also truncations of this region down to –572 bp) was transcriptionally active in ECs (human umbilical vein endothelial cell [HUVEC] and EA.hy926) but not in non-ECs (HepG2). Whereas these results suggested the presence of important regulatory sequences within PP that were responsible for EC-specific gene expression in vitro, it remained unclear whether PP was sufficient to confer cell-specific transcription in vivo. In this study we have identified an additional regulatory element 5.5 kb upstream of the hEPCR translation start site, which is essential for driving hEPCR expression in ECs and primitive hemopoietic cells, cell types that share a common precursor.
Materials and methods
DNaseI hypersensitive site mapping
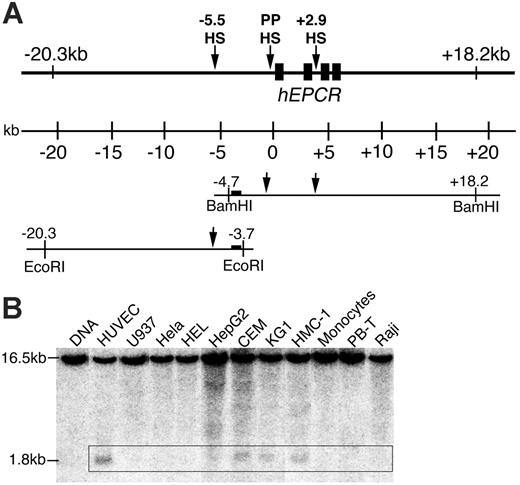
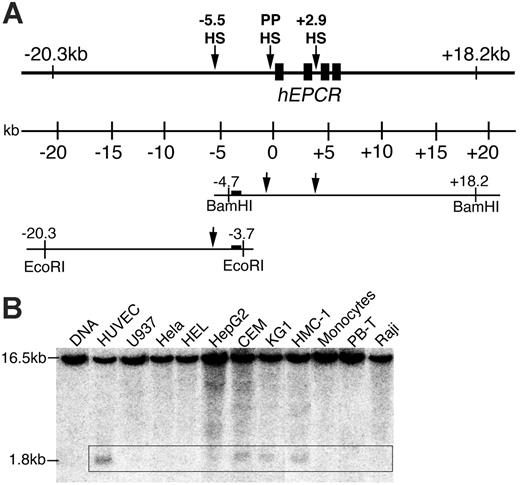
DNaseI hypersensitive (DH) sites were assayed in cultured human cells and cell lines (HUVEC, U937, HeLa, HEL, HepG2, CEM, KG1, HMC-1, peripheral blood T [PB-T] cells, peripheral blood monocytes, and Raji) as previously described.19 In this study, additional DH site mapping was performed between EcoRI sites at –3.8 and –20.3 kb, using the same probes as previously used for BamHI (Figure 1A).
Sequence analysis
Genomic sequences from around the human and murine EPCR loci were obtained from the Ensembl genome browser.20 Sequence alignment was performed by the MAVID server (http://baboon.math.berkeley.edu/mavid/) and analyzed using GeneDoc (http://www.psc.edu/biomed/genedoc/). Putative transcription factor binding sites within –5.5HS were identified using Transfac,21 , Alibaba 2.1 (http://www.gene-regulation.com/pub/programs/alibaba2/index.html), and Transcription Element Search System (http://www.cbil.upenn.edu/tess/index.html) programs.
In vivo dimethylsulphate (DMS) footprinting
In vivo DNA footprinting was carried out as previously described.15 Briefly, naked genomic DNA or cultured cells (HUVEC and HepG2) were treated with DMS, following which methylated guanines were digested with piperidine.22 DMS reactivity was visualized by ligation-mediated polymerase chain reaction (PCR), using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). Experiments were performed in triplicate. The sequences of primers are provided in Table S1 (available at the Blood website; see the Supplemental Materials link at the top of the online article).
Plasmid constructs
pSEAP2-basic (pSB) and pSEAP2-promoter (pSP) vectors (Clontech, Palo Alto, CA), both containing the secreted alkaline phosphatase (SEAP) reporter gene, were used for in vitro reporter gene analysis. pSB does not contain any eukaryotic promoter, whereas pSP contains the SV40 promoter (SV40P) upstream of SEAP. The previously generated PP-pSB construct, containing –2336 to –9 bp of the hEPCR PP cloned upstream of SEAP, was also used.15 DNA sequences under investigation were PCR-amplified from either genomic DNA or the PAC clone 212-C5 (which contains hEPCR and its 5′-flanking region) using primers that introduced XhoI or SalI restriction sites for cloning. Mutation of transcription factor binding sites (Ets no. 1 C-5609T, GATA A-5598G, Tal1 C-5586G/T-5582C, Ets no. 2 C-5547T) was performed using the Quick Change XL site-directed mutagenesis kit (Stratagene). Deletion and insertion mutants were generated by inverse PCR.
For hEPCR promoter transgenes, various lengths (–1194 to –9 bp, –2336 to –9 bp, or –6 kb to –9 bp) of the hEPCR 5′-flanking region or –5.5HS/SV40P were PCR-amplified from PAC 212-C5 and cloned upstream of LacZ. Endofree Plasmid Maxiprep kits (Qiagen, Hilden, Germany) were used for large-scale preparations of all reporter constructs.
Cell culture, transfection, and reporter assays
HUVECs (PromoCell, Heidelberg, Germany), EA.hy926 (Dr Cora-Jean Edgell, University of North Carolina, Chapel Hill, NC), MS1, and HepG2 (European Collection of Cell Cultures, Salisbury, United Kingdom) cells were maintained as previously described.15 For reporter gene assays, cells were grown in 24-well plates and transfected with the SEAP reporter constructs using Lipofectamine 2000 transfection reagent (Invitrogen, Karlsruhe, Germany). For normalization, cells were cotransfected with pGL3-control vector (Promega, Mannheim, Germany) encoding firefly luciferase under the control of the SV40 promoter and enhancer.15 All transfections were performed in duplicate. Values reported are the mean of at least 6 independent experiments. Statistical significance of variation in reporter activity was determined by 2-tailed paired t test. All reported changes were statistically significant (P < .05).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed essentially as previously described.23 Briefly, EAhy926, HepG2, or MS1 cells (106 cells per ChIP) were treated with 0.4% formaldehyde for 10 minutes, cells and nuclei were lysed, and crosslinked chromatin was sonicated to approximately 500 bp. The DNA-protein complexes were incubated overnight with antibodies against Fli1, Elf1, Ets1, Ets2, Erg, GATA2 (Santa Cruz Biotechnology, Santa Cruz, CA), Tal1 (a generous gift from Dr C. Porcher, University of Oxford, United Kingdom), or acetyl H3 (Upstate, Charlottesville, VA). Immunoprecipitations were performed using Protein G-Sepharose beads (Roche, Hertfordshire, United Kingdom). Crosslinking between DNA and protein was reversed at 67°C for 5 hours, the protein was digested with proteinase K (Sigma, Poole, United Kingdom), and the DNA was extracted. Enrichment was determined by real-time PCR using SYBR Green intercalant dye (Applied Biosystems, Warrington, United Kingdom) according to the manufacturer's instructions. Primers were designed to amplify the –5.5HSCR, the homologous murine –8.3 kb region, or control regions (–14.5 kb from hEPCR or –14 kb from mEPCR) (Figure S1).
Antisense transcription analysis
Total RNA was isolated from EA.hy926 and HepG2 cells using the RNeasy mini kit (Qiagen). Reverse transcriptase (RT)–PCR was performed using the SuperScript III First-Strand Synthesis System (Invitrogen) and primers that specifically annealed to either sense or antisense sequences in hEPCR exons 1 and 2. Thereafter nested PCR was performed on all first strands using the same primer pair. The identity of amplified sequences was confirmed by sequencing. The 5′ origin of the hEPCR antisense transcripts was determined by 5′-RACE (Invitrogen). The sequences of primers are presented in Figure S1.
Transgene preparation and transgenic mice
For generation of transgenic mice, transgene sequences were excised from vectors, purified, and microinjected at 4 ng/mL in 10 mM Tris (pH 7.4), 0.1 mM EDTA into pronuclei of fertilized eggs from C57B1/10 × CBA/Ca mice using standard techniques.24 This study was approved by the Imperial College Central Ethical Review Process Committee. For analysis of reporter gene expression during embryogenesis, recipient females were killed at E11.5 to E13.5, and the embryos were whole-mount stained using X-gal. Thereafter, X-gal–positive embryos were dehydrated, wax-embedded, sectioned, dewaxed, and counterstained with eosin (BDH, Poole, United Kingdom). For generating transgenic lines, the offspring following microinjections were biopsied 3 to 4 weeks after birth and genotyped for integration of the transgene using PCR. Transgenic founders were crossed with wild-type mice to generate nonchimeric transgenic mice. For tissue analysis, adult transgenic mice were killed by cervical dislocation, and organs were harvested. Tissues were rinsed in PBS/2 mM MgCl2, fixed in 4% paraformaldehyde/PBS at 4°C for 3 hours, cryoprotected in 30% sucrose/PBS/MgCl2 at 4°C for 4 hours, mounted in OCT (BDH), snap-frozen in liquid nitrogen-cooled isopentane, and stored at –70°C. Cryosections (10 μm) were taken for X-gal staining.
Results
Proximal promoter (PP) alone is incapable of directing cell-specific gene expression in vivo
To assess PP transcriptional activity in vivo, transgenic mice were generated in which LacZ expression was directed by PP (–2336 to –9 bp relative to the translation start site) or truncations thereof. Of 17 separate transgenic lines, 13 exhibited germ line transmission of the transgene (Table 1). In 8 of these, no β-galactosidase activity was detected in adult transgenic tissues. In the 5 remaining lines, LacZ expression was ectopic and nonreproducible. Similarly, LacZ expression was either ectopic or absent from embryonic transgenic tissues. Failure of PP to direct EPCR-like transcription in transgenic mice suggested that additional DNA elements outside of this 2.3 kb region are required for EC specificity in vivo.
DNaseI hypersensitive sites around the hEPCR locus
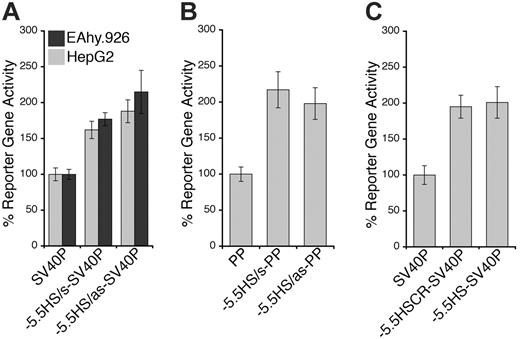
Previously, DH site mapping between –4.7 and +18.2 kb relative to the hEPCR ATG start codon identified DH sites within PP (PPHS) and the second intron (+2.9HS) of hEPCR (Figure 1A).15 Whereas PPHS hypersensitivity was indiscriminate of EPCR expression, +2.9HS was only weakly hypersensitive in HUVECs (which express EPCR) but strongly so in HepG2 cells (which do not express EPCR). An additional potential regulatory region located at –5.5 kb (–5.5HS) was identified by DH site mapping up to –20.3 kb (Figure 1B). Interestingly, –5.5HS was only hypersensitive in endothelial and some immature hematopoietic cell lines but not in any other cell lines or primary cell types tested, suggesting that the function of this region is confined to ECs and immature hematopoietic cells.
DNaseI hypersensitive (DH) site mapping around the hEPCR locus. (A) Schematic representation of the hEPCR locus. DH site mapping was performed using BamHI sites at –4.7 and +18.2 kb and EcoRI sites at –3.7 and –20.3 kb, as shown. Black rectangles represent hEPCR exons 1 to 4. Region used as probe is shown as a black box. Arrows indicate DH regions in the hEPCR proximal promoter (PPHS), intron 2 (+2.9HS), and at –5.5 kb (–5.5HS). (B) DH site mapping of EcoRI-digested DNA from different cell types. The EcoRI fragment shown in panel A was detected as a 16.5-kb band. In endothelial cells (HUVECs) and immature hematopoietic cell lines (CEM, KG1, HMC-1), a 1.8-kb band (boxed) was detected, indicative of a DH site at –5.5 kb (–5.5HS). Lane 1 indicates DNA undigested with DNaseI.
DNaseI hypersensitive (DH) site mapping around the hEPCR locus. (A) Schematic representation of the hEPCR locus. DH site mapping was performed using BamHI sites at –4.7 and +18.2 kb and EcoRI sites at –3.7 and –20.3 kb, as shown. Black rectangles represent hEPCR exons 1 to 4. Region used as probe is shown as a black box. Arrows indicate DH regions in the hEPCR proximal promoter (PPHS), intron 2 (+2.9HS), and at –5.5 kb (–5.5HS). (B) DH site mapping of EcoRI-digested DNA from different cell types. The EcoRI fragment shown in panel A was detected as a 16.5-kb band. In endothelial cells (HUVECs) and immature hematopoietic cell lines (CEM, KG1, HMC-1), a 1.8-kb band (boxed) was detected, indicative of a DH site at –5.5 kb (–5.5HS). Lane 1 indicates DNA undigested with DNaseI.
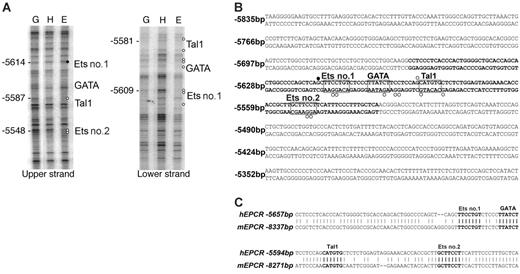
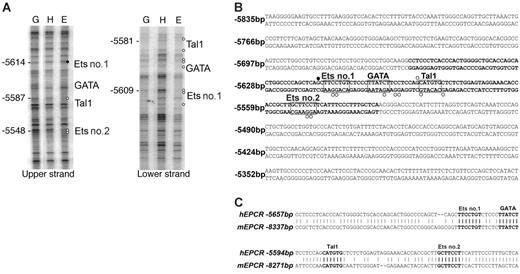
A comparative genomics approach was used to analyze approximately 45 kb of genomic sequence, encompassing 20 kb either side of hEPCR. Apart from the EPCR coding regions and the PPHS sequence, only 2 nonrepetitive sequences were found to be conserved between human and mouse. Significantly, one 130 bp conserved region (CR) (–5657 to –5527 bp) was located within –5.5HS. This region, termed –5.5HSCR, exhibited 75% sequence identity with its murine counterpart (located –8.3 kb relative to the mEPCR translation start site). In conjunction with its hypersensitivity, this suggested a functional role for this sequence. The second conserved region was a very short (∼ 40 bp) sequence located within intron 1 of hEPCR and did not correspond to a DH site. The +2.9HS was not conserved between species.
–5.5HS acts as a classic enhancer in vitro
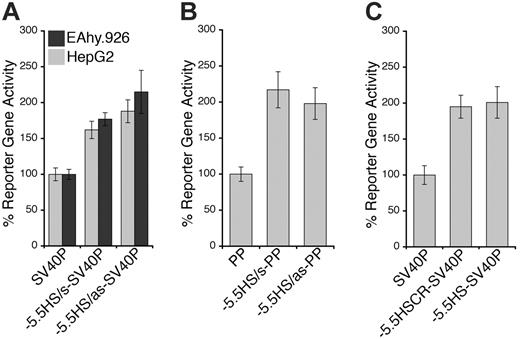
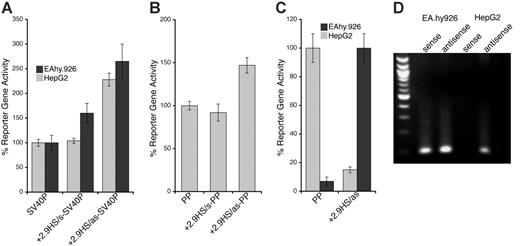
Because both DH site mapping and comparative genomics suggested that –5.5HS might be a transcriptional regulatory element, we investigated its function in vitro. For this, the –5.5HS sequence (∼ 500 bp) was cloned, in either the sense (s) or antisense (as) orientation, upstream of SV40P linked to SEAP. In EA.hy926 cells, –5.5HS/s and –5.5HS/as enhanced SV40P-directed transcription by 1.6- and 1.9-fold, respectively (Figure 2A). Similar data were obtained using HUVECs (not shown). In HepG2 cells, SV40P-driven transcription was increased 1.8- and 2.1-fold by –5.5HS/s and –5.5HS/as, respectively. In EA.hy926 cells, –5.5HS/s and –5.5HS/as also enhanced PP-directed transcription by 2.2- and 2-fold, respectively (Figure 2B). –5.5HS did not affect the transcriptional activity of PP in HepG2 cells (not shown), because PP was inactive in these cells. These results demonstrate that –5.5HS acts as a classic enhancer in vitro.
–5.5HSCR is responsible for –5.5HS activity
To delineate the region(s) within –5.5HS responsible for its enhancer activity, we assessed the function of various –5.5HS fragments. Whereas the fragments encompassing the 130 bp –5.5HSCR were all similarly active to the approximately 500 bp –5.5HS (Figure 2C), those lacking –5.5HSCR lost all enhancer activity (not shown). We thus surmised that its function was primarily attributable to cis-elements contained within this conserved region.
Enhancer activity of –5.5HS. Graphic representation of relative normalized reporter gene activity in cells transfected with constructs as marked. Data from EA.hy926 (▦) and HepG2 (▪) cells are shown. Results for the –5.5HS cloned in both the sense and antisense orientation (–5.5HS/s and –5.5HS/as, respectively) upstream of (A) the SV40 promoter (SV40P), and (B) the hEPCR proximal promoter (PP) are shown. (C) Comparison of the enhancer function of –5.5HS with that of the conserved region within –5.5HS (–5.5HSCR) coupled to SV40P. Data shown are ± SEM (n > 6).
Enhancer activity of –5.5HS. Graphic representation of relative normalized reporter gene activity in cells transfected with constructs as marked. Data from EA.hy926 (▦) and HepG2 (▪) cells are shown. Results for the –5.5HS cloned in both the sense and antisense orientation (–5.5HS/s and –5.5HS/as, respectively) upstream of (A) the SV40 promoter (SV40P), and (B) the hEPCR proximal promoter (PP) are shown. (C) Comparison of the enhancer function of –5.5HS with that of the conserved region within –5.5HS (–5.5HSCR) coupled to SV40P. Data shown are ± SEM (n > 6).
–5.5HSCR contains cis-elements specifically occupied in ECs
Cell-specific occupancy of cis-elements within the –5.5HSCR by trans-acting factors was determined by in vivo DMS footprinting of both DNA strands of –5.5HSCR in different cell types. The elements that were footprinted in ECs, but not in HepG2 cells (Figure 3A), corresponded to putative binding sites for Ets, GATA, and Tal1 (also known as SCL) transcription factors (Figure 3B). These motifs were also highly conserved in the murine sequence (Figure 3C).
Cis-elements within –5.5HSCR are specifically occupied in endothelial cells. (A) Analysis of both DNA strands of the –5.5HSCR sequence by in vivo DMS footprinting. DMS methylation of naked genomic DNA (G) is compared with in vivo methylation of HepG2 (H) and HUVEC (E) DNA. Open circles represent at least 2-fold protection, and closed circles represent at least 2-fold enhancement of DMS reactivity of E relative to H and G. Potential transcription factor binding sites corresponding to the footprinted regions are indicated at the right of each gel image. (B) DNA sequence of both strands of –5.5HS (from –5835 to –5283 bp relative to the hEPCR ATG start codon) encompassing –5.5HSCR (bold). Guanines specifically footprinted in ECs are denoted by circles. Putative binding sites for 2 Ets (nos. 1-2), a GATA, and a Tal1 transcription factor are boxed and labeled. (C) Alignment of upper DNA strand of –5.5HSCR with its murine counterpart (located –8.3 kb relative to mEPCR).
Cis-elements within –5.5HSCR are specifically occupied in endothelial cells. (A) Analysis of both DNA strands of the –5.5HSCR sequence by in vivo DMS footprinting. DMS methylation of naked genomic DNA (G) is compared with in vivo methylation of HepG2 (H) and HUVEC (E) DNA. Open circles represent at least 2-fold protection, and closed circles represent at least 2-fold enhancement of DMS reactivity of E relative to H and G. Potential transcription factor binding sites corresponding to the footprinted regions are indicated at the right of each gel image. (B) DNA sequence of both strands of –5.5HS (from –5835 to –5283 bp relative to the hEPCR ATG start codon) encompassing –5.5HSCR (bold). Guanines specifically footprinted in ECs are denoted by circles. Putative binding sites for 2 Ets (nos. 1-2), a GATA, and a Tal1 transcription factor are boxed and labeled. (C) Alignment of upper DNA strand of –5.5HSCR with its murine counterpart (located –8.3 kb relative to mEPCR).
Ets, GATA, and Tal1 motifs are essential for –5.5HS function
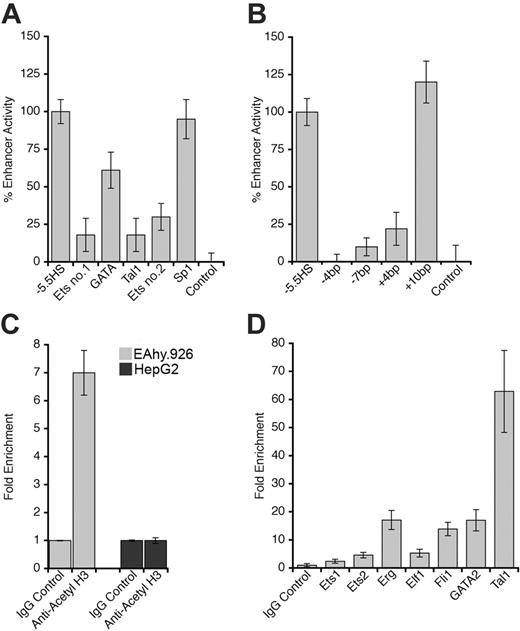
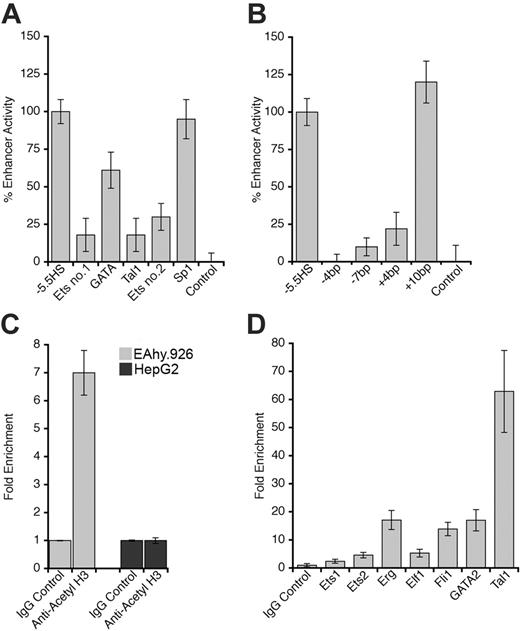
The footprinted Ets, GATA, and Tal1 motifs were each disrupted by site-directed mutagenesis in the –5.5HS-SV40P-SEAP construct. Individual disruption of any of these motifs reduced –5.5HS activity by 40% to 80% (Figure 4A). An Sp1 consensus motif, present within –5.5HSCR but not footprinted in ECs, was similarly disrupted without any loss of enhancer function (Figure 4A).
The GATA and Tal1 motifs within –5.5HSCR are separated by 9 bp (∼ 1 helix turn). Homologous and similarly spaced motifs have previously been shown to be functionally interdependent through the binding of a transcriptional transactivating GATA-1/Ldb1/Lmo2/Tal1 complex, which is known to be essential for early stages of hemopoiesis and angiogenesis.25,26 For this reason, the spacing between the GATA and Tal1 motifs was either reduced by 4 or 7 bp (ie, –one-third or –two-thirds helix turn), or increased by 4 or 10 bp (ie, +one-third or +1 helix turn). Whereas the addition of one helix turn between the GATA and Tal1 motifs did not affect –5.5HS function (Figure 4B), those spacing alterations that entailed a perturbation of the relative orientation between these motifs either ablated, or significantly impaired, its enhancer activity. These observations suggest that the GATA and Tal1 motifs act in synergy and may be involved in the assembly of a transcriptional complex that depends on their relative spatial orientation.
Chromatin structure influences –5.5HS function
Although –5.5HS was hypersensitive in ECs and contained functionally important transcription factor binding sites that were specifically footprinted in ECs, it exhibited enhancer activity in both ECs and HepG2 cells in transient transfection experiments. To reconcile these findings, we hypothesized that chromatin structure might influence –5.5HS function. ChIP experiments using an anti–acetyl H3 antibody revealed that histone acetylation around –5.5HSCR was increased approximately 7-fold in EA.hy926 relative to HepG2 cells (Figure 4C), which corroborates the DH data, and is consistent with a chromatin-dependent function of –5.5HS.
Characterization of –5.5HS enhancer function. (A-B) One hundred percent –5.5HS enhancer activity was defined as the difference in normalized reporter gene activity measured in EA.hy926 cells transfected with either a construct containing wild-type –5.5HS upstream of SV40P (–5.5HS) or the same construct without –5.5HS (control). Normalized enhancer activity of –5.5HS constructs in which (A) the Ets, GATA, or Tal1 binding sites in Figure 3 were disrupted or (B) the spacing between the GATAand Tal1 motifs was altered (by the number of bp indicated) are shown graphically. Data are presented ± SEM (n = 6). (C) Chromatin immunoprecipitation (ChIP) experiments were performed on EA.hy926 and HepG2 cells using primers that amplified the –5.5HSCR. The fold enrichments in immunoprecipitates obtained with anti–acetyl H3 and control IgG antibodies are shown ± SEM. (D) ChIP experiments were performed from the murine endothelial cell line, MS1, using anti-Ets1, Ets2, Elf1, Fli1, Erg, GATA2, and Tal1 antibodies, or control IgG antibodies. The primers used amplified the murine region homologous to the –5.5HSCR (located –8.3 kb from mEPCR). The fold enrichment in immunoprecipitates obtained with antitranscription factor antibodies relative to control IgG antibodies are shown ± SEM.
Characterization of –5.5HS enhancer function. (A-B) One hundred percent –5.5HS enhancer activity was defined as the difference in normalized reporter gene activity measured in EA.hy926 cells transfected with either a construct containing wild-type –5.5HS upstream of SV40P (–5.5HS) or the same construct without –5.5HS (control). Normalized enhancer activity of –5.5HS constructs in which (A) the Ets, GATA, or Tal1 binding sites in Figure 3 were disrupted or (B) the spacing between the GATAand Tal1 motifs was altered (by the number of bp indicated) are shown graphically. Data are presented ± SEM (n = 6). (C) Chromatin immunoprecipitation (ChIP) experiments were performed on EA.hy926 and HepG2 cells using primers that amplified the –5.5HSCR. The fold enrichments in immunoprecipitates obtained with anti–acetyl H3 and control IgG antibodies are shown ± SEM. (D) ChIP experiments were performed from the murine endothelial cell line, MS1, using anti-Ets1, Ets2, Elf1, Fli1, Erg, GATA2, and Tal1 antibodies, or control IgG antibodies. The primers used amplified the murine region homologous to the –5.5HSCR (located –8.3 kb from mEPCR). The fold enrichment in immunoprecipitates obtained with antitranscription factor antibodies relative to control IgG antibodies are shown ± SEM.
Functional characterization of +2.9HS. (A-B) Enhancer function of the +2.9HS. The sense (+2.9HS/s) and antisense (+2.9HS/as) sequences of +2.9HS were coupled to either the SV40P (A) or the hEPCR PP (B). Normalized reporter gene activity obtained following transfection of constructs into EA.hy926 (▦) and HepG2 (▪) cells are presented, ± SEM (n = 6). (C) Promoter function of +2.9HS/as compared with that of PP in EA.hy926 and HepG2 cells. +2.9HS/as and PP were cloned upstream of a reporter gene in a construct without any other promoter. Normalized reporter gene activity is presented relative to that of PP in EA.hy926 cells, ± SEM (n = 6). (D) Detection of antisense hEPCR transcripts. RT-PCR was performed on total RNA from EA.hy926 or HepG2 cells using strand-specific primers that specifically amplified transcripts spanning hEPCR exons 1 and 2. Nested PCR and electrophoresis was used to visualize amplified sequences. The identity of the amplified RNA transcripts (sense and antisense) was confirmed by sequencing.
Functional characterization of +2.9HS. (A-B) Enhancer function of the +2.9HS. The sense (+2.9HS/s) and antisense (+2.9HS/as) sequences of +2.9HS were coupled to either the SV40P (A) or the hEPCR PP (B). Normalized reporter gene activity obtained following transfection of constructs into EA.hy926 (▦) and HepG2 (▪) cells are presented, ± SEM (n = 6). (C) Promoter function of +2.9HS/as compared with that of PP in EA.hy926 and HepG2 cells. +2.9HS/as and PP were cloned upstream of a reporter gene in a construct without any other promoter. Normalized reporter gene activity is presented relative to that of PP in EA.hy926 cells, ± SEM (n = 6). (D) Detection of antisense hEPCR transcripts. RT-PCR was performed on total RNA from EA.hy926 or HepG2 cells using strand-specific primers that specifically amplified transcripts spanning hEPCR exons 1 and 2. Nested PCR and electrophoresis was used to visualize amplified sequences. The identity of the amplified RNA transcripts (sense and antisense) was confirmed by sequencing.
Transcription factor binding to the –5.5HSCR was also investigated by ChIP assays on both human and murine EC lines (EA.hy926 and MS1). Whereas binding of certain Ets and GATA factors (Fli-1 and GATA-2) was suggested in EAhy926 cells, binding of other Ets (Ets-1 or 2, Elf-1, and Erg) or Tal1 transcription factors could not be demonstrated. In the murine MS1 cells, however, the –8.3 kb element homologous to the –5.5HSCR was enriched 5-fold in Ets-2 and Elf-1, 14-fold in Fli-1, 17-fold in Erg and GATA-2, and 63-fold in Tal1 immunoprecipitates (Figure 4D). The extent of histone acetylation around the –8.3 kb element was also significantly (∼ 6-fold) increased, confirming a state of open chromatin (not shown) for this region.
Potential role of +2.9HS in hEPCR regulation
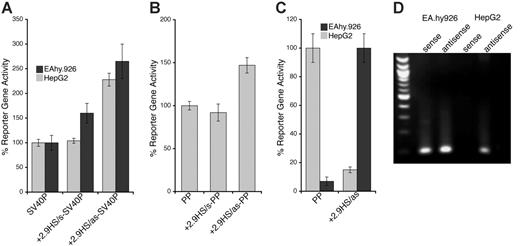
The transcriptional function of the nonconserved +2.9HS, which was strongly hypersensitive in HepG2 cells but only weakly so in HUVECs, was examined in transfection studies. In the sense orientation, the +2.9HS sequence (+2.9HS/s) did not augment SV40P-directed transcription in EA.hy926 cells and did so only marginally in HepG2 cells (Figure 5A). In contrast, the antisense sequence (+2.9HS/as) enhanced transcription by 2.3- and 2.6-fold in EA.hy926 and HepG2 cells, respectively (Figure 5A). Similarly, +2.9HS/as imparted a 1.5-fold increase in PP-directed transcription in EA.hy926 cells, whereas +2.9HS/s did not affect PP activity (Figure 5B). This orientation dependence demonstrated that +2.9HS was not a classic enhancer. We hypothesized that it might negatively regulate hEPCR expression through autonomous initiation of antisense transcription. We assessed whether +2.9HS/as was capable of directing transcription. A SEAP-encoding construct in which +2.9HS/as was the only control element was generated. The promoter activity of +2.9HS/as was strong in HepG2 cells (comparable to that of PP in EA.hy926 cells) but weak in EA.hy926 cells (Figure 5C). Furthermore, hEPCR antisense RNA transcripts were identified in both EA.hy926 and HepG2 cells by strand-specific reverse transcription PCR (Figure 5D). By using 5′-RACE, these antisense transcripts were found to originate close to +2.9HS near the end of exon 2 (Figure S1), implying that +2.9HS may represent the initiation site of this transcript. These observations therefore suggest that +2.9HS may direct antisense transcription of hEPCR and could be involved in hEPCR silencing.
Six kilobases of the hEPCR 5′-flanking region (encompassing -5.5HS and PP) directs reporter gene expression to the endothelium in vivo
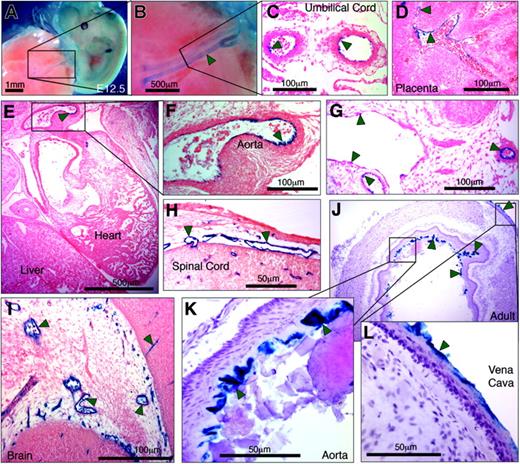
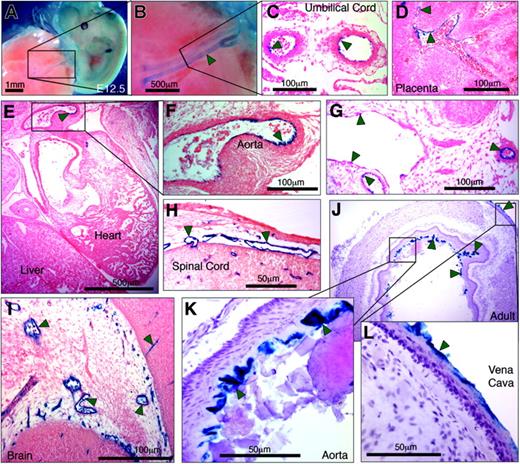
Having shown that by itself PP was unable to direct LacZ expression to the endothelium of transgenic mice, we evaluated whether an extended hEPCR promoter fragment that included –5.5HS would be capable of driving EC-specific transcription. Transgenes were generated in which LacZ expression was directed by 6 kb of the hEPCR 5′-flanking region (encompassing both PP and –5.5HS). Prior to generating transgenic mouse lines, LacZ expression in transgenic embryos (E11.5-13.5) was analyzed. Three of 6 transgenic embryos exhibited strong β-galactosidase activity in the umbilical vessels, following whole-mount X-gal staining (Figure 6A-B; Table 1). Tissue sections taken from these embryos confirmed LacZ expression by the ECs of the umbilical vessels (Figure 6B) and also revealed β-galactosidase activity in the ECs of certain large vessels (including aorta, vena cava, and large embryonic placental vessels) (Figure 6D-G). Reporter gene activity was also specifically detected in ECs of certain smaller vessels in and around the brain, the developing spinal cord, and between rib primordia (Figure 6H-I). In the remaining 3 transgenic embryos, no LacZ expression was detected in any location. Thereafter, transgenic lines were generated to assess the transcriptional function of –5.5HS with PP in adult mice. Of the 12 independent lines generated, 10 exhibited germ line transmission of the transgene (Table 1). In 60% (6 of 10) of these, no LacZ expression was detected. In all of the remaining 4 lines, however, β-galactosidase activity was specifically and consistently targeted toward the ECs of the aorta and vena cava (Figure 6J-L).
Six kilobases of the hEPCR 5′-flanking region transcriptionally targets ECs in transgenic mice. Transgenic mice containing 6 kb of the hEPCR 5′-flanking region coupled to LacZ were generated. Embryos (A-I) and adult tissues (J-L) were analyzed. Embryos were harvested at E12.5 and whole-mount stained with X-gal (A-B). Thereafter stained embryos were wax embedded, sectioned, and counterstained with eosin (C-I). Sites of LacZ expression are dark blue (green arrows). In whole-mount stained embryos, staining in the umbilical vessels was apparent (A-B), which was confirmed as endothelial in tissue sections (C). Transgene expression was also specifically detected in ECs of large placental veins (D), aorta (E-F), and certain other larger veins and arteries (G). EC-specific expression was also seen in small vessels surrounding the spinal cord (H) and in the developing brain (I). Tissues from adult transgenic mice were stained with X-gal and counterstained with hematoxylin (J-L). Reporter gene expression was specifically detected in the endothelium lining the aorta and vena cava (J-L). Microscope images were captured using a Nikon 55i microscope coupled to a DSL1 camera and EclipseNET software (Nikon, Knighton-upon-Thames, United Kingdom). Objectives used were 1×/0.75 NA (panels A, B), 20×/0.75 NA (panels C, D, F-I, K, L), and 2×/0.75 (panels E, J).
Six kilobases of the hEPCR 5′-flanking region transcriptionally targets ECs in transgenic mice. Transgenic mice containing 6 kb of the hEPCR 5′-flanking region coupled to LacZ were generated. Embryos (A-I) and adult tissues (J-L) were analyzed. Embryos were harvested at E12.5 and whole-mount stained with X-gal (A-B). Thereafter stained embryos were wax embedded, sectioned, and counterstained with eosin (C-I). Sites of LacZ expression are dark blue (green arrows). In whole-mount stained embryos, staining in the umbilical vessels was apparent (A-B), which was confirmed as endothelial in tissue sections (C). Transgene expression was also specifically detected in ECs of large placental veins (D), aorta (E-F), and certain other larger veins and arteries (G). EC-specific expression was also seen in small vessels surrounding the spinal cord (H) and in the developing brain (I). Tissues from adult transgenic mice were stained with X-gal and counterstained with hematoxylin (J-L). Reporter gene expression was specifically detected in the endothelium lining the aorta and vena cava (J-L). Microscope images were captured using a Nikon 55i microscope coupled to a DSL1 camera and EclipseNET software (Nikon, Knighton-upon-Thames, United Kingdom). Objectives used were 1×/0.75 NA (panels A, B), 20×/0.75 NA (panels C, D, F-I, K, L), and 2×/0.75 (panels E, J).
–5.5HS acts as an autonomous EC-specific enhancer in vivo
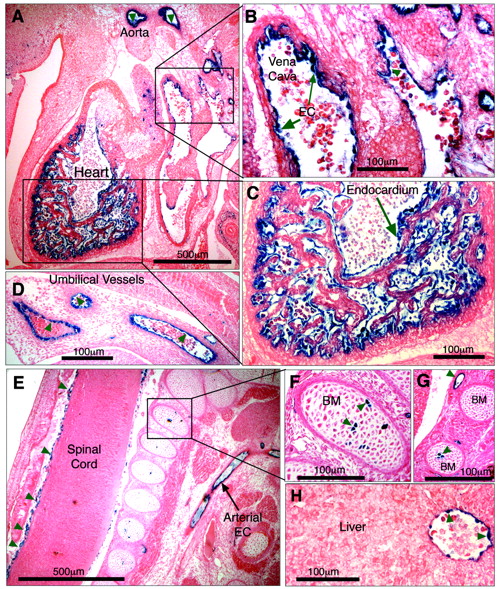
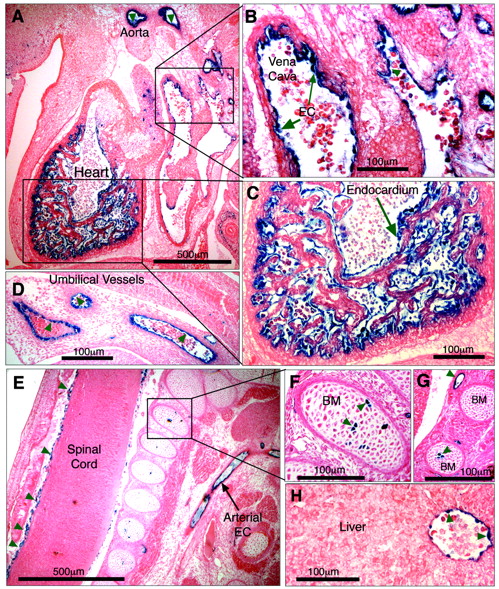
To determine whether it was –5.5HS that conferred EC-specificity to the 6-kb hEPCR promoter fragment, a transgene was generated in which the –5.5HS sequence was placed upstream of LacZ driven by the heterologous SV40P. Previously, it has been demonstrated that the SV40P does not drive specific transcription in transgenic embryos in the absence of a specific enhancer.27 Four transgenic embryos were generated (Table 1). Tissue sections of all of these revealed β-galactosidase activity in ECs of various vascular beds, including large vessels such as the aorta, the vena cava, the hepatic vein, and the umbilical vessels (Figure 7A-H). Reporter gene activity was also detected in smaller vessels surrounding the spinal cord (Figure 7E) and in the developing vascular networks surrounding the brain. LacZ expression was seen in the endothelium of all transgenic embryos, suggesting that its EC specificity was independent of the site of transgene integration. These data confirm that –5.5HS is able to autonomously direct transcription to the ECs in vivo. In addition, 1 of the 4 transgenic embryos also presented strong X-gal staining in the endocardium lining the chambers of the developing heart (Figure 7A,C). Two also presented strong LacZ expression in a small number of cells within the bone marrow (Figure 7E-G; Table 1).
Parallel analyses of the function of the 40-bp conserved sequence in intron 1 failed to reveal any role for this element in specific gene regulation (data not shown).
–5.5HS functions as an autonomous endothelial enhancer in transgenic mice. Transgenic embryos generated with LacZ under the control of –5.5HS/SV40P were harvested at approximately E12.5, whole-mount stained with X-gal, sectioned, and counterstained with eosin. Sites of LacZ expression are dark blue (green arrows). In 4 of 4 transgenic embryos reporter gene expression was specifically detected in ECs, including those of the aorta and vena cava (A-B). In 1 embryo, very strong staining of the endocardium lining the chambers of the developing heart was seen (A,C). Transgene expression was also specifically detected in ECs of umbilical vessels (D), small vessels surrounding the spinal cord (E), and in the hepatic vein (H). In 2 of 4 embryos, strong staining in a small number of bone marrow (BM) cells was seen (E-G). Microscope images were captured using a Nikon 55i microscope coupled to a DSL1 camera and EclipseNET software (Nikon). Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA). Objectives used were as follows: 4×/0.75 NA (A, E) and 20×/0.75 (B-D, F-H).
–5.5HS functions as an autonomous endothelial enhancer in transgenic mice. Transgenic embryos generated with LacZ under the control of –5.5HS/SV40P were harvested at approximately E12.5, whole-mount stained with X-gal, sectioned, and counterstained with eosin. Sites of LacZ expression are dark blue (green arrows). In 4 of 4 transgenic embryos reporter gene expression was specifically detected in ECs, including those of the aorta and vena cava (A-B). In 1 embryo, very strong staining of the endocardium lining the chambers of the developing heart was seen (A,C). Transgene expression was also specifically detected in ECs of umbilical vessels (D), small vessels surrounding the spinal cord (E), and in the hepatic vein (H). In 2 of 4 embryos, strong staining in a small number of bone marrow (BM) cells was seen (E-G). Microscope images were captured using a Nikon 55i microscope coupled to a DSL1 camera and EclipseNET software (Nikon). Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA). Objectives used were as follows: 4×/0.75 NA (A, E) and 20×/0.75 (B-D, F-H).
Discussion
Despite considerable progress in our knowledge of EC gene regulation and vascular bed specificity,28-31 there has been incomplete understanding of the mechanisms targeting the expression of genes such as EPCR to large-vessel ECs. In a study by Gu et al,14 transgenic mice were generated that expressed the green fluorescent protein under the control of mEPCR promoter fragments (extending up to –1080 bp). However, that study failed to observe any fluorescence of this reporter protein that might allow the assessment of cellular specificity in vivo.14 The present study investigates the transcriptional regulation of hEPCR and is the first to elucidate the mechanisms underlying its EC-specific expression. Our survey of the chromatin structure around the hEPCR locus (from –20.3 to +18.2 kb relative to the hEPCR ATG start codon) identified 3 DH regions: PPHS, +2.9HS, and –5.5HS. Although the PP, which encompasses the PPHS, was previously shown to mediate EC-specific transcription in transfected ECs,15 this region alone was insufficient to direct cell-specific reporter gene expression in transgenic mice. In this study, we demonstrate that EC-specific expression of hEPCR in vivo requires the cooperation of –5.5HS.
Although we have not identified a clear function for the intronic +2.9HS, this element was more hypersensitive to DNaseI in cells that do not express EPCR (HepG2) than in cells that do (HUVECs). In its antisense orientation, +2.9HS exhibited autonomous promoter activity, which was greater in HepG2 than in EA.hy926 cells. Given the increasingly recognized role of antisense transcripts in gene regulation,32 we hypothesize that +2.9HS could help silence hEPCR expression in EPCR-negative cells or modulate its levels in ECs, by directing antisense transcription of hEPCR from intron 2 toward exon 1. Our hypothesis is supported by the detection of hEPCR antisense transcripts originating near +2.9HS. Because the +2.9HS sequence is not conserved between humans and mice, this limits the possibility of probing its functionality in a murine model.
The –5.5HS was only hypersensitive to DNaseI in ECs and immature hematopoietic cells. In transient transfection experiments, –5.5HS behaved as a classic enhancer, whose activity was not cell specific. This does not, however, preclude a cell-specific function for –5.5HS in vivo, because this enhancer could require chromosomal/chromatin integration to exert its cellular specificity, as suggested by its hypersensitivity. Histone acetylation around the –5.5HSCR, reflecting disruption of higher-order chromatin structure, was increased in cells that express EPCR (EA.hy926) compared with cells that do not (HepG2), corroborating the contention that –5.5HS function is chromatin dependent. Other examples of chromatin-dependent enhancers involved in EC- or HSC-specific gene regulation include those of the Flk1,33 Tal1 (also known as Scl),34 and CD3435 genes.
The –5.5HS encompasses approximately 130 bp (–5.5HSCR) that is conserved between humans and mice and that was both necessary and sufficient for –5.5HS enhancer function. Its activity relied on the interplay of transcription factors acting on Ets, GATA, and Tal1-binding motifs. Disruption of any of these motifs resulted in marked loss of enhancer function, suggesting that the bound transcription factors may be components of an oligomeric complex. This observation is reminiscent of the Tal1 enhanceosome whose specific function in ECs and HSCs requires a combination of Ets and GATA family transcription factors.34 Perturbation of the spatial orientation between the GATA and Tal1 motifs almost completely abrogated –5.5HS activity, whereas addition of 10 bp (1 helix turn) between these motifs did not affect enhancer function. These findings are consistent with the concept that the GATA-Tal1 bipartite DNA motif is involved in the assembly of an enhanceosome that can only form with the GATA and Tal1 motifs in a specific relative orientation on the DNA helix. A GATA-Tal1 bipartite DNA motif analogous to that present in the –5.5HS has previously been shown to be capable of binding an oligomeric transactivation complex essential for both hematopoiesis and angiogenesis.25,26
Ets, GATA, and Tal1 transcription factors are expressed in both ECs and HSCs,36-38 suggesting that they might contribute to a common mechanism of hEPCR regulation. In vivo DMS footprinting of the –5.5HSCR revealed that the Ets, GATA, and Tal1 motifs were occupied by trans-acting factors in ECs but not in HepG2 cells. ChIP assays for these factors performed with the murine MS1 EC line demonstrated binding of the Ets (Ets2, Erg, Elf1, and Fli1), GATA2, and Tal1 transcription factors to their cognate motifs in the murine region homologous to –5.5HSCR.
The hEPCR PP failed to drive cell-specific transcription in vivo. However, using 6 kb of the hEPCR 5′-flanking region that included both PP and –5.5HS, strong, specific, and reproducible reporter gene expression was directed to the large-vessel endothelium. This included the aorta, umbilical vessels, and selected small vessels surrounding the developing spinal cord and brain. The contrast between these findings and the findings obtained with PP alone strongly suggested that –5.5HS played a critical role in targeting EC-specific transgene expression. In each of the 4 independent adult transgenic lines expressing LacZ, β-galactosidase activity was limited to the endothelium of the aorta and vena cava, and in no other location. Although LacZ was not expressed in all vascular beds known to express EPCR, the 6-kb promoter fragment was capable of discriminating between large and small vessels, a functionality that has not been demonstrated for any other promoter fragment. The more restricted transgene expression in adult mice possibly reflects differences in gene regulation between embryonic and adult tissues.39
To confirm that it was the –5.5HS that conferred EC specificity to the 6-kb hEPCR promoter, –5.5HS was coupled to the heterologous SV40P. In transgenic embryos, high-level reporter gene expression was reproducibly directed to ECs, demonstrating that, by itself, the –5.5HS enhancer contains the regulatory sequences necessary to impart vascular specificity and thus functions as an autonomous EC-specific enhancer. Other enhancers have been shown to autonomously direct transcription to the endothelium of transgenic embryos,29,34,40-42 but –5.5HS is the first human enhancer capable of targeting transcription to both embryonic and adult ECs. Moreover, to our knowledge, –5.5HS is the first regulatory element capable of specifically targeting transcription to large-vessel ECs. As such, it may provide a valuable tool for the development of a gene expression unit that specifically targets large vessels, which are the principal site of atherosclerosis and many vascular diseases.
A recent study by Balazs et al13 identified abundant EPCR expression on primitive HSCs and demonstrated it to be an excellent marker for the identification of such cells from murine bone marrow. We hypothesize that –5.5HS contains the regulatory elements necessary to target gene expression not only to ECs but potentially also to HSCs. The hypersensitivity of –5.5HS in immature hematopoietic cells, and the detection of β-galactosidase in small numbers of cells within the bone marrow of transgenic mice generated with the –5.5HS-SV40P-LacZ construct, both support this hypothesis. An ontogenic rationale is available for the proposed dual endothelial-hematopoietic function of –5.5HS because the development of blood and endothelium are known to be intertwined.43 In the early embryo, the fate of differentiation of the hemangioblast into either an HSC or an EC is thought to be dictated by the combinatorial effects of the Tal1 and Flk1 genes.44 Together with these genes, the Fli1 and PRH genes contribute to the transcriptional regulation of embryonic HSC and EC formation.42 Interestingly, the Tal1, Flk1, Fli1, and PRH genes are all governed by autonomous enhancers with dual HSC/EC activity.34,40-42 Furthermore, the activity of each of these enhancers is dependent on cis-elements similar to those present within –5.5HS, namely Ets, GATA, and Tal1 motifs. In light of this, we propose a model in which –5.5HS targets hEPCR expression to primitive stem cells with dual specification potential, and in which hEPCR expression is progressively lost as these cells embark into hematopoietic specification. Of the autonomous HSC/EC enhancers, only the murine Tal1 +19-kb enhancer has been shown capable of targeting transgene expression to adult HSCs.45 The Tal1 +19-kb enhancer is not, however, entirely HSC/EC specific, because it also targets transgene expression to mast cells, megakaryocytes, and thymocytes.45 By comparison, –5.5HS may be more specific for primitive cells and thereby potentially represents a more specific HSC gene targeting tool.
Prepublished online as Blood First Edition Paper, April 20, 2006; DOI 10.1182/blood-2006-02-001461.
Supported by grants from the British Heart Foundation, the Quebec Research Society Foundation, the Canadian Haemophilia Society, the Association of Haemophilia Clinic Directors of Canada, and Novo Nordisk Canada.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Rachel Simmonds, Dr George Bou-Gharios, Dr Alain Chan, and Dr Bertie Gottgens for their generous help and advice with this work.