Abstract
C/EBPα is required for generation of granulocyte-monocyte progenitors, but the subsequent role of C/EBPα in myeloid lineage commitment remains uncertain. We transduced murine marrow cells with C/EBPα-estradiol receptor (ER) or empty vector and subjected these to lineage depletion just prior to culture in estradiol with myeloid cytokines. This protocol limits biases due to lineage-specific effects on developmental kinetics, proliferation, and apoptosis. Also, lowering the dose of estradiol reduced activated C/EBPα-ER to near the physiologic range. C/EBPα-ER increased Mac1+/Gr1–/MPO–/low monocytes 1.9-fold while reducing Mac1+/Gr1+/MPOhi granulocytes 2.5-fold at 48 hours, even in 0.01 μM estradiol. This pattern was confirmed morphologically and by quantitative polymerase chain reaction (PCR) assay of lineage markers. To directly assess effects on immature progenitors, transduced cells were cultured for 1 day with and then in methylcellulose without estradiol. A 2-fold increase in monocytic compared with granulocytic colonies was observed in IL-3/IL-6/SCF or GM-CSF, but not G-CSF, even in 0.01 μM estradiol. C/EBPα-ER induced PU.1 mRNA, and PU.1-ER stimulated monocytic development, suggesting that transcriptional induction of PU.1 by C/EBPα contributes to monopoiesis. A C/EBPα variant incapable of zippering with c-Jun did not induce monopoiesis, and a variant unable to bind NF-κB p50 stimulated granulopoiesis, suggesting their cooperation with C/EBPα during monocytic commitment.
Introduction
C/EBPα and its family members C/EBPβ, C/EBPδ, and C/EBPϵ are expressed predominantly in myeloid cells within hematopoiesis, with C/EBPα the most prominent isoform in immature myeloid cells.1-3 C/EBPs homodimerize or heterodimerize via C-terminal, α-helical leucine zipper (LZ) domains and then contact DNA via the adjacent basic region (BR).4 C/EBPα transactivates lineage-specific genes, inhibits cell cycle progression via interaction with E2F1 and additional mechanisms, and stimulates cell survival by inducing bcl-2 in cooperation with NF-κB.5-7 C/EBPα–/– hematopoietic cells derived from either fetal liver or adult marrow do not effectively generate granulocyte-monocyte progenitors (GMPs) from the common myeloid progenitor (CMP).8,9
However, the role of C/EBPα in the further commitment of the GMP to the granulocytic versus the monocytic myeloid lineages remains uncertain. Exogenous C/EBPα directs granulocytic maturation of several myeloid cell lines or erythroid progenitors but induces monocytic development from B-cell progenitors.3,10-12 Importantly, neither myeloid cell lines nor erythroid or lymphoid progenitors represent the ideal cellular context to investigate the role of C/EBPα in myelopoiesis because each may lack critical cytokine signals and cooperating transcription factors. KRAB-C/EBPα-estradiol receptor (ER), containing the C/EBPα DNA-binding domain linked to the KRAB transrepression domain and the ER ligand-binding domain, markedly suppresses both granulocytic and monocytic marrow colony formation by murine myeloid progenitors.13 However, this fusion protein may bind and so actively repress genes not physiologically regulated by C/EBPα.
To evaluate the role of C/EBPα in monocyte versus granulocyte lineage determination, we have transduced murine myeloid progenitors with C/EBPα-ER, a fusion protein regulated by estradiol (E2).14 Marrow cells were subjected to lineage depletion after transduction and selection and just prior to addition of E2 and culture in cytokines permissive for myeloid but not erythroid or lymphoid progenitors. Maturation in liquid culture was assessed at 48 hours, the earliest time at which the majority of cells acquired lineage surface markers. This experimental design attempts to ensure that observed effects of C/EBPα on myeloid progenitors are independent of biases introduced during transduction or selection due to lineage-specific effects on developmental kinetics or to the strong antiproliferative effect of constitutively expressed C/EBPα. Reducing the E2 dose 30- to 100-fold produced a level of activated C/EBPα-ER near that of endogenous C/EBPα, and exposure of transduced cells to E2 for only 24 hours prior to plating in methylcellulose in its absence further avoided experimental bias. Morphologic, fluorescence-activated cell sorting (FACS), quantitative polymerase chain reaction (PCR), and colony-forming unit (CFU) assays each support the key conclusion that C/EBPα increases monocytic commitment of myeloid progenitors in IL-3/IL-6/SCF or GM-CSF. Only granulocytic development was evident in G-CSF, underscoring the role of cytokine signaling in lineage choice, potentially via effects on factors cooperating with C/EBPα or on C/EBPα itself.
Additional findings support a model in which C/EBPα cooperates with additional factors to induce monopoiesis. First, consistent with its ability to bind and activate the PU.1 promoter and to induce endogenous PU.1 in cell lines,10,15 C/EBPα induced PU.1 mRNA 2-fold in normal myeloid progenitors. Genetic analyses suggest that elevation of PU.1 supports monocytic over granulocytic development. Lack of one PU.1 allele favors neutrophil development from embryonic stem (ES) cells in vitro and favors neutrophil development in vivo in the absence of the G-CSF receptor (G-CSFR); deletion of the PU.1 distal enhancer located at –14 kb results in 20% of normal PU.1 expression and loss of monopoiesis with preservation of granulopoiesis; and Cre-mediated deletion of PU.1 in adult mice preserves granulocytes at the expense of monocytes.16-18 However, the latter 2 studies demonstrate loss of the M-CSF receptor (M-CSFR) with preservation of the G-CSFR, leaving open the question of whether increased PU.1 favors monopoiesis independent of its effect on receptor expression. We find that PU.1-ER(T) induces monocytic over granulocytic development in our in vitro assay, consistent with the conclusion that induction of PU.1 by C/EBPα contributes to early monopoiesis. We also find that a C/EBPα variant that homodimerizes but cannot zipper with AP-1 proteins does not direct monopoiesis and that a second variant that does not bind NF-κB p50 actually favors granulopoiesis. These results suggest that signal transduction pathways that activate these factors cooperate with C/EBPα to specify monocytic commitment and that in the presence of different signals and transcription factors C/EBPα contributes to granulopoiesis.
Materials and methods
Viral vectors and packaging
pBabePuro-C/EBPαER, its L12V, BR3, and GZ variants, and pBabePuro-PU.1-ER(T) have been described.10,19 The C/EBPα-ER cDNA was transferred as an EcoR1 fragment into the MIGR1 retroviral vector (kindly provided by W. Pear, University of Pennsylvania), which encodes GFP downstream of an internal ribosome entry site. These vectors were packaged by transient transfection with pEcokat in 293T cells.7 293T cells were grown in Dulbecco modified Eagle medium with 10% heat-inactivated fetal bovine serum (HI-FBS).
Transduction and culture
Female 7- to 10-week-old C57BL/6 mice were injected intraperitoneally with 150 mg/kg 5-fluorouracil (5-FU). Six days later, marrow was collected, subjected to red cell lysis with NH4Cl, and cultured in Iscove modified Dulbecco medium (IMDM) with 10% HI-FBS, 10 ng/mL murine IL-3, 10 ng/mL murine IL-6, and 50 ng/mL murine stem cell factor (SCF; PeproTech, Rocky Hill, NJ), 5 × 105 cells/mL, at 37°C, 5% CO2. Then 1 mL retroviral supernatant was added to each 2 mL of cells with 4 μg/mL Polybrene for 2 additional days. Cells transduced with pBabePuro or its derivatives were then diluted to 105/mL in the same media with 2 μg/mL puromycin. Forty-eight hours later, at which point more than 99% of untransduced cells were dead, 30% to 50% of transduced cells were viable with each vector. Dead cells were removed by centrifugation through Lympholyte-M polysucrose, (Cedarlane Labs, Hornby, ON, Canada). Viable cells were subjected to lineage depletion using immunomagnetic beads and a cocktail of lineage antibodies, B220, CD5, Mac1, Ter119, Gr-1, and 7-4 (Stem Cell Technologies, Vancouver, BC, Canada), yielding 5% to 10% of total mononuclear cells. Cells were either plated in methylcellulose at 5000 cells/mL with IMDM and 15% HI-FBS (Methocult 3234, Stem Cell Technologies) or in liquid culture at 2 × 105 cells/mL with IMDM and 10% HI-FBS. The medium was supplemented with IL-3/IL-6/SCF or 10 ng/mL murine GM-CSF, with or without E2 or 4HT, as indicated. Ethanol vehicle was added to cultures not receiving E2 or 4HT. Myeloid CFUs were enumerated on day 8. Marrow cells were also transduced with MIGR1-based vectors for 3 days and then cultured for an additional day prior to lineage depletion and culture with or without E2. To assess more immature progenitors, erythropoietin (Epo; 5 U/mL), thrombopoietin (Tpo; 10 ng/mL), IL-11 (20 ng/mL), and FL (20 ng/mL) were included in the methylcellulose cultures with GM-CSF and SCF. M-CSF was used at 10 ng/mL and G-CSF at 100 ng/mL with SCF. To assess progenitor engraftment, 8 × 104 lineage-depleted, pBabePuro-transduced Ly5.1 cells were injected into irradiated (900 cGy) Ly5.2 recipient mice with 2 × 105 Ly5.2 carrier mononuclear marrow cells.
FACS analysis and morphology
Cultured cells were stained with PE–anti-Mac1 and FITC–anti–Gr-1, PE–anti–c-kit and FITC–anti–Sca-1, PE–anti-FcγR and FITC–anti-CD34, PE–anti–IL7-Rα, PE–anti-Ter119, PE–anti-B220, or PE–anti-CD3 (BD PharMingen, San Diego, CA). Alternatively, cells blocked with rat anti–mouse FcγII/III receptor and IgG1 antisera were stained with PE–anti-Mac1, exposed to Fix and Perm reagents (Caltag, Burlingame, CA), and stained with anti–murine MPO antibody clone 8F4 (HyCult Biotechnology, Uden, The Netherlands) followed by FITC-conjugated rat anti–mouse IgG1. Marrow cells isolated from mice given transplants were stained with FITC–anti-Ly5.1(CD45.2) and PE-CY5–anti-Ly5.2(CD45.1) with PE–anti-Mac1 and APC–anti–Gr-1 or PE–anti-Ter119. Photomicrographs of cells were taken using a Zeiss Axiophot microscope (Carl Zeiss, Thornwood, NY), a Kontron Electronik Progress 3012 camera (Kontron, Munich, Germany), and a 63×/1.40 NA oil objective.
Western blotting and RNA analysis
Total cellular protein extracts prepared using Laemmli sample buffer and corresponding to 5 × 105 cells were electropheresed on 9% polyacrylamide gels and subjected to Western blotting as described.7 We used ERα (HC-20) and C/EBPα (14AA) antisera (Santa Cruz Biotechnology, Santa Cruz, CA) and actin monoclonal antibody Ab-1 (Calbiochem, San Diego, CA). RNA samples were prepared from 1 to 5 × 106 cells using TRIzol reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was prepared using AMV reverse transcriptase (Promega, Madison, WI) and random primers at 42°C for 1 hour, or without enzyme as a control. Quantitative PCR was carried out using 100 ng of each cDNA and a range of GAPDH plasmid standards using iQ SYBR Green supermix (Bio-Rad, Hercules, CA). Oligonucleotides used were: GAPDH-F, 5′-TGGCCTCCAAGGAGTAAGAA; GAPDH-R, 5′-GGTCTGGGATGGAAATTGTG; MPO-F, 5′-GAGGCCCGGAAGATTGTAG; MPO-R, 5′-AGGATCGGTACTGCGGTAGG; NE-F, 5′-GAACGGTCTAAATTTCCGGTCA; NE-R, 5′-AAGGTCTGTCGAGTGCGCTC; C/EBPϵ-F, 5′-CAGCCCTTGCGTGTCCT; C/EBPϵ-R, 5′-CACGTCGCAGTCGGTACTCC; CD14-F, 5′-CGCAGCCTGGAATACCTT; CD14-R, 5′-GCTCCGAATAGAATCCGACT; MS-F, 5′-ACAGCCAGCCCTACGA; MS-R, 5′-TGGTGGTGGCAGGGTTATGA; PU.1-F, 5′-CAGAAGGGCAACCGCAAGAA; PU.1-R, 5′-GCCGCTGAACTGGTAGGTGA; MCSFR-F, 5′-ACCACAAGCAACGCGACCT; MCSFR-R, 5′-TGGGCCGGATCTTTGACATA.
The Student t test was used for statistical comparisons.
Results
A protocol for assessing the immediate effects of C/EBPα on myelopoiesis
We have developed a novel strategy for assessing the direct effect of C/EBPα on myeloid lineage determination by combining use of E2-regulated CEBPα-ER fusion proteins with lineage depletion only after the 5-day period of retroviral transduction and selection and just before splitting the cultures with or without E2 in myeloid cytokines (Figure 1A). The approach of lineage depletion after transduction was taken in lieu of transducing lineage-negative or more purified progenitor subsets because such a population would likely mature during the transduction and selection periods into a similar or even more mature mixed population. In addition, expression of C/EBPα in a regulated manner avoids the marked cell cycle inhibition and consequent experimental biases that result from stable expression of C/EBPα.
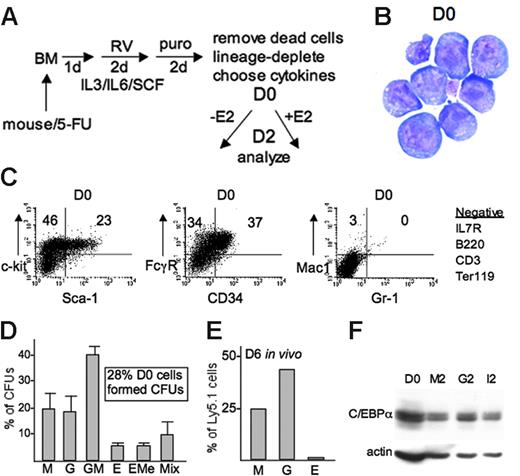
Protocol for assessing rapid effects on myelopoiesis and the starting cell population obtained. (A) The protocol used to obtain a cell population containing myeloid progenitors transduced with C/EBPα-ER or PU.1-ER(T) but lacking mature myeloid cells. Cells were split on day 0 (D0) with or without E2 and analyzed on D2. (B) Wright-Giemsa stain of cytospun D0 cells. (C) D0 cells were analyzed by FACS for expression of the indicated surface markers. (D) CFUs arising from D0 cells plated in SCF, IL-11, IL-3, GM-CSF, Epo, and Tpo were enumerated (mean values and SE from 2 determinations with independently transduced cells are shown). (E) Percentage of transduced D0 Ly5.1 marrow cells that were monocytes (M, Mac1+Gr-1–), granulocytes (G, Mac1+Gr-1+), or erythroid cells (E, Ter119+) 6 days after transplantation into Ly5.2 mice (cells pooled from 5 recipients). (F) Protein extracts from D0 cells or equal numbers of D0 cells cultured for 2 days in M-CSF/SCF (M2), G-CSF/SCF (G2), or IL-3/IL-6/SCF (I2) were subjected to Western blotting for endogenous C/EBPα or β-actin.
Protocol for assessing rapid effects on myelopoiesis and the starting cell population obtained. (A) The protocol used to obtain a cell population containing myeloid progenitors transduced with C/EBPα-ER or PU.1-ER(T) but lacking mature myeloid cells. Cells were split on day 0 (D0) with or without E2 and analyzed on D2. (B) Wright-Giemsa stain of cytospun D0 cells. (C) D0 cells were analyzed by FACS for expression of the indicated surface markers. (D) CFUs arising from D0 cells plated in SCF, IL-11, IL-3, GM-CSF, Epo, and Tpo were enumerated (mean values and SE from 2 determinations with independently transduced cells are shown). (E) Percentage of transduced D0 Ly5.1 marrow cells that were monocytes (M, Mac1+Gr-1–), granulocytes (G, Mac1+Gr-1+), or erythroid cells (E, Ter119+) 6 days after transplantation into Ly5.2 mice (cells pooled from 5 recipients). (F) Protein extracts from D0 cells or equal numbers of D0 cells cultured for 2 days in M-CSF/SCF (M2), G-CSF/SCF (G2), or IL-3/IL-6/SCF (I2) were subjected to Western blotting for endogenous C/EBPα or β-actin.
To further characterize the day 0 (D0) population obtained after transduction and subsequent lineage depletion, the cells were subjected to cytospin, FACS, CFU, and in vivo transplantation assays (Figure 1B-E). The cells are morphologic blasts; 23% are c-kit+Sca-1+ hematopoietic stem cells (HSCs), and 46% are a c-kit+Sca-1– progenitor population. Thirty-seven percent of the cells are FcγR+CD34+, indicating that the majority of the progenitors are myeloid (FcγRlo CMPs and mainly FcγRhi GMPs). The starting population was essentially negative for Mac1+Gr-1– monocytes, Mac1+Gr-1+ granulocytes, IL-7R+ B-cell progenitors, B220+ B-lymphoid cells, CD3+ T-lymphoid cells, and Ter119+ erythroid cells. In a cocktail of multiple cytokines, 28% of the D0 population formed colonies. These were mainly CFU-GMs, CFU-Gs, CFU-Ms, all of which may derive from a cell that at the time of plating maintained myeloid lineage plasticity. CFU-Es, CFU-E/Mega, and CFU-Mix were not obtained in IL-3/IL-6/SCF (not shown). Finally, on transplantation into irradiated, syngeneic recipients, the D0 population effectively reconstituted monocytes and granulocytes by day 6 (D6), with few erythroid cells evident.
C/EBPα stimulates monocytic maturation of immature myeloid cells
Endogenous C/EBPα protein was detected in pBabePuro-transduced D0 cells cultured for 2 days in either M-CSF/SCF or G-CSF/SCF, consistent with the possibility that C/EBPα plays a role in both monopoiesis and granulopoiesis (Figure 1F). To identify effects of C/EBPα on granulocyte versus monocyte lineage development, we transduced marrow cells isolated from mice treated with 5-FU with the pBabePuro-C/EBPαER retroviral vector or with empty, control vector for 2 days and then cultured the cells with puromycin for an additional 2 days. A diagram of C/EBPα-ER (αER) is shown, along with additional mutants to be described (Figure 2A). Thirty percent to 50% of transduced cells survived puromycin selection, whereas more than 99% of untransduced cells exposed to puromycin died by 48 hours. Viable cells were isolated by density centrifugation, subjected to lineage depletion, and cultured with or without E2 in IL-3/IL-6/SCF and monitored for myeloid maturation. This cytokine cocktail stimulates immature myeloid cell survival and proliferation. Twenty-five percent to 60% of the cells acquired the Mac1 or Gr-1 myeloid lineage markers (or both) by day 1 (D1) and 75% to 95% by day 2 (D2) in most experiments, although in some 3 days were required. We therefore focused on D2 to assess lineage commitment. In the absence of E2, viable cell numbers typically increased 10-fold over 48 hours, and activation of C/EBPα-ER with 1 μM E2 slowed accumulation 2- to 3-fold by D2 without inducing evident cell death. By D3 after E2 addition, cell accumulation was reduced 7-fold, on average. C/EBPα-ER expression after 48 hours in E2 was verified using an ER antiserum (Figure 2B top panel, lanes 1-4), and detection with a C/EBPα antiserum indicated that C/EBPα-ER is expressed approximately 3-fold higher than endogenous C/EBPα in the same extract on D0 and D2 (Figure 2B lower left panels).
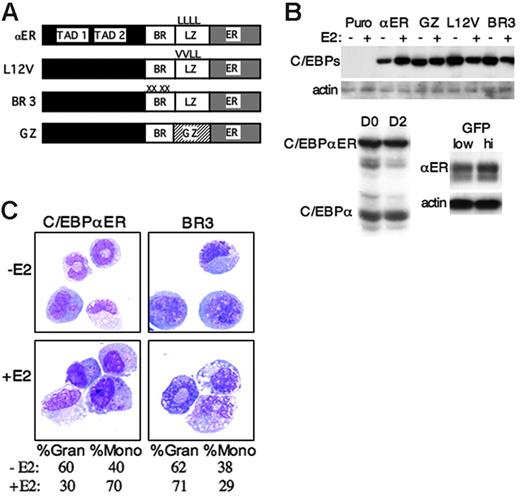
C/EBPα-ER favors monocytic morphologic development. (A) Diagram of C/EBPα-ER and 3 mutant variants. TAD indicates transactivation domain; BR, basic region; LZ, leucine zipper; GZ, GCN4 leucine zipper; ER, estradiol receptor ligand-binding domain. (B) Western analysis of each variant 2 days after culture of lineage-depleted, transduced marrow cells with or without E2, detected with ER antiserum (top). Western blot of C/EBPα-ER and endogenous C/EBPα, without E2 on D0 and D2, detected with C/EBPα antiserum (bottom left). Detection of C/EBPα-ER in low versus high GFP cells after transduction with MIGR1-C/EBPαER, using ER antiserum (bottom right). (C) Marrow cells transduced with pBabePuro-C/EBPαER (αER) pBabePuro-C/EBPαBR3 (BR3), lineage-depleted and then cultured with or without E2 for 2 days were visualized by Wright-Giemsa staining. The differentials obtained from counting 100 cells per culture are shown below the micrographs. %Gran includes granulocytes, bands, metamyelocytes, myelocytes, and promyelocytes. %Mono represents the number of monocytes.
C/EBPα-ER favors monocytic morphologic development. (A) Diagram of C/EBPα-ER and 3 mutant variants. TAD indicates transactivation domain; BR, basic region; LZ, leucine zipper; GZ, GCN4 leucine zipper; ER, estradiol receptor ligand-binding domain. (B) Western analysis of each variant 2 days after culture of lineage-depleted, transduced marrow cells with or without E2, detected with ER antiserum (top). Western blot of C/EBPα-ER and endogenous C/EBPα, without E2 on D0 and D2, detected with C/EBPα antiserum (bottom left). Detection of C/EBPα-ER in low versus high GFP cells after transduction with MIGR1-C/EBPαER, using ER antiserum (bottom right). (C) Marrow cells transduced with pBabePuro-C/EBPαER (αER) pBabePuro-C/EBPαBR3 (BR3), lineage-depleted and then cultured with or without E2 for 2 days were visualized by Wright-Giemsa staining. The differentials obtained from counting 100 cells per culture are shown below the micrographs. %Gran includes granulocytes, bands, metamyelocytes, myelocytes, and promyelocytes. %Mono represents the number of monocytes.
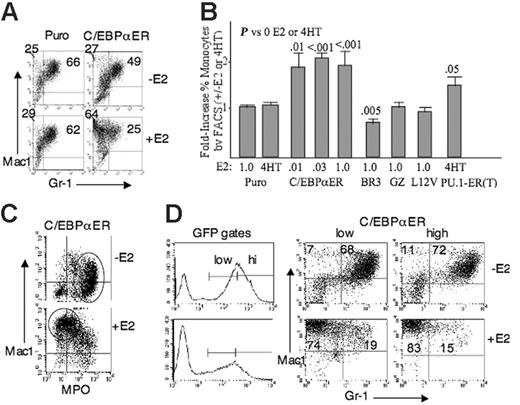
C/EBPα-ER favors monocytic development as assessed by FACS. (A) Marrow cells transduced with pBabePuro or pBabePuro-C/EBPαER, lineage-depleted, and cultured with or without 1 μME2 for 2 days were subjected to FACS for Mac1 and Gr-1. (B) Results from 3 experiments with the indicated vectors. Puro was evaluated in both E2 and 4HT, and C/EBPα-ER was evaluated in several E2 doses. The fold increase in monocytic cells resulting from E2 was obtained as: (% monocytic cells with E2)/(% monocytic cells without E2). For, the C/EBPα-ER data in panel A, this would be (64/64 + 25)/(27/27 + 49). Mean values and SE from 3 determinations are shown. (C) FACS analysis for Mac1 and MPO using C/EBPα-ER–transduced cells. (D) Marrow cells transduced with MIGR1-C/EBPαER, lineage-depleted and then cultured with or without E2 were analyzed for Mac1 and Gr-1 expression within the indicated low-GFP and high-GFP gates on day 3.
C/EBPα-ER favors monocytic development as assessed by FACS. (A) Marrow cells transduced with pBabePuro or pBabePuro-C/EBPαER, lineage-depleted, and cultured with or without 1 μME2 for 2 days were subjected to FACS for Mac1 and Gr-1. (B) Results from 3 experiments with the indicated vectors. Puro was evaluated in both E2 and 4HT, and C/EBPα-ER was evaluated in several E2 doses. The fold increase in monocytic cells resulting from E2 was obtained as: (% monocytic cells with E2)/(% monocytic cells without E2). For, the C/EBPα-ER data in panel A, this would be (64/64 + 25)/(27/27 + 49). Mean values and SE from 3 determinations are shown. (C) FACS analysis for Mac1 and MPO using C/EBPα-ER–transduced cells. (D) Marrow cells transduced with MIGR1-C/EBPαER, lineage-depleted and then cultured with or without E2 were analyzed for Mac1 and Gr-1 expression within the indicated low-GFP and high-GFP gates on day 3.
Activation of C/EBPα-ER favored monocytic morphologic maturation in liquid culture (Figure 2C left panels), with the proportion of monocytes increasing 1.7-fold. This was further supported by FACS analysis of surface markers (Figure 3A). In the experiment shown, Mac1+/Gr-1– monocytic cells increased from 27% to 64%, whereas Mac1+/Gr-1+ granulocytic cells decreased from 49% to 25%. In contrast, E2 only increased the proportion of monocytic cells from 25% to 29%. Results from 3 independent experiments demonstrated a statistically significant 1.9-fold increase in monocytic cells on activation of C/EBPα-ER with 1 μM E2 (Figure 3B), corresponding to a 2.5-fold decrease in granulocytic cells. Due to our concern that C/EBPα-ER is expressed above physiologic levels, this experiment was repeated using lower doses of E2. Estradiol (1 μM) reduced the yield of myeloid CFUs obtained from C/EBPα-ER–transduced marrow 6-fold, but 0.01 or 0.03 μM E2 suppressed CFU yield only 1.7-fold, on average (not shown). These findings, together with the difference in expression between endogenous and exogenous C/EBPα, indicate that the activity of C/EBPα-ER in 0.01 or 0.03 μM E2 is near the physiologic range. Each of these doses of E2 stimulated the same degree of monocytic development from C/EBPα-ER transduced, lineage-negative progenitors as did 1 μM E2, based on Mac1/Gr-1 analysis (Figure 3B).
A FACS assay for intracellular murine myeloperoxidase (MPO) was optimized and combined with surface analysis of Mac1 as an additional assessment of the effect of C/EBPα-ER on lineage outcome (Figure 3C). Addition of E2 induced a striking shift from Mac1–/low/MPOhi granulocytic cells (circled in –E2 panel) to Mac1hi/MPO–/low monocytic cells (circled in +E2 panel), a 5-fold increase in the proportion of monocytes in the experiment shown. Similar findings were seen in a second experiment.
To assess the effect of exogenous C/EBPα-ER using an alternative selection strategy, marrow from 5-FU–treated mice was transduced with the MIGR1-C/EBPαER, lineage-depleted, and cultured with or without E2. FACS analysis for Mac1 and Gr-1 was carried out 3 days later, gating on low-GFP and high-GFP cells (Figure 3D). C/EBPα-ER increased the proportion of monocytic myeloid cells 7.9-fold in the low-GFP and 6.5-fold in the high-GFP subset. These increases were 3.9- and 4.4-fold in a second experiment, and exposure of MIGR1 vector-transduced cells to E2 had no effect (not shown). Western blotting of cells selected by flow cytometry indicated that C/EBPα-ER levels were approximately 3-fold higher in the high-GFP compared with the low-GFP fractions (Figure 1B bottom right panel). Thus, use of GFP rather than puromycin selection confirms our conclusion that C/EBPα-ER favors monocytic maturation of immature myeloid progenitors, even when expressed near endogenous levels.
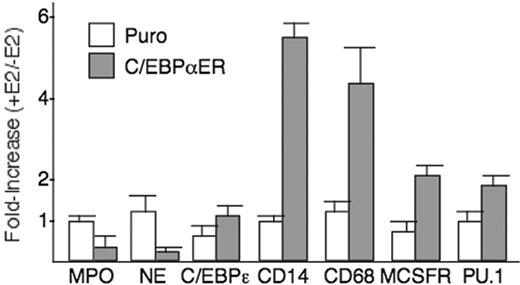
To assess the effect of C/EBPα-ER on additional myeloid genes, total RNAs were prepared from cells transduced with pBabePuro-C/EBPαER or pBabePuro after culture with or without E2 for 48 hours. These samples were analyzed by quantitative reverse transcription PCR (RT-PCR) for the granulocytic markers MPO and neutrophil elastase (NE) and for the monocytic markers, CD14, CD68, and M-CSFR, and for the C/EBPϵ and PU.1 transcription factors (Figure 4). Activation of C/EBPα-ER suppressed MPO expression 3-fold and NE 5-fold and induced CD14, CD68, and M-CSFR expression 6-, 5-, and 2-fold, respectively, whereas exposure of vector-transduced cells to E2 had little effect. PU.1 mRNA was induced 2-fold, whereas C/EBPϵ was unaffected. An almost identical pattern was seen with an independently prepared set of RNA samples from C/EBPα-ER transduced cells cultured with or without E2 for 2 days (not shown).
C/EBPα stimulates monocytic commitment of immature myeloid cells
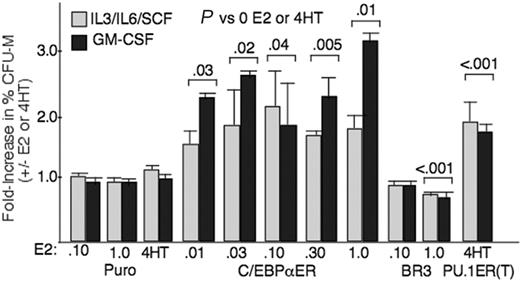
To assess the effect of C/EBPα on the lineage commitment of more immature progenitors, murine marrow mononuclear cells isolated from mice exposed to 5-FU were transduced with pBabePuro or pBabePuro-C/EBPαER, selected with puromycin, lineage-depleted, cultured with or without a range of E2 concentrations for 24 hours, and then plated in the absence of E22to avoid cell cycle inhibition during colony growth. Culture in M-CSF/SCF gave rise to only monocytic CFU-Ms, and culture in G-CSF/SCF only produced granulocytic CFU-Gs (not shown). The ratio of CFU-Ms to the sum of CFU-Ms and CFU-Gs, in the presence versus the absence of E2, is shown for culture in IL-3/IL-6/SCF or GM-CSF (Figure 5). E2 had no effect on the distribution of progenitors transduced with the control vector. In contrast, C/EBPα-ER significantly increased the formation of CFU-Ms by approximately 2-fold when activated by as low as 0.01 μME2.
C/EBPα-ER favors monocytic development as assessed by RNA analysis. Total cellular RNAs were isolated from marrow cells transduced with pBabePuro or pBabePuro-C/EBPαER followed by lineage-depleted, and culture with or without 1 μM E2 for 2 days. After first-strand cDNA synthesis, quantitative PCR was carried out in 3 separate runs for the indicated mRNAs and GAPDH. Fold increase was calculated as the ratio of normalized signal with E2/without E2. Mean values and SE are shown.
C/EBPα-ER favors monocytic development as assessed by RNA analysis. Total cellular RNAs were isolated from marrow cells transduced with pBabePuro or pBabePuro-C/EBPαER followed by lineage-depleted, and culture with or without 1 μM E2 for 2 days. After first-strand cDNA synthesis, quantitative PCR was carried out in 3 separate runs for the indicated mRNAs and GAPDH. Fold increase was calculated as the ratio of normalized signal with E2/without E2. Mean values and SE are shown.
C/EBPα-ER favors monocytic commitment of immature progenitors in 2 cytokine conditions. Marrow cells transduced with Puro, C/EBPα-ER, C/EBPαBR3-ER, or PU.1-ER(T) in IL-3/IL-6/SCF were lineage-depleted, cultured for 1 day at the indicated doses of E2 (or in 4HT for PU.1-ER), and plated in methylcellulose in IL-3/IL-6/SCF or GM-CSF without E2 or 4HT. The ratio of CFU-Ms to CFU-Ms + CFU-Gs was assessed at 7 to 8 days. The fold increase in percent CFU-Ms is shown for each condition as mean and SE from 3 determinations. P values, combining results in IL-3/IL-6/SCF and GM-CSF, are indicated.
C/EBPα-ER favors monocytic commitment of immature progenitors in 2 cytokine conditions. Marrow cells transduced with Puro, C/EBPα-ER, C/EBPαBR3-ER, or PU.1-ER(T) in IL-3/IL-6/SCF were lineage-depleted, cultured for 1 day at the indicated doses of E2 (or in 4HT for PU.1-ER), and plated in methylcellulose in IL-3/IL-6/SCF or GM-CSF without E2 or 4HT. The ratio of CFU-Ms to CFU-Ms + CFU-Gs was assessed at 7 to 8 days. The fold increase in percent CFU-Ms is shown for each condition as mean and SE from 3 determinations. P values, combining results in IL-3/IL-6/SCF and GM-CSF, are indicated.
Activities of C/EBPα variants implicate protein interactions in monocytic specification
Three C/EBPα-ER variants were evaluated for their effects on in vitro myeloid development (Figure 2A). C/EBPαBR3 carries mutations in 4 BR residues, interfering with DNA binding but not LZ interactions and preventing interaction with NF-κB p50.4,7 In C/EBPαL12V-ER, 2 leucines in the LZ are changed to valine, preventing dimerization and DNA binding.4 Finally, in C/EBPαGZ-ER, the GCN4 LZ replaces the C/EBPα LZ.20 C/EBPα zippers with c-Jun and other AP-1 proteins and may bind novel sites, whereas C/EBPαGZ homodimerizes and only binds C/EBPα sites.21-23 These 3 C/EBPα variants, linked to ER, were expressed at levels similar to C/EBPα-ER in transduced murine myeloid progenitors (Figure 2B). Each was evaluated for its effect on the myeloid maturation of lineage-depleted cells using FACS (Figure 3B). C/EBPαBR3-ER significantly reduced monocytic and increased granulocytic development, an effect opposite to that obtained with C/EBPα-ER. This pattern was confirmed morphologically (Figure 2C right panels) and was evident also when myeloid CFUs were assayed in methylcellulose (Figure 5). Both the L12V mutations and the GCN4 zipper swap prevented induction of monopoiesis by C/EBPα-ER (Figure 3B).
PU.1-ER(T) directs monopoiesis from myeloid progenitors
Having found that C/EBPα induces endogenous PU.1 mRNA in myeloid progenitors, we assessed the effect of 4HT activation of PU.1-ER(T) on the myeloid lineage development of transduced and then lineage-depleted murine marrow cells. Activation of PU.1-ER(T) increased Mac1+/Gr-1– monocytic cells 1.5-fold in liquid culture (Figure 3B) and increased the formation of CFU-Ms in methylcellulose assay 1.8-fold, in either IL-3/IL-6/SCF or GM-CSF (Figure 5), whereas 4HT exposure of vector-transduced cells had no effect. These findings, based on direct, regulated expression of PU.1 in primary progenitors, extend earlier genetic analyses implicating increased PU.1 as directing monocytic as opposed to granulocytic lineage commitment.16-18
Discussion
A key conclusion of this study is that C/EBPα has the capacity to direct monocytic development from myeloid progenitors. In assessing the effects of exogenous C/EBPα on myelopoiesis, we have used several strategies to minimize bias due to cell cycle inhibition or differentiation induction during transduction: Lineage-negative cells were isolated after transduction and just prior to assay, results in liquid culture were assessed at early times, an inducible form of C/EBPα was expressed, and low-dose E2 allowed activation of C/EBPα-ER in the physiologic range. Lack of maturation markers on the majority of cells on D1 after E2 addition indicated that results on D2 were not biased by rapid maturation and death of a subset of cells. C/EBPα-ER expressed from pBabePuro increased the proportion of monocytes evident at D2 in liquid culture by nearly 2-fold, and activation of C/EBPα-ER with low-dose E2 for 24 hours was sufficient to similarly increase the commitment of stem/progenitor cells to CFU-Ms at the expense of CFU-Gs.
The D0 population contained more than 50% c-kit+ HSCs and progenitors. The majority of these were myeloid, because 80% of the CFUs obtained using multiple cytokines were CFU-Gs, CFU-Ms, or CFU-GMs. Lack of IL-7R, B220, and CD3 indicates absence of lymphoid progenitors and precursors, and less than 10% of the CFUs were committed to the erythroid or megakaryocytic lineages. In addition, abundant myeloid but negligible erythroid marrow engraftment was evident 6 days after in vivo transplantation of D0 cells, reflecting the behavior of GMPs.24 Thus, the 2- to 3-fold increase in the proportion of monocytic cells in liquid culture or of CFU-Ms in methylcellulose mainly reflects activity of C/EBPα-ER in myeloid progenitors. Whether C/EBPα has a differential capacity to induce HSCs, CMPs, or GMPs toward the monocytic or granulocytic lineages remains an open question. We did not attempt isolation of specific progenitor subsets prior to transduction because a mixed cell population would still be present once E2 is added as a result of ongoing differentiation during transduction and selection. For example, it was noted that CMPs differentiate into GMPs and megakaryocyte-erythroid progenitors (MEPs) after 2 to 3 days in culture, and cultured GMPs generate a significant number of CFU-Ms and CFU-Gs.24 Cre-mediated deletion of a floxed C/EBPα allele in GMPs did not prevent their further maturation, potentially reflecting compensation by other C/EBPs during terminal maturation and ongoing lineage commitment during the transduction period.9 Indeed, we found that activation of C/EBPα-ER for only 24 hours was sufficient to increase the proportion of CFU-Ms obtained, suggesting that there is a temporal window during which this decision is made. The effect of exogenous C/EBPα on specific progenitor subsets will be more effectively addressed in future studies by transgenic expression of C/EBPα-ER combined with their isolation and rapid analysis.
By D3 of C/EBPα-ER activation, severe inhibition of proliferation was generally observed in our study. Similar findings were evident in preliminary efforts to express C/EBPα constitutively. In one approach, lineage depletion was carried out after transduction with pBabePuro-C/EBPα and drug selection. Alternatively, lineage depletion was carried out prior to transduction. In both experiments, a striking 6- to 7-fold decrease in cell accumulation was evident over the 48-hour period after puromycin selection, compared to pBabePuro-transduced cells, highlighting the difficulty of drawing reliable conclusions using an unregulated version of C/EBPα. Nevertheless, monocytic cells were increased 2.6- or 1.6-fold by these approaches (not shown).
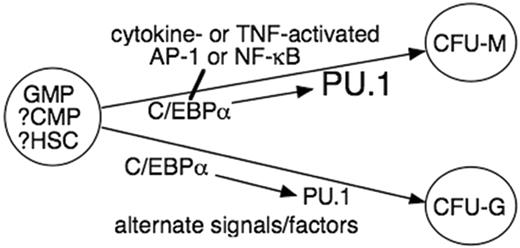
Model for the role of C/EBPα in myeloid lineage commitment. C/EBPα directs CMP to the GMP stage, perhaps via induction of PU.1.8,9,15 We propose that C/EBPα then interacts with cytokine/TNF-α–activated AP-1 or NF-κB p50 to direct monocytic commitment via further elevation of PU.1. In the presence of alternative signals, C/EBPα contributes to granulopoiesis, but PU.1 level or activity is not further induced. Our findings leave open the possibility that increased C/EBPα in CMPs or HSCs also directly induces monocytic or granulocytic commitment dependent on their expression of requisite cooperating factors.
Model for the role of C/EBPα in myeloid lineage commitment. C/EBPα directs CMP to the GMP stage, perhaps via induction of PU.1.8,9,15 We propose that C/EBPα then interacts with cytokine/TNF-α–activated AP-1 or NF-κB p50 to direct monocytic commitment via further elevation of PU.1. In the presence of alternative signals, C/EBPα contributes to granulopoiesis, but PU.1 level or activity is not further induced. Our findings leave open the possibility that increased C/EBPα in CMPs or HSCs also directly induces monocytic or granulocytic commitment dependent on their expression of requisite cooperating factors.
The findings presented in this study, combined with earlier work by us and others, suggest a model of myelopoiesis (Figure 6) wherein C/EBPα induces CMPs or the recently identified Flt3+ lymphoid-myeloid LMPP stem cell25 to develop into GMPs and then, in appropriate cytokine conditions, directs monocytic commitment via further transcriptional induction of PU.1. Neonatal C/EBPα–/– mice lack granulocytes but retain monocytes, although CFU-Ms are reduced 2-fold despite expression of M-CSFR, and both Mac1+/Gr-1+ and Mac1+/Gr-1– cells are markedly reduced in their fetal and newborn livers.26 Further evaluation of these mice demonstrates that their fetal livers lack GMPs but retain CMPs and that the CMPs cannot generate CFU-GMs, CFU-Gs, or CFU-Ms.9 Also, when a floxed C/EBPα allele was deleted in neonatal mice by induction of Mx1-Cre, both monocytes and granulocytes were greatly reduced in blood and marrow 3 weeks later.9 C/EBPα–/– fetal liver cells from independently generated C/EBPα-null mice lack macrophages and CFU-Ms.8 Finally, transduction of adult marrow with KRAB-C/EBPα-ER reduces CFU-Gs, CFU-Ms, and CFU-GMs but not erythroid burst-forming units (BFU-Es).13 Taken together, and consistent with our findings, these loss-of-function studies suggest a role for C/EBPα in the development of both the monocyte and granulocyte lineages.
The effect of exogenous C/EBPα on normal lymphoid and erythroid progenitors has also been evaluated. C/EBPα reprograms murine CD19+ B-cell progenitors into macrophages, either alone or in cooperation with PU.1, but was not effective in a B-cell line lacking PU.1.12 Notably, granulocytes were not observed in this study, indicating that lymphoid cells lack a factor required to obtain this lineage, and in addition C/EBPα was not expressed inducibly. C/EBPα-ER reprograms human CD71+ erythroid progenitors into granulocytes in vitro, and transduction with C/EBPα inhibits murine erythroid development and favors monocyte development in vivo.11,27 Because GATA-1 and PU.1 are antagonistic,28,29 it is interesting to speculate that exogenous C/EBPα converts erythroid cells into granulocytes only if there is an excess of GATA-1 that can then block PU.1-mediated monopoiesis. Considered together, these reports are again consistent with our conclusion that C/EBPα has the capacity to induce monocytic development, although the present study is the first to focus on the effect of exogenous C/EBPα in myeloid progenitors.
C/EBPα-ER induces PU.1 mRNA and protein in myeloid cell lines,10 C/EBPα binds and activates the PU.1 promoter,15 and C/EBPα binds 3 elements in the PU.1 distal enhancer in vitro (C. Yeamans and A.D.F., unpublished data, November 2005). Increased PU.116-18 or activation of PU.1-ER(T), as assessed herein, favors monocytic over granulocytic development. Therefore, induction of PU.1 is apparently one mechanism whereby C/EBPα directs the monocytic commitment of myeloid progenitors. As indicated in our model, cytokine signals that act on C/EBPα directly or on cooperating transcription factors might in turn regulate the degree to which C/EBPα induces PU.1 and thus monocyte versus granulocyte lineage commitment. For example, C/EBPα activity is regulated by phosphorylation of S248 or S21,30,31 and cytokine activated c-Jun, JunB, or NF-κB might influence C/EBPα via direct interaction.
The BR3 variant favored granulopoiesis rather than monopoiesis. C/EBPαBR3 does not interact with NF-κB p50 or cooperate with C/EBPα to induce bcl-2 transcription, suggesting that C/EBPα: p50 interaction potentiates monocytic commitment via induction of PU.1 or other genetic targets. Of note, the GM-CSFR α subunit directly activates NF-κB via interaction with IKKβ.32 Alternatively, C/EBPαBR3 may dominantly inhibit endogenous C/EBPs, although in that case both monocytic and granulocytic development might have been inhibited. The GZ variant, which can homodimerize but cannot zipper with AP-1 proteins, did not favor granulopoiesis. AP-1 proteins direct monocytic maturation of myeloid cell lines and can directly zipper with C/EBPα and bind novel DNA elements, suggesting that C/EBPα:AP-1 interaction via zippering also favors monocytic commitment.21-23,33,34
C/EBPα but not C/EBPαBR3 slowed the rapid, terminal macrophage maturation of 32DPKCδ cells in response to phorbol ester, an effect opposite to findings here with immature primary myeloid progenitors.19 Results using 32DPKCδ cells may reflect later events normally mediated by C/EBPβ in response to specific cytokine/TNF-α signals that activate NF-κB p50.
The role of C/EBPα in granulopoiesis versus monopoiesis might be considered analogous to the role of GATA-1 in erythroid versus megakaryocytic lineage choice, contributing to either lineage dependent on cooperative signals and factors. Future investigations will further define the multiple regulatory events that converge on C/EBPα and PU.1 to guide myeloid lineage commitment. Our novel approach to the study of in vitro myeloid development should facilitate such studies.
Prepublished online as Blood First Edition Paper, April 27, 2006; DOI 10.1182/blood-2005-12-008763.
Supported by National Institutes of Health grants HL62274, HL082948, and CA070970, the Children's Cancer Foundation (A.D.F.), and by Fellow Award from the National Foundation for Cancer Research (C.I.C.).
D.W. and J.D'C. designed and performed the experiments and analyzed the data. C.I.C. and A.D.F. designed the research, analyzed the data, and wrote the paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Jon Adler for assistance with the in vivo transplantation protocol.