Abstract
Activating mutations of the transmembrane receptor NOTCH1 are common in precursor T-cell lymphoblastic leukemia (T-ALL). We systematically analyzed the impact of activating NOTCH1 mutations on early treatment response and long-term outcome in 157 patients with T-ALL of the pediatric ALL–Berlin-Frankfurt-Munster (BFM) 2000 study. We confirm previous results that NOTCH1 mutations occur in more than 50% of T-ALL in children. In 82 patients (82/157; 52.2%), activating NOTCH1 mutations were identified either in the heterodimerization (55/82; 67.1%), in the PEST (13/82; 15.9%), or in both domains (14/82; 17.0%). The presence of NOTCH1 mutations was significantly correlated with a good prednisone response and favorable minimal residual disease (MRD) kinetics, which was independent from sex, age, white blood cell count, and T-cell immunophenotype at the time of diagnosis. Furthermore, activating NOTCH1 mutations specified a large subgroup of patients with an excellent prognosis. These findings indicate that in the context of the ALL-BFM 2000 treatment strategy, NOTCH1 mutations predict a more rapid early treatment response and a favorable long-term outcome in children with T-ALL.
Introduction
The outcome of children with acute lymphoblastic leukemia (ALL) has improved dramatically in the past 3 decades.1 Despite this progress, approximately 25% of patients develop recurrent disease, and the prognosis of relapsed ALL remains poor.2 Furthermore, surviving patients often experience significant toxicity.3 Therefore, individualized treatment strategies are warranted that improve survival in high-risk patients and decrease toxicity and late effects in standard-risk patients.
Pediatric ALLs are commonly subdivided by their antigenic profile and, more recently, by their gene expression profiles.1,4 Precursor B-cell lymphoblastic leukemias account for 85% to 90% of all pediatric ALLs, and a number of clinically important genetic markers have been identified (eg, t(1;19), t(4;11), t(9;22), t(12;21), and hyperdiploidy).5 By contrast, in precursor T-cell lymphoblastic leukemias (T-ALLs) that account for 10% to 15% of all pediatric ALLs no solid prognostic genetic markers have been shown to be of clinical relevance,5,6 although an overexpression of HOX11, TAL1, or LYL1 has been reported to confer a favorable or an unfavorable prognosis in a small number of patients.7,8
Rare cases of T-ALL (< 1%) harbor chromosomal translocations t(7;9) that involve NOTCH1, a gene encoding a single-pass, heterodimeric transmembrane receptor.9 NOTCH1 is essential for the commitment of pluripotent progenitors to the T-cell fate and for the subsequent assembly of pre–T-cell–receptor complexes in immature thymocytes.10-12 The NOTCH1 heterodimer consists of noncovalently associated extracellular (NEC) and transmembrane (NTM) subunits.13 A heterodimerization domain (HD) is responsible for stable subunit association. The HD comprises a 103–amino acid region of the NEC (HD-N) and a 65–amino acid region of the NTM (HD-C) domains. The NEC subunit contains 36 iterated EGF-like repeats that bind DSL (Delta-Serrate-Lag1) ligands, 3 Notch/Lin-12 repeats, and HD-N. The NTM subunit contains HD-C, a RAM (RBP-Jκ [recombination-signal-sequence-binding protein for Jκ genes] associated molecule) domain, 7 ankyrin repeats, a proline-rich and glutamine-rich transcriptional activation domain (TAD), and a PEST (rich in the amino acids proline [P], glutamic acid [E], serine [S], and threonine [T]) domain, where the intracellular receptor fragment is proteolytically inactivated.14,15
Recently, Weng et al16 demonstrated activating NOTCH1 mutations in more than 50% of pediatric T-ALLs. These mutations involve the heterodimerization domain, the C-terminal PEST domain, or both, which result in aberrant up-regulation of NOTCH1-dependent signal transduction.14,17,18 Mutations of HD-N and HD-C enhance production of the active intracellular NOTCH1 (ICN1) by destabilizing the noncovalent intersubunit association. Mutations of the PEST domain increase NOTCH1 signaling by enhancing the stability of ICN1.14,19,20
We have now analyzed the NOTCH1 mutation status of 157 pediatric T-ALL patients, who were enrolled into the ALL-BFM 2000 study. The results confirm the high prevalence of activating NOTCH1 aberrations in pediatric T-ALL16 and, for the first time, demonstrate that activating NOTCH1 mutations predict a more rapid early treatment response and a favorable long-term outcome in patients treated on the ALL–Berlin-Frankfurt-Munster (BFM) 2000 protocol.
Patients, materials, and methods
Patients and cell samples
All children with T-ALL analyzed here were enrolled into and treated according to the multicenter ALL-BFM 2000 protocol.3,21 The present study was approved by the institutional review board of the Hanover Medical School and other participating institutions. Informed consent was obtained in accordance with the Declaration of Helsinki. Mononuclear cells (MNCs) were isolated from bone marrow (BM) samples and stored in liquid nitrogen or at –80°C until DNA extraction. All BM samples displayed a blast percentage of 80% or more.
A total of 157 patients with T-ALL were included in the present study, 117 were male and 40 female, with a median age at diagnosis of 8.8 years (range: 2.0-18.0 years). The patients were selected from a total of 345 T-ALL patients of the ALL-BFM 2000 study according to the availability of sufficient amounts of DNA for molecular analysis. This subgroup of patients did not differ from the entire group in their basic clinical and immunologic characteristics (Table S1, available at the Blood website; see the Supplemental Tables link at the top of the online article). Immunophenotyping of ALL was carried out as previously described,22 and the criteria for the subclassification of T-ALL were adopted from the guidelines proposed by the European Group for the Immunological Characterization of Leukemias (EGIL).23 The presence of TEL/AML1, BCR/ABL, and MLL/AF4 fusion transcripts was excluded as described previously.24,25 Treatment efficiency was analyzed according to the early in vivo response to prednisone, defined as the cytoreduction (number of peripheral blood blasts per microliter on day 8) to a 7-day prednisone treatment prophase.26 Based on the prednisone response, patients were classified into good (< 1000 blasts/microliter) and poor (≥ 1000 blasts/microliter) responders. The patients were further stratified according to minimal residual disease (MRD) kinetics that were assessed at 2 different time points (TPs), at days 33 (TP1) and 78 (TP2) of treatment.27-29 As previously described, allele-specific oligonucleotide–polymerase chain reaction (PCR) protocols were used for quantitative detection of leukemic clone-specific immunoglobulin and T-cell–receptor gene rearrangements on a LightCycler instrument (Roche Diagnostics, Mannheim, Germany).30 At both time points, a favorable MRD status (< 10–4) was defined as the absence of detectable leukemic cells in 104 cells, whereas an unfavorable MRD status (≥ 10–4) was defined by the presence of at least one leukemic cell in 104 cells. Controls consisted of 50 BM samples from children with precursor B-cell ALL. Complete remission (CR) was defined as the absence of leukemia blasts in the peripheral blood and cerebrospinal fluid, less than 5% lymphoblasts in marrow aspiration smears, and no evidence of localized disease. Relapse was defined as recurrence of lymphoblasts or localized leukemic infiltrates at any site.
Genomic DNA preparation
Genomic DNA was prepared from BM samples (2-5 mL) of patients with primary T-ALL and precursor B-cell ALL using the QIAamp DNA blood midi kit (Qiagen, Hilden, Germany) and stored at –20°C until use.
Detection of NOTCH1 mutations
Mutations in the N-terminal region of the heterodimerization domain (HD-N) (exon 26), the C-terminal region of the heterodimerization domain (HD-C) (exon 27), the transcriptional activation domain (TAD) (exon 34), and the PEST domain (exon 34) of the NOTCH1 gene were identified by sequencing of PCR-amplified DNA fragments16 with the ABI PRISM 3100 DNA Analyzer (Applied Biosystems, Foster City, CA).
Statistical analyses
Proportional differences between groups were analyzed by chi-squared (χ2) or Fisher exact tests. The association between NOTCH1 mutations and prednisone response or MRD was examined by use of unconditional logistic regression analysis to calculate relative risks (RRs) and their 95% confidence intervals (CIs). For these analyses, MRD loads smaller than 10–4 were defined as negative. The Kaplan-Meier method was used to estimate survival rates. Differences were compared with the 2-sided log-rank test. Event-free survival was calculated from diagnosis to the time of analysis or to the first event. Failure to achieve remission (early death or resistant leukemia), relapse, death during continuous complete remission, and second malignancy were evaluated as events. Gray test was used to analyze differences in the cumulative incidence of relapse between patients with NOTCH1 mutations and those without. Relative risks were estimated according to the Cox proportional-hazards model. A P value of .05 or less (2-sided) was considered to indicate statistical significance.
Results
NOTCH1 mutations are common in childhood T-ALL
Primary bone marrow samples were obtained from 157 children and adolescents with T-ALL (median age of 8.8 years; range: 2.0-18.0 years), who were enrolled in the ALL-BFM 2000 study between 1999 and 2004. Heterozygous NOTCH1 mutations were identified in 82 (52.2%) of 157 T-ALL samples. In 55 (67.1%) of 82 samples mutations were detected in the HD domain, in 13 (15.9%) of 82 in the PEST domain, and in 14 (17.0%) of 82 in both domains. Fifty-one of the mutations identified have not been reported previously (Figures 1, 2), whereas 31 have been described by Weng et al.16 In contrast, NOTCH1 mutations were not observed in the analyzed control population, consisting of bone marrow samples from 50 patients with precursor B-cell ALL of the ALL-BFM 2000 study, confirming that NOTCH1 mutations are characteristic for T-ALL.16
Activating NOTCH1 mutations are predicted to affect the secondary structure of the protein
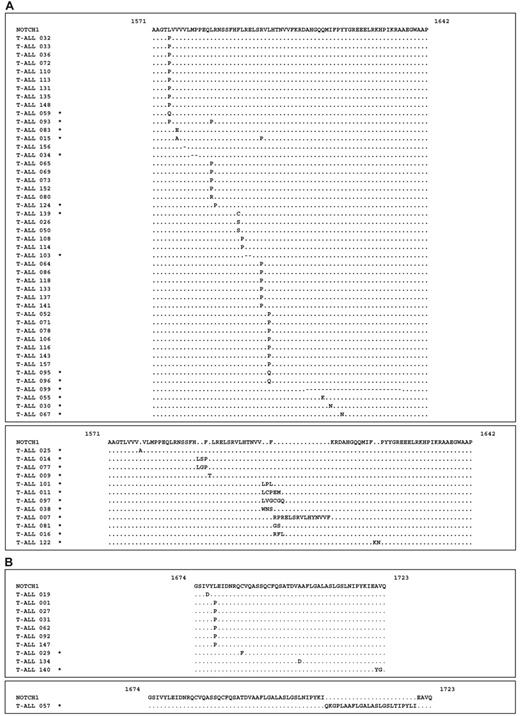
Most of the HD domain mutations were located within the HD-N region (n = 58), whereas HD-C mutations were less common (n = 11). The HD-N domain mutations were clustered in “hot spots” spanning amino acid residues 1574 to 1602 (n = 48) and 1606 to 1638 (n = 12) (Figure 1A). The HD-C domain mutations were located in a region spanning amino acid residues 1674 to 1723 (Figure 1B). Thirty-one of 58 of the HD-N mutations and 6 of 11 of the HD-C mutations introduced Pro-residues into the protein (Figure 1A-B). Although the structure of the HD domain of NOTCH1 has not yet been determined, Pro-residues are known to disrupt α-helical structures, and these mutations may potentially disturb the 3-dimensional structure of the protein. Fourteen of 58 of the HD-N mutations were short in-frame insertions or deletions of up to 6 residues. In 2 samples, a larger in-frame deletion of 75 nucleotides and a larger in-frame insertion of 45 nucleotides within HD-N was identified (Figure 1A). In one patient, a larger in-frame insertion of 72 nucleotides of HD-C was detected, consisting of a 14-bp random insertion and a 58-bp partial duplication of the HD-C domain (Figure 1B).
NOTCH1 mutations in patients with precursor T-cell ALL of the ALL-BFM 2000 study. (A) NOTCH1 HD-N domain mutations. Missense and deletion mutations (top panel) and insertions (bottom panel) located between positions 1571 and 1642. The mutated amino acid residues are indicated. Novel NOTCH1 mutations are indicated by an asterisk next to the sample identification number. Deletions are indicated by dashes (–). (B) NOTCH1 HD-C domain mutations. Missense mutations (top panel) and insertion (bottom panel) located between positions 1674 and 1723. The mutated amino acid residues are indicated.
NOTCH1 mutations in patients with precursor T-cell ALL of the ALL-BFM 2000 study. (A) NOTCH1 HD-N domain mutations. Missense and deletion mutations (top panel) and insertions (bottom panel) located between positions 1571 and 1642. The mutated amino acid residues are indicated. Novel NOTCH1 mutations are indicated by an asterisk next to the sample identification number. Deletions are indicated by dashes (–). (B) NOTCH1 HD-C domain mutations. Missense mutations (top panel) and insertion (bottom panel) located between positions 1674 and 1723. The mutated amino acid residues are indicated.
NOTCH1 PEST domain mutations. Nonsense mutations are indicated by an asterisk behind the last amino acid. Frame-shift mutations with the elongated abnormal reading frame are indicated. All numbers correspond to position of amino acid residues in human pro-NOTCH1 protein, including the signal peptide. Novel NOTCH1 mutations are indicated by an asterisk next to the sample identification number.
NOTCH1 PEST domain mutations. Nonsense mutations are indicated by an asterisk behind the last amino acid. Frame-shift mutations with the elongated abnormal reading frame are indicated. All numbers correspond to position of amino acid residues in human pro-NOTCH1 protein, including the signal peptide. Novel NOTCH1 mutations are indicated by an asterisk next to the sample identification number.
In contrast to the missense and in-frame deletions and insertions of the HD domain, the PEST domain mutations were nonsense or frame-shift mutations that create premature termination codons (PTCs) (Figure 2). All of these PTCs were located in the last exon of the NOTCH1 gene and are thus not expected to activate nonsense-mediated decay.31-33 Therefore, it is likely that these PTC-mutated mRNAs result in the production of C-terminally truncated proteins.
T-ALLs with NOTCH1 mutations respond favorably to ALL-BFM 2000 treatment
The clinical and biologic characteristics of the study group were typical for childhood T-ALL1 and did not differ between the groups with or without NOTCH1 mutations (Table 1). Remarkably, however, most T-ALLs with the common cortical immunophenotype were NOTCH1 mutated, whereas in samples with the rarer pro-T, pre-T, and mature-T immunophenotypes NOTCH1 mutations were less common (P = .019; Table 1). This bias of NOTCH1-mutated T-ALL suggests that an activated NOTCH1 pathway may predispose for malignant transformation at the intermediate step of T-cell differentiation.
Prednisone response has been identified as a strong prognostic factor in childhood ALL26 and is used for the stratification of risk groups and of treatment intensity in the ALL-BFM 2000 study. Of interest, NOTCH1 mutations were more common in the prednisone good-responder than in the poor-responder subgroup (P = .001) (Table 2). In a logistic regression analysis, T-ALL patients with NOTCH1 mutations were 3 times less likely to show a poor prednisone response (relative risk = 0.33, 95% CI = 0.17-0.65; P = .001) than patients with wild-type NOTCH1. Furthermore, in a multivariate analysis, including variables known to be associated with prednisone response (sex, age at diagnosis, presenting WBC count at diagnosis, and T-cell immunophenotype), the NOTCH1 mutation status retained its significant effect on early response to treatment (relative risk = 0.42, 95% CI = 0.19-0.91; P = .028).
In the ALL-BFM 2000 study, treatment response is further assessed by the measurement of MRD after 33 days and 78 days of treatment.27,28 At both time points, most of the T-ALLs with a favorable MRD status (< 10–4) were NOTCH1 mutated, whereas in most with an unfavorable MRD response (≥ 10–4) NOTCH1 mutations were not identified. While this difference was statistically highly significant at both time points (Table 2), NOTCH1 mutations appeared to more robustly predict a favorable MRD response at day 78 (P = .001) than at day 33 (P = .019). In a logistic regression analysis, patients with NOTCH1 mutations had a 3.9-fold lower risk of an unfavorable MRD result at day 78 compared with patients with wild-type NOTCH1 (relative risk = 0.26, 95% CI = 0.13-0.53; P = .001). In multivariate analyses, including variables known to be associated with treatment response (sex, age at diagnosis, WBC count at diagnosis, T-cell immunophenotype), the NOTCH1 mutation status retained its significant effect on treatment response as measured by MRD at day 78 (relative risk = 0.35, 95% CI = 0.16-0.77; P = .009). The effect of activating NOTCH1 mutations on treatment response did not differ between the subgroups with HD, PEST, or HD + PEST mutations (Table S2). Therefore, the mechanism of NOTCH1 activation does not appear to play a clinically relevant role.
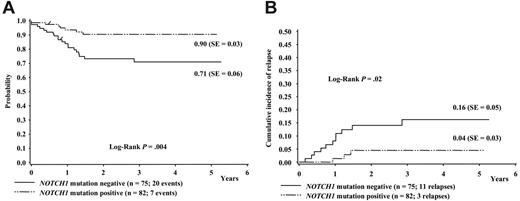
Considering the favorable effect of NOTCH1 mutations on response to treatment, it was interesting to analyze the effect on long-term prognosis. The median length of follow-up of the 157 T-ALL patients analyzed was 3.35 years (range, 0.5-5.3 years). Figure 3A shows a Kaplan-Meier estimate of event-free survival according to NOTCH1 mutation status. Patients with NOTCH1 mutations showed a significantly better relapse-free survival compared with those without mutations (90% vs 71%, P = .004). Figure 3B shows the cumulative incidence of relapse in our study sample according to NOTCH1 mutation status. The cumulative incidence of relapse was significantly higher in patients without mutations than in those with NOTCH1 mutations (16% vs 4%, P = .02). Although it is possible that the differences between groups may change with longer follow-up, this does not appear likely because the EFS and cumulative event curves reach a plateau at approximately 2 years, while the median period of follow-up of the 157 patients studied extends to 3.35 years. In a Cox regression analysis, this corresponded to a univariate relative risk of 0.24 (95% CI = 0.07-0.84; P = .03). In a multivariate analysis, this favorable long-term outcome was, of course, influenced by the early outcome measures (prednisone response and MRD kinetics) but independent from sex, age at diagnosis, and WBC count at diagnosis. Of importance, when the influence of NOTCH1 mutation status was analyzed in a multivariate analysis controlled for immunophenotype, the NOTCH1 mutation status retained its significant effect on long-term event-free survival (relative risk = 0.23, 95% CI = 0.06-0.86; P = .029), which shows that the NOTCH1 status is an independent prognostic factor on event-free survival in the context of ALL-BFM 2000 therapy.
Effect of NOTCH1 mutation status on long-term prognosis in T-ALL. One-hundred fifty-seven children with precursor T-ALL of the ALL-BFM 2000 study were classified according to either the presence or the absence of NOTCH1 mutations. (A) Kaplan-Meier estimate of event-free survival (EFS) at 4 years. (B) Cumulative incidence of relapse.
Effect of NOTCH1 mutation status on long-term prognosis in T-ALL. One-hundred fifty-seven children with precursor T-ALL of the ALL-BFM 2000 study were classified according to either the presence or the absence of NOTCH1 mutations. (A) Kaplan-Meier estimate of event-free survival (EFS) at 4 years. (B) Cumulative incidence of relapse.
Discussion
The concept of risk-adapted, individualized therapy in childhood ALL is one of the true success stories of modern clinical oncology.1 In precursor B-cell ALLs, common genetic risk factors such as numeric chromosome abnormalities and the presence of MLL/AF4, BCR/ABL, E2A/PBX1, or TEL/AML1 fusion transcripts5 are components of risk stratification algorithms of current treatment protocols.
The important finding of this paper is the highly significant correlation between the presence of activating NOTCH1 mutations and a favorable early treatment response (assessed by prednisone response and MRD kinetics) and long-term treatment outcome. These favorable effects were independent of other clinical prognostic factors such as age, sex, initial WBC count, or T-cell immunophenotype at the time of diagnosis.34 In addition, the effect of NOTCH1 on treatment response and prognosis was also independent from the localization of the NOTCH1 mutations or the predicted mechanism of activation (Table S2).
In pediatric T-ALL, the risk-adapted treatment stratification relies on treatment response parameters (prednisone and MRD kinetics). Currently, there are no genetic markers that reliably predict treatment response or outcome. The overexpression of HOX11, TAL1, and LYL1 has been reported to indicate a favorable or an unfavorable prognosis in a small number of patients.7,8 Although RNA expression profiles may provide useful prognostic markers in the future, the quantification of mRNA in clinical samples is technically demanding and has been shown to critically depend on the preanalytic handling of the bone marrow.35,36 The common cortical T-cell immunophenotype has previously been reported in early ALL-BFM, COALL, and POG protocols to be associated with a favorable clinical outcome when compared with other immunophenotypic profiles34,37,38 and with increased in vitro susceptibility to apoptosis-inducing drugs.39 The cortical immunophenotype can also be seen to have a favorable effect on early treatment response in this series and thus represents a confounding variable in the analysis of the NOTCH1 effect. However, the prognostic value of NOTCH1 mutations retained its independent value in a multivariate analysis.
Activating NOTCH1 mutations are thus likely to turn out to be clinically most useful genetic prognostic markers in T-ALL. The particular value of the NOTCH1 mutations appears to be reflected by the early identification of a large subgroup of patients with an excellent prognosis (Figure 3) who may benefit from a reduction of treatment intensity. Furthermore, NOTCH1 mutations will likely contribute to the reliability of the high risk profile of patients who require more intensive therapy. Technically, NOTCH1 mutation analysis is advantageous because it relies on robust and well-established DNA technology and, therefore, is more reliable than markers that depend on the quantification of RNA.
NOTCH1 receptors are involved in the regulation of various cellular differentiation events. The diverse functions are mediated through a conserved signal transduction pathway including regulated proteolysis of NOTCH1 receptors.40 Binding of ligands to the extracellular subunit (NEC) initiates a cascade of proteolytic cleavages of the transmembrane subunit (NTM) and releases an intracellular NOTCH1 fragment (ICN1). The final step in this cascade is catalyzed by the γ-secretase multiprotein protease complex.41 Finally, ICN1 translocates to the nucleus and participates in a transcriptional activation complex that is responsible for the up-regulation of NOTCH1 target genes.42
In hematopoiesis, NOTCH1 is important for the commitment of stem cells to develop into functional T cells and particularly for the assembly of pre–T-cell–receptor complexes in immature thymocytes.43,44 Less than 1% of T-ALLs harbor the chromosomal translocation t(7;9) that involves NOTCH1,9 which results in a deregulated expression of intracellular forms of the NOTCH1 protein. When expressed in the bone marrow stem cells of mice, constitutively activated forms of the NOTCH1 receptor are potent inducers of T-ALL.45,46 Furthermore, it has been shown that the expression of the active receptor fragment ICN1 in bone marrow is sufficient to induce T-cell leukemia in 100% of the mice that underwent transplantation.47
The NOTCH1 mutations identified here are localized in the heterodimerization and in the PEST domains. Mutations in these domains have previously been documented to result in an aberrant up-regulation of the NOTCH1-dependent signal transduction.14,16 The types of mutations detected within the HD domain may potentially destabilize the noncovalent intersubunit association, and finally enhance the production of ICN1.15 In contrast, the nonsense and frame-shift mutations within the 3′ terminal exon of the NOTCH1 gene are expected to result in the synthesis of C-terminal truncations of the NOTCH1 protein.31-33 The C-terminal part of NOTCH1 contains the PEST domain, a motif implicated in the regulation of protein turnover, which characterizes proteins with very short half-lives.48 The (partial) deletion of this region increases the stability of the protein and thus up-regulates NOTCH1 signaling. While some of the mutations identified here are identical to those described and functionally tested previously, others are novel mutations (Figures 1, 2). Therefore, the question arises of whether these novel mutations are also functionally relevant or whether they represent polymorphisms. The novel PEST domain mutations can reasonably be predicted to cause gain of function, because they result in the removal of the functionally critical C-terminal amino acid sequence that controls protein stability. The novel HD-domain mutations are similar to those that have been described previously in that they can be predicted to cause a substantial change of the secondary structure by involving nonconservative amino acid substitutions, in-frame deletions, and insertions (Figures 1, 2). Furthermore, the separate analysis of known and novel mutations regarding early treatment response shows that the patient groups with bona fide activating NOTCH1 mutations are clinically indistinguishable from those with novel HD-domain mutations (Table S3). Therefore, it appears likely that these novel mutations as a group cause relevant activation of the NOTCH1 receptor function, although it cannot be excluded that some of these mutations may represent polymorphisms. In a clinical setting, it will be interesting to prospectively study the effect of these novel mutations on ICN1 activity in cells.
The high frequency of these activating mutations implies a central role for NOTCH1 signaling in the pathogenesis of T-ALL. This notion is strengthened by the exclusive occurrence of these types of NOTCH1 mutations in T-ALL, but not in precursor B-cell ALL. It will now be interesting to identify the NOTCH1-dependent target genes and the molecular mechanism(s) that lead to the development of T-ALL. It is likely that the transforming activity of NOTCH1 is related to its central role in normal hematopoiesis. An increase in NOTCH1 signaling causes atypical T-cell differentiation and expands the pool of immature T-cell progenitors.49 This effect of NOTCH1 activation is mirrored by the predominance of the cortical immunophenotype (Table 1), although NOTCH1 mutations can also be identified, albeit less frequently, in T-ALL with noncortical immunophenotypes. The characterization of the NOTCH1 pathway in T-ALL will hopefully identify target molecules for novel and specific therapeutic strategies. Already, inhibitors of γ-secretase have been tested in T-ALL cell lines and induce cell cycle arrest.50
In conclusion, this study of children and adolescents with T-ALL demonstrated that activating NOTCH1 mutations are correlated with favorable early treatment response and an improved long-term prognosis. These findings indicate that NOTCH1-mutated T-ALLs are more sensitive to the ALL-BFM 2000 treatment strategy than T-ALLs without NOTCH1 mutations. It will now be interesting to validate the role of activating NOTCH1 mutations as early prognostic markers in a prospective setting and to evaluate this genetic marker in an improved risk-adapted strategy for the treatment of T-ALL.
Prepublished online as Blood First Edition Paper, April 13, 2006; DOI 10.1182/blood-2005-12-4956.
Supported by a grant from the German National Genome Network (N1KR-S19T11).
S.B. contributed to the design of the study, supervised the experimental work, interpreted and analyzed the data, and wrote the paper; M. Stanulla contributed to the acquisition, analysis, and interpretation of the clinical data, performed the statistical analysis, and revised the paper; T.F. performed the minimal residual disease (MRD) analyses; M. Schrappe is the clinical chair of the ALL–Berlin-Frankfurt-Munster (BFM) 2000 study and revised the paper; W.-D.L. supplied leukemic cell samples, performed immunophenotypic characterization of precursor T-cell leukemias by flow cytometry, and revised the paper; G.T. and M.H. obtained the primary molecular data; M.U.M. contributed to the design of the study and supervised the experimental work; and A.E.K. contributed to the design of the study, supervised the experimental work, and wrote the paper.
The online version of this article contains a data supplement.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We acknowledge all participants of the ALL-BFM 2000 study. We thank Sven Danckwardt for helpful discussions and Martin Zimmermann for contributing to the statistical analysis.