Abstract
We recently reported that Swedish VH3-21-using chronic lymphocytic leukemia (CLL) patients showed restricted immunoglobulin gene features and poor prognosis despite VH mutation status. To investigate this further, we analyzed the VH and VL gene rearrangements in 90 VH3-21+ patients from Sweden, Germany, Italy, United States, Finland, and Australia and correlated these data with survival and other prognostic markers. Sixty-three percent exhibited mutated VH genes and 37% unmutated VH genes. Fifty (56%) patients displayed a short and homologous heavy-chain CDR3, many of these with the amino acid motif DANGMDV. Also, a highly biased Vλ2-14 use was evident in 72% of patients with a restricted light-chain CDR3, QVWDS(S/G)SDHPWV. Combined restricted heavy- and light-chain CDR3s were found in patients from all included countries. Although VH3-21+ CLLs have a remarkably predominant λ expression, analyses of kappa deleting element indicated a conserved light-chain rearrangement order. The overall survival was poor in the VH3-21+ cohort (median survival, 88 months), with no significant difference in relation to mutation status or CDR3 homology. High ZAP-70 and CD38 expression was found in both mutated and unmutated VH3-21+ cases as well as a slight increase of 11q-aberrations. In summary, highly restricted B-cell receptors and worse outcome characterize VH3-21+ CLLs independent of geographic origin and mutation status.
Introduction
In chronic lymphocytic leukemia (CLL), preferential use of individual VH genes has been revealed, for instance, the VH1-69, VH3-07, and VH4-34 genes.1-5 VH1-69 gene rearrangements were shown early on to display characteristic features in CLL, such as unmutated VH genes, a long heavy-chain complementary determining region 3 (HCDR3), and preferential use of certain diversity (D) and joining (JH) genes compared with normal B cells.1-3,6-8 Recently, several CLL subsets using certain VH genes have been characterized with homologous HCDR3s, which also showed restricted variable light-chain (VL) gene use.5,9,10 These findings have led to the speculation that antigen selection could play a role in the pathogenesis of CLL,5,10,11 where foreign antigens or autoantigens may stimulate proliferation of B cells with specific B-cell-receptor (BCR) features, increasing the risk for malignant transformation.12
We recently reported preferential use of the VH3-21 gene in CLL.13 Of interest, many of the VH3-21+ CLLs (both Ig unmutated and mutated) showed rearrangements with short (7 codons) and homologous HCDR3s and had a predominant λ expression with biased Vλ2-14 gene use.13,14 These data indicate a common antigenic epitope being recognized by the individual VH3-21+ tumors. Furthermore, VH3-21-using patients had short overall survival, similar to patients with unmutated VH genes, despite the fact that two thirds of this group had mutated VH genes.14 We therefore suggested that the VH3-21+ CLL patient group should be considered as a separate entity, independent of mutation status, a finding that was further supported by the distinct gene-expression profile recently demonstrated in VH3-21+ CLL compared with unmutated and mutated CLL using other VH genes.15 Furthermore, a recent study of VH3-21+ CLLs from several Mediterranean countries showed rearrangements identical to the ones previously reported by us, although they had a lower frequency of VH3-21-using patients (∼ 3%).16 In this study, VH3-21+ patients with a homologous HCDR3 had a worse prognosis compared with patients with a nonhomologous HCDR3.16
In the present study, we investigated whether VH3-21+ CLLs have similar molecular characteristics of the VH/VL gene rearrangements, independent of geographic origin, in an extended joint material of VH3-21+ CLLs (90 cases) from Sweden, Germany, Italy, United States, Finland, and Australia. We also analyzed the prognostic impact of VH3-21 gene use and its relation to other prognostic markers (eg, CD38 and ZAP-70 expression and genomic aberrations). Since most VH3-21+ cases display a strikingly biased Vλ gene use, we investigated the rearrangement order of the light-chain loci and assessed the kappa deleting element (KDE) rearrangement status in λ-expressing VH3-21+ CLLs.
Patients, materials, and methods
Patients and materials
Tumor samples from 90 CLL patients with VH3-21 gene use were obtained from larger CLL cohorts partly included in previous studies13,14,17 from the University hospitals of Uppsala (n = 17), Umeå (n = 7), Linköping (n = 7), Huddinge (n = 1), Sweden; Tampere (n = 3), Finland; Ulm (n = 37), Germany; Modena (n = 9), Italy; North Shore, Manhasset (n = 7), NY; and Sydney (n = 2), Australia. All VH3-21-using CLL patients from the respective cohorts were included. The tumor material was obtained mainly from peripheral blood and bone marrow, but also from lymph node and spleen in a few cases. The CLL diagnosis was based on morphologic and immunophenotypic features according to the World Health Organization (WHO) classification.18 Light-chain expression was known in 67 cases; 10 cases expressed κ light chains and 57 cases expressed λ light chains. The median age at diagnosis was 62 years and the male-female ratio was 2:1. Survival data, obtained from medical records and cancer registries, were available in 64 of 90 patients with a median follow-up of 51 months. Informed consent was provided according to the Declaration of Helsinki, and the study was approved by the ethics committee of Uppsala University (Ups 01-082), Umeå University (05-102M), Linköping University (98-319, 02-459, and 97-252), University of Ulm, Tampere University Hospital (288.94171), North Shore-LIJ Health System (Human Leukemia Tissue Bank), and the Wentworth Health Area Service, University of Sydney (2001/070).
CD38 and ZAP-70 expression
Cytogenetic analysis
Fluorescence in situ hybridization (FISH) analysis of the following aberrations was performed as described previously22 : 11q-, +12, 13q-, and 17p-.
Analysis of VH and VL gene rearrangements
The polymerase chain reaction (PCR) amplification of the VH and VL gene rearrangements was performed with either genomic DNA or cDNA using family-specific VH (either framework region 1 [FR1] or VH leader primers), Vλ, Vκ primers together with consensus J primers as previously described.2,11,14,17 In 9 cases, VH gene rearrangements were amplified with consensus FR2 and JH primers.23 The PCR conditions for the VH family PCR amplification were as earlier outlined.2,11,14,17,24 The VL family PCR analysis was either carried out as reported earlier11,25 or in a 50-μL reaction containing 200 ng DNA, 0.2 mM of each dNTP, 2.0 mM MgCl2, 2.5 U Platinum Taq (Invitrogen, Paisley, United Kingdom), 0.125 μM of each primer, and 1 × PCR Rxn buffer (Invitrogen). The Vλ/Jλ gene rearrangements were amplified with an initial 2-minute denaturation at 95°C followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 61°C, and 30 seconds at 72°C, ending with a 5-minute elongation at 72°C. The Vκ family PCR was performed at 95°C for 2 minutes, 65°C for 1 minute, and 72°C for 1 minute followed by 40 cycles of 30 seconds at 95°C, 30 seconds at 61°C, and 45 seconds at 72°C, ending with 72°C for 5 minutes.
The sequencing of the VH gene rearrangements was performed as described previously.11,14,17 The VL PCR products were direct sequenced using the Big Dye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Foster City, CA) or cloning was performed, as described earlier.14 For the VL gene rearrangement, a minimum of 5 colonies was subsequently sequenced from each cloned sample. All VL sequences were analyzed using an automated DNA sequencer (ABI 377 or ABI 3700; Applied Biosystems).
The obtained VHDJH and VLJL sequences were submitted to 3 different Ig databases (GenBank, IMGT, and V-Base) and aligned to the closest germ-line sequence as detailed previously.26 VH and VL sequences with less than 98% homology to the germ-line sequence were considered mutated. At least 7 consecutive nucleotides aligned to the most homologous germ-line sequence were required to identify the D gene segment. All in-frame sequences from the VH and VL gene rearrangements were converted into amino acid sequences. The HCDR3 length was calculated between codons 95 to 102 and the light-chain CDR3 (LCDR3) between codons 89 to 97, as described by Kabat.27 Alignments of the HCDR3s and LCDR3s were performed by using Clustal X version 1.83 multiple sequence alignment software (UBC Bioinformatics Centre, Vancouver, BC, Canada) for Windows. High CDR3 homology was defined as 1 or fewer amino acid deviations from the most frequently occurring CDR3 sequence, whereas sequences with less homology (eg, 2-3 deviations from the most homologous sequence) were referred to as CDR3s with moderate homology. The VL genes were named according to the nomenclature used in the GenBank database in order to avoid confusion between the VH and VL genes since Vλ2-14 (GenBank) is named Vλ3-21 using the IMGT nomenclature.
Analysis of KDE rearrangements
Analysis of the KDE rearrangements was performed on 42 samples with known λ expression using primers as previously described.28 Amplification of germ-line KDE was carried out using conditions identical to those of the Vκ/Jκ PCR, as described in the previous section. The same conditions were also applied for amplification of the rearrangement between the KDE and the recombination signal sequence (RSS) in the Jκ-Cκ intron region (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article), with the exception of a lower MgCl2 concentration (1.5 mM) and a shorter elongation step (30 seconds). Primers detecting KDE-Vκ gene rearrangements were included in the Vκ-PCR for the λ-expressing cases.
Statistical analyses
Overall survival was defined as the time from diagnosis until last follow-up or death. Kaplan-Meier survival analysis and log-rank test were carried out to investigate survival differences between subsets. All statistical calculations were performed with Statistica 6.0 (Stat Soft, Tulsa, OK).
Results
Characterization of VH3-21+ gene rearrangements
Of 90 VH3-21+ CLL cases, all but 2 (cases 36G and 89S) had VH3-21+ rearrangements that were in-frame and potentially functional. In 57 (63%) cases, more than 2% somatic hypermutation was detected within the VH gene, with a median mutation rate of 3.1% (range, 2.1%-10%), whereas 33 (37%) cases showed unmutated VH genes. The D gene segment was recognized in 31 of 90 VH3-21+ rearrangements; the most common D genes were D3-3 and D3-10, detected in 5 rearrangements each. In the remaining 59 VH3-21+ rearrangements, it was not possible to assign the D gene; the reason for this remains unknown but could hypothetically be due to extensive deletion of nucleotides during VDJ recombination. The most used JH gene was JH6, which was amplified in 60 rearrangements (67%), followed by JH4 (16 rearrangements, 18%). All VH3-21 sequencing data are shown in Table S1.
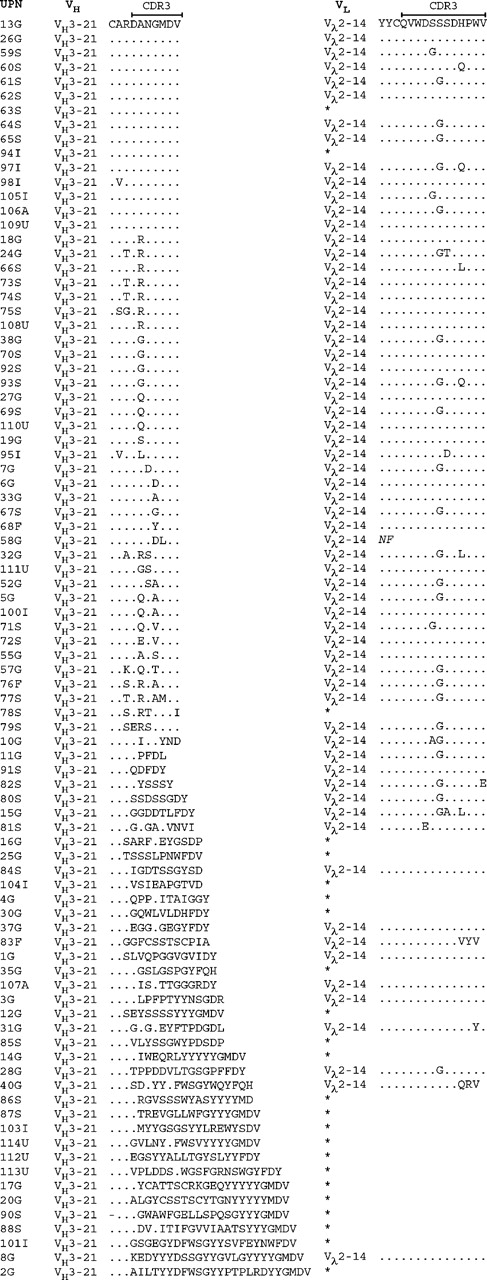
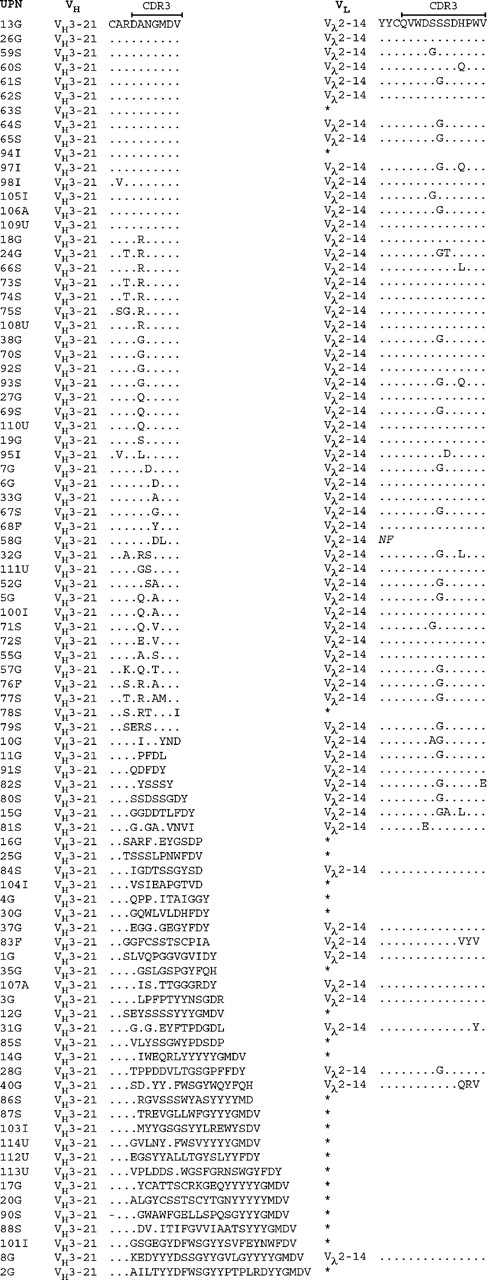
At the amino acid level, the functional VH3-21+ rearrangements displayed HCDR3s with a median length of 7 codons (range, 5-25 codons). Fifty (56%) of these, including cases from all countries, showed an HCDR3 with an unidentified D gene together with a JH6 gene. Fifteen patients had an identical amino acid sequence expressing the Asp-Ala-Asn-Gly-Met-Asp-Val (DANGMDV) motif, and the HCDR3 was highly homologous (1 amino acid difference) in another 21 of the patients. In a further 14 patients, moderate homology of the HCDR3 was shown, corresponding to 2 to 3 deviations from the DANGMDV sequence. Of the VH3-21+ cases with high/moderate HCDR3 homology, 74% (n = 37) had a mutated VH gene with a mutation frequency ranging between 2.2% and 10% (median, 3.4%). The HCDR3 sequences for all functional VH3-21 gene rearrangement are displayed in Figure 1.
A summary of the characteristics of the VH3-21+ rearrangements from the different countries is shown in Table 1.
VL gene rearrangements
Of 90 VH3-21+ cases, at least 1 VL gene rearrangement was demonstrated in each case, and in total 89 Vκ (26 functional and 63 nonfunctional) and 88 Vλ (78 functional and 10 nonfunctional) rearrangements were amplified and sequenced. Sixty-eight cases showed Vκ rearrangements and 77 Vλ gene rearrangements (55 cases with both Vκ and Vλ gene rearrangements). All VL gene sequencing data are shown in Table S1.
Functionality. At least one functional VL gene rearrangement was demonstrated in 82 of 90 cases. In the 10 cases with κ expression, all showed functional Vκ rearrangements, and in the 57 cases with λ expression, a functional Vλ rearrangement was detected in 52 of these. In the remaining 23 cases without known light-chain expression, 16 cases had a functional Vλ rearrangement, 1 case had a functional Vκ rearrangement, 4 cases showed both functional Vκ and Vλ rearrangements, and 2 cases had no functional rearrangement amplified.
VL gene mutation status. The majority of the Vκ genes were unmutated, and only 8 of 89 rearrangements harbored somatic hypermutation, with a median mutation rate of 2.9% (range, 2.3%-5.8%). Somatically mutated Vλ genes were detected in 20 of 88 rearrangements, with a mutation frequency of a median of 3.4% (range, 2.4%-8.1%).
VL/JL gene use. The most frequently rearranged Vκ genes were VκB3 (n = 16) and VκA27 (n = 12). Jκ4 and Jκ1 were the most commonly used Jκ genes, detected in 31 and 19 rearrangements, respectively. The most frequently rearranged Vλ gene was Vλ2-14, which was shown in 65 (72%) cases, followed by Vλ2-1 detected in 4 (4%) cases. The Jλ3 gene was found in 68 (76%) patients and the Jλ1 and Jλ2 genes in 10 and 6 patients, respectively.
The combination of the Vλ2-14/Jλ3 genes was demonstrated in 61 cases from all included countries (Germany, n = 24; Sweden, n = 24; Italy, n = 5; United States, n = 4; Australia, n = 2; and Finland, n = 2). The LCDR3 of the VH2-14+ cases had a median length of 12 codons (range, 11-12 codons) and showed high sequence homology in the cases with functional rearrangements. The most common LCDR3 sequence, detected in 45 cases, was Gln-Val-Trp-Asp-Ser-(Ser/Gly)-Ser-Asp-His-Pro-Trp-Val (QVWDS[S/G]SDHPWV), where the sixth amino acid could be either a serine (S) or a glycine (G). In 14 cases, 1 additional difference from either of these 2 sequences was displayed, and 2 to 3 additional differences were demonstrated in another 4 cases. The LCDR3 amino acid sequences of the Vλ2-14+ rearrangements are shown in Figure 1.
Amino acid sequences of the HCDR3 and LCRD3 in the VH3-21-using cases. The HCDR3 sequences are aligned to the most commonly occurring HCDR3 sequence (DANGMDV). Amino acids identical to the top sequence are indicated with a dot (.) and missing amino acids are indicated with a dash (-). Only the Vλ2-14 LCDR3 sequences are outlined, and an asterisk (*) indicates that the case used a VL gene other than Vλ2-14. Geographic origin of the patient is indicated in the case number: Germany (G), Sweden (S), Italy (I), United States (U), Finland (F), and Australia (A).
Amino acid sequences of the HCDR3 and LCRD3 in the VH3-21-using cases. The HCDR3 sequences are aligned to the most commonly occurring HCDR3 sequence (DANGMDV). Amino acids identical to the top sequence are indicated with a dot (.) and missing amino acids are indicated with a dash (-). Only the Vλ2-14 LCDR3 sequences are outlined, and an asterisk (*) indicates that the case used a VL gene other than Vλ2-14. Geographic origin of the patient is indicated in the case number: Germany (G), Sweden (S), Italy (I), United States (U), Finland (F), and Australia (A).
Combined VH and VL gene use
The specific combination of VH3-21/D-/JH6 together with Vλ2-14/Jλ3 was found in 51% of the patients (n = 46), of which 34 cases displayed highly homologous HCDR3 and LCDR3 sequences and 12 patients moderate HCDR3 homology and highly homologous LCDR3s (Figure 1; Table 1). The conserved Ig sequences were detected in patients from all 6 included countries: Sweden (n = 19), Germany (n = 15), Italy (n = 5), United States (n = 4), Finland (n = 2), and Australia (n = 1).
Analysis of kappa deleting element (KDE) rearrangements
Forty-two of the cases, with known λ light-chain expression, were examined for the presence of a rearranged KDE, as well as the germ-line configuration of KDE. Rearrangement of the KDE, either to the Jκ-Cκ intron RSS or to a Vκ RSS (Figure S1), was detected in 41 (98%) of 42 cases, on one (38%) or both (60%) alleles. A nonfunctional Vκ gene rearrangement together with 1 or 2 KDE rearrangements was demonstrated in 15 and 16 patients, respectively. PCR product showing germ-line KDE configuration was found in all analyzed samples. The KDE data are shown in Table S2.
Relation to other prognostic markers
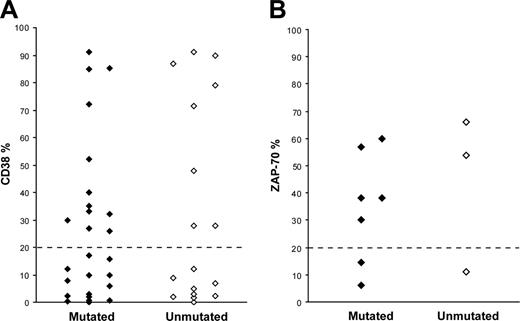
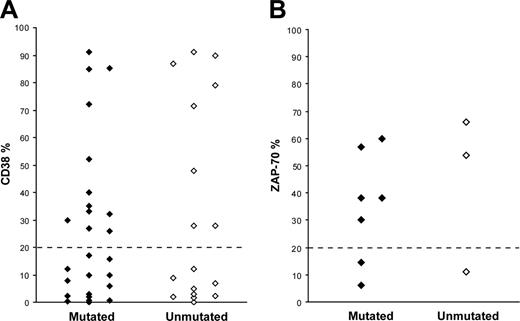
CD38 expression data were available for 43 patients; in 20 cases (12 mutated and 8 unmutated) the CD38 expression level was more than 20%, whereas 23 cases (14 mutated and 9 unmutated) showed less than 20% CD38 expression (Figure 2A). ZAP-70 expression was assessed in 10 patients, of which 7 cases (5 mutated and 2 unmutated) demonstrated more than 20% ZAP-70+ cells, while the remaining 3 cases (2 mutated and 1 unmutated) had less than 20% ZAP-70+ cells (Figure 2B). FISH analysis was performed in 55 cases (34 mutated and 21 unmutated); 13q-was the most common aberration (n = 25, 18 mutated and 7 unmutated), followed by 11q-(n = 15, 8 mutated and 7 unmutated), 17p-(n = 5, 3 mutated and 2 unmutated), and +12 (3 unmutated). In 7 cases, none of the analyzed cytogenetic aberrations was detected.
CD38 and ZAP-70 expression in VH3-21+ CLL. (A) CD38 expression in 26 mutated and 17 unmutated VH3-21+ CLL patients. (B) ZAP-70 expression in 7 mutated and 3 unmutated VH3-21+ CLL patients.
CD38 and ZAP-70 expression in VH3-21+ CLL. (A) CD38 expression in 26 mutated and 17 unmutated VH3-21+ CLL patients. (B) ZAP-70 expression in 7 mutated and 3 unmutated VH3-21+ CLL patients.
Survival analysis
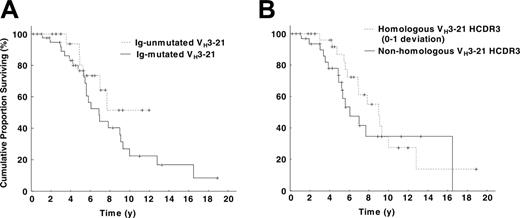
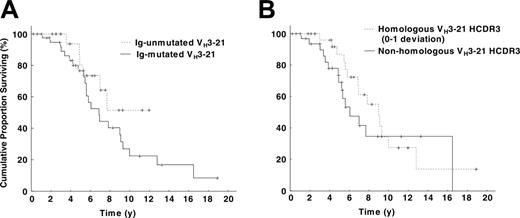
Survival data were available for 64 patients from Sweden (n = 32), Germany (n = 14), Italy (n = 9), United States (n = 4), Finland (n = 3), and Australia (n = 2), and the overall survival was 88 months (range, 37-198 months). No difference in survival was found between mutated (41 cases) and unmutated (23 cases) VH3-21+ patients, with a median survival of 79 months for the mutated, whereas the median survival was not reached for the unmutated cohort (log-rank test, P = .17; Figure 3A). Also, no significant difference in overall survival was evident when comparing survival in patients with highly homologous HCDR3 (30 cases) versus nonhomologous HCDR3 (34 cases) (104 vs 70 months, P = .32; Figure 3B), short (41 cases) versus long (23 cases) HCDR3 (92 vs 65 months, P = .60), and Vλ2-14 use (48 cases) versus use of other VL genes (16 cases) (85 vs 77 months, P = .81). Different borders for the HCDR3 homology were also applied (0, 0-1, 0-2, 0-3 amino acid deviation from the DANGMDV sequence) but did not reveal any differences in overall survival.
Kaplan-Meier survival curve analysis in VH3-21+ CLL. (A) Comparison of VH3-21+ patients with mutated and unmutated VH rearrangements. No significant difference could be detected between the 2 subsets (log-rank test, P = .17); the median survival was 74 months for patients with mutated VH3-21 genes, whereas the median survival was not reached for patients with unmutated VH3-21 genes. (B) Comparison of VH3-21+ patients with highly homologous HCDR3s versus nonhomologous HCDR3s. The highly homologous HCDR3s had a median overall survival of 104 months, while patients with nonhomologous HCDR3s had a median survival of 70 months (log-rank test, P = .32).
Kaplan-Meier survival curve analysis in VH3-21+ CLL. (A) Comparison of VH3-21+ patients with mutated and unmutated VH rearrangements. No significant difference could be detected between the 2 subsets (log-rank test, P = .17); the median survival was 74 months for patients with mutated VH3-21 genes, whereas the median survival was not reached for patients with unmutated VH3-21 genes. (B) Comparison of VH3-21+ patients with highly homologous HCDR3s versus nonhomologous HCDR3s. The highly homologous HCDR3s had a median overall survival of 104 months, while patients with nonhomologous HCDR3s had a median survival of 70 months (log-rank test, P = .32).
Discussion
We have previously identified a VH3-21-using subset of CLL patients that displayed peculiar characteristics of the Ig genes and had a poor outcome.13,14 To further study the Ig gene features in VH3-21+ CLL and the prognostic impact of VH3-21 use, we extended the VH3-21+ material and included CLL patients from 6 different countries around the world (ie, Germany, Sweden, Italy, United States, Finland, and Australia). In parallel with our previous studies,13,14 we could demonstrate the characteristic Ig gene rearrangements from all included countries. A large proportion (56%) of VH3-21+ CLL patients showed gene rearrangements with very short and homologous HCDR3s, where many of these also had the amino acid motif, DANGMDV. Furthermore, a majority (72%) of VH3-21+ patients displayed preferential VL gene use of the Vλ2-14 gene. The LCDR3s were very similar in the majority (91%) of the VH3-21+/Vλ2-14+ patients and showed a conserved amino acid sequence, QVWDS(S/G)SDHPWV. The combination of homologous HCDR3/LCDR3 was detected in samples from all countries. Hence, according to our data, conserved VH3-21+ BCRs are most probably a general phenomenon worldwide, which is further supported by the recent study of Mediterranean VH3-21+ patients that showed cases with similarly restricted Ig gene characteristics.16
The uniformity of the BCRs found in the VH3-21 CLL cases strongly indicates that antigen selection is involved in CLL development via recognition of common antigenic epitopes. The fact that very similar VH3-21+/Vλ2-14+ rearrangements can be found in different countries in a considerable number of patients strengthens the theory of antigen selection, since this is unlikely to have occurred by chance. One current theory proposes that an antigen most likely promotes proliferation of a VH3-21+ B cell followed by clonal expansion and acquisition of genetic events before leukemia transformation (see the recent review by Chiorazzi et al12 ). Although, to date the potential antigen(s) is unknown, previous29-31 and recent studies32 suggest that autoantigens may be involved in this process. Further research into finding such causative antigen(s) is necessary.
An important finding, correlating to our previous studies,13,14 was the short overall survival (median survival, 88 months) of the VH3-21+ patients. In CLL, the VH gene mutation status is known to be of prognostic value, with an improved survival for patients with somatically hypermutated VH genes.4,19 In this VH3-21 cohort, 63% of patients had somatically hypermutated VH genes but still had a poor prognosis similar to that of the unmutated VH3-21+ CLLs and unmutated CLLs using other VH genes,13,14,17 verifying that the mutation status is not applicable for this subset of patients. We also compared different subsets such as homologous HCDR3 versus nonhomologous HCDR3 and Vλ2-14 use versus use of other VL genes, but could not find any significant differences in overall survival. This is in contrast to the study of Mediterranean VH3-21+ CLL where the aggressive disease was attributed mainly to cases with homologous HCDR3s.16 The low number of VH3-21 cases analyzed (16 cases) in the Mediterranean cohort may explain this discrepancy. Furthermore, analysis of other prognostic markers showed that a considerable fraction of VH3-21+ patients displayed CD38 and ZAP-70 expression, both mutated and unmutated cases.21 A slightly higher number of 11q-aberrations was also found both in mutated and unmutated VH3-21+ CLLs (24% and 33%, respectively).
Studies from Sweden, Northern Ireland, and the United Kingdom have indicated a higher frequency (∼ 9%-10%) of VH3-21+ patients13,14,33,34 compared with CLL studies from other European countries and the United States (between 0% and 3%).2,4,16,35 What could be the reason for the difference in VH3-21 frequencies in different materials? We believe that the selection of patient material may be one major cause influencing the results of the VH gene analyses. The German and Swedish CLL cohorts used have been collected from different referral centers, leading to a patient cohort that in general has a worse prognosis compared with patient materials collected at local hospitals. Indeed, in our previous studies, the median overall survival of the CLL patient cohorts was significantly shorter than other studies with more stage A disease patients.4,13,14,17,19 Furthermore, most published studies on VH mutation status/VH gene use do not represent population-based material and have selected materials, to some extent at least. On the other hand, the fact that the frequency of VH1-69 gene rearrangements appears more or less similar in different parts of the world supports the concept that regional differences in VH3-21 frequency indeed exist. The frequency of VH3-21+ CLLs varied in this present material with the highest frequency in the Swedish cohort with decreasing frequencies as follows: Finland > Germany/Italy > United States/Australia. Hypothetically, a variation in exposure for a potential antigen(s) in different parts of the world could be an explanation for this finding. In the future, population-based investigations of VH gene use in large series of CLL patients may reveal the true frequencies of different VH genes.
Since the VH3-21+ CLL subset is characterized by a strikingly biased expression of λ light chains, this led us to ask if these VH3-21+ cases have undergone the traditional light-chain rearrangement pathway (Igκ, Igκ, Igλ). In most of the 42 VH3-21+ λ-expressing patients analyzed, the amplified Vκ gene rearrangements were nonfunctional due to KDE and/or out-of-frame Vκ rearrangements. This indicates that the light-chain rearrangements in the VH3-21+ cases have followed the ordered recombination and have gone through several rearranging attempts before producing a functional light-chain. The reason for the multiple rearrangement events occurring in VH3-21+ CLLs is unknown, but it may be speculated that negative selection has acted on BCRs with a VH3-21/Vκ rearrangement.
In conclusion, homologous VH3-21/Vλ2-14 rearrangements are found in CLLs from various centers around the world, and the very similar HCDR3/LCDR3s strongly indicate that antigen selection is involved in development of VH3-21+ CLL. VH3-21 gene use was also associated with increased ZAP-70 and CD38 expression and a more severe disease course, regardless of mutation status or HCDR3 homology.
Prepublished online as Blood First Edition Paper, November 29, 2005; DOI 10.1182/blood-2005-06-2227.
Supported by grants from the Swedish Cancer Society, Lion's Cancer Research Foundation in Uppsala and Umeå, Sweden, the research foundation of the Department of Oncology at Uppsala University, Sweden, the Sander Stiftung (2001.04.02) and Fresenius Stiftung, Germany, the National Health and Medical Research Council of Australia, and the Leukemia Foundation of Australia.
M.T. and A.K. contributed equally to this work.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.