Abstract
Although the expression of Pitx2, a bicoid family homeodomain transcription factor, is highly regulated during hematopoiesis, its function during this process was not documented; we thus studied hematopoiesis in Pitx2-null mice. We found that Pitx2–/– embryos display hypoplastic livers with reduced numbers of hematopoietic cells, but these cells had normal hematopoietic potential, as evidenced by colony-forming assays, immature progenitor cell assays, and long-term repopulation assays. Because the microenvironment is also crucial to the development of normal hematopoiesis, we established Pitx2–/– and Pitx2+/+ stromas from fetal liver and studied their hematopoietic supportive capacity. We showed that the frequency of cobblestone area-forming cells was 4-fold decreased when using Pitx2–/– stromal cells compared with Pitx2+/+ stromal cells, whatever the Pitx2 genotype of hematopoietic cells tested in this assay. This defect was rescued by expression of Pitx2 into Pitx2–/– fetal liver stromal cells, demonstrating a major and direct role of Pitx2 in the hematopoietic supportive capacity of fetal liver stroma. Finally, we showed a reduced capacity of MS5 stromal cells expressing Pitx2 RNAi to support human hematopoiesis. Altogether these data showed that Pitx2 has major functions in the hematopoietic supportive capacity of fetal liver and adult bone marrow stromal cells.
Introduction
Pitx2, a bicoid-type homeobox protein, plays an important role in the development of multiple organs.1 Pitx2 is expressed in many tissues during development, including derivatives of the first branchial arch, the eyes, the brain, the pituitary gland, the mandible, the heart, the lungs, and the limbs.1-9
The Pitx2 gene is expressed as 3 mRNA isoforms derived by either alternative splicing, for Pitx2a and Pitx2b, or from an internal promoter located upstream exon 4 for Pitx2c (Figure 1A). Exons 5 and 6, which encode the homeodomain and the COOH terminus of the protein, are common to all isoforms. Among these 3 isoforms, Pitx2c shows an asymmetrical pattern of expression during development on the left side of the embryo at the lateral plate mesoderm stage as well as later during development in the heart, gut, and lung. In contrast, all 3 isoforms are expressed symmetrically in the head mesoderm and the lateral body wall. Therefore, only the c isoform of Pitx2 was found to be involved in the determination of left-right asymmetry during development.10-16
Mutations in the homeodomain of Pitx2 are one cause of Rieger syndrome in humans, an autosomal-dominant disorder.3,17,18 It is characterized by craniofacial and umbilical stump abnormalities. The phenotype of mice heterozygous for a null Pitx2 allele mimics human Rieger syndrome and is characterized by eye and tooth abnormalities.4 Homozygous null embryos do not survive beyond embryonic day 14.5 after coitus due to severe cardiac and other developmental defects. Organogenesis of the eyes, teeth, and pituitary gland is also severely impaired in these embryos.4,5,9
Recently, Pitx2 has been identified as playing a possible role in hematopoiesis. Using differential display, Pitx2 cDNA was found to be underexpressed in Mll (All1)–/– embryonic stem (ES) cells. The Mll transcription factor, a human homolog of the Drosophila trithorax, is known to be involved in human acute leukemias. Moreover, down-regulation of Pitx2 mRNA was correlated with the presence of Mll gene translocations in human leukemia cell lines,19 and the Pitx2 gene has been found to be frequently methylated in its CpG islands in acute myeloid leukemia cells.20
In addition, Pitx2c cDNA was detected in a cDNA library derived from immature lin–/Hoechstlow/rhodaminelow murine bone marrow hematopoietic cells after subtraction with cDNA of more mature lin+ cells. Finally, expression of Pitx2c mRNA was very rapidly down-regulated when immature hematopoietic cells were stimulated to differentiate by cytokine stimulation.21 Taken together, these data suggest that Pitx2 could play an important role in hematopoietic stem cells.
To study the role of Pitx2 in hematopoiesis, we analyzed the hematopoietic tissue of Pitx2 knock-out mice. Pitx2-null embryos do not survive beyond 14.5 days after coitus, so adult bone marrow was not available for study. However, from 12 days after coitus to birth, fetal liver is the predominant site of definitive hematopoiesis. Therefore, we studied hematopoiesis in fetal liver of Pitx2-null embryos versus Pitx2+/+ littermates at 13.5 days after coitus. We showed that Pitx2-null embryos exhibited a clear deficiency in the number of their fetal liver hematopoietic cells. In contrast, these cells had normal hematopoietic potential in functional assays. We then demonstrated that the quantitative hematopoietic cell defect likely resulted from deficient hematopoietic supportive capacity of the Pitx2–/– fetal liver stromal cells. Furthermore, this impaired hematopoietic supportive capacity was reversed after lentiviral vector transduction of Pitx2 cDNA into Pitx2–/– fetal liver cells. Finally, expression of RNAi directed against Pitx2 mRNA in the MS5 stromal cell line reduced its capacity to support hematopoiesis. These results demonstrate a direct role of Pitx2 gene expression in the hematopoietic supportive capacity of fetal liver stromal cells and provide new insights into the regulation of hematopoiesis during fetal liver organogenesis.
Materials and methods
Pitx2-null mice
Generation of Pitx2 knock-out mice has been previously described.4 The homologous recombination event deletes part of the homeodomain-coding sequences (exon 5 in Figure 1A) resulting in the inactivation of all 3 Pitx2 isoforms. Pitx2+/– mice were maintained as a colony in which they had been backcrossed with C57BL/6J mice for at least 10 generations to ensure a stable genetic background. Genotypes of newborn mice were determined by amplification of genomic DNA isolated from tail biopsies, as described by Gage et al.4
Fetal liver cells
Pitx2–/+ male and female mice were mated overnight. When vaginal plugs were observed the following morning, 0.5 days after coitus was defined as noon of that day. At 13.5 days after coitus, pregnant mice were killed and the embryos were microdissected to remove livers. Murine fetal liver cells were suspended in α–minimal essential medium (MEM) containing 10% fetal calf serum (FCS) with a 23-gauge needle, and cells were enumerated. The genotype of the embryos was determined by polymerase chain reaction (PCR)4 of DNA extracted from other tissues.
Primary cells from normal fetal liver embryos were incubated with antimouse fluorescein isothiocyanate–coupled CD45 (CD45-FITC) and phycoerythrin-coupled TER119 (TER119-PE) (both from Pharmingen, San Diego, CA) and sorted on an EPICS ELITE (Beckman-Coulter, Villepinte, France) to isolate 104 cells of each of the following cell populations: CD45+/TER119–, CD45–/TER119+, CD45–/TER119. Alternatively, cells from fetal liver embryos were incubated for 15 minutes at 4°C with magnetic beads coated with monoclonal rat antimouse CD45 antibodies and purified by magnetic column (Miltenyi Biotec, Paris, France). Purity was verified by fluorescence-activated cell sorter (FACS) analysis to be more than 90%.
RNA extraction and RT-PCR
Total RNA was prepared from suspensions of liver cells or from cultured cells using Trizol (Life Technologies, Rockville, MD) and following the manufacturer's instructions. One microgram of total RNA was converted to cDNA using hexaprimers, Superscript II reverse transcriptase (RT) (Invitrogen, San Diego, CA), and dNTPs. For RT-PCR reactions, murine Pitx2 (mPitx2) cDNA was amplified for 40 cycles at an annealing temperature of 60°C (Pitx2c) and 52°C (Pitx2a and Pitx2b) using the following primers: mPitx2c, forward (fwd): 5′-TCGTCCATGAACTGCATGAAAG-3′; mPitx2c, reverse (rev): 5′-ATGTCATCGTAGGGCTGCATG-3′; mPitx2a and mPitx2b fwd: 5′-CGCAAACTAGTGTCGGCC-3′; and mPitx2a and mPitx2b rev: 5′-CTTTCTCCATTTGGCCCG-3′. As a control, Hprt cDNA was amplified for 30 cycles at an annealing temperature of 55°C, with the following primers: Hprt fwd: 5′-GCTGGTGAAAAGGACCTCT-3′; Hprt rev: 5′-CACAGGACTAGAACACCTGC-3′.
Real-time quantitative PCR
Real-time PCR was performed using a LightCycler rapid thermal cycler system (Roche Diagnostics, Lewes, United Kingdom) according to the manufacturer's instructions. Reactions were performed at a final concentration of 4 mM MgCl2 for Hprt and 3 mM MgCl2 for Pitx2 using the LightCycler-DNA Master SYBR Green I reaction kit (Roche Diagnostics). The annealing temperature was 60°C (Hprt) or 55°C (Pitx2a, Pitx2b, and Pitx2c). The following primers that recognize all Pitx2 cDNA isoforms were used: fwd 5′-AGCTGTGCAAGAATGGCTTT-3′; rev 5′-CACCATGCTGGACGACATAC-3′ for murine Pitx2 and fwd 5′-GCTCCTCAAAAGGACCTCT-3′; rev 5′-CACAGGACTAGAACACCTGC-3′ for murine Hprt, used as an internal standard to normalize cDNA input. All samples were run in duplicate and in 2 separate experiments. Melting curve analysis and quantification were carried out by using fluorimetric online detection with the LightCycler (Roche Diagnostics).
Immunofluorescence cell marking and flow cytometry
Single-cell suspensions from fetal liver at 13.5 days after coitus were stained with monoclonal antibodies: Ter119-FITC, CD45-FITC, allophycocyanin (APC)–coupled CD45, c-Kit–PE, and Sca1-FITC. Single-cell suspensions from peripheral blood, bone marrow, spleen, and thymus of mice that had previously received transplants were also stained with monoclonal antibodies: CD11b-FITC, Ly-6G–FITC, CD41-PE, CD19-PE, CD8-PE, CD4-FITC, CD4-PE, CD45.1-PE, CD45.2-FITC, TER119-PE. All antibodies were purchased from Pharmingen. Cells were analyzed on a FACSCalibur flow cytometer using CellQuest software (Becton Dickinson, Meylan, France).
Progenitor cell assays
Sorted CD45+ cells from 13.5 days after coitus murine fetal liver were plated at different concentrations in methylcellulose semisolid medium containing recombinant cytokines (MethoCult GF M3434; Stem Cell Technologies, Vancouver, BC, Canada). Cultures were incubated at 37°C in a 5% CO2 humidified atmosphere, and colonies were enumerated after 2 days for erythroid colony-forming units (CFUe's) and 10 days for erythroid burst-forming units (BFUe's) and granulocyte-macrophage colony-forming cells (GM-CFCs).
Long-term hematopoiesis repopulation assay
Unsorted total fetal liver cells (2 × 105 cells per mouse) were injected intravenously into lethally irradiated (9.5 Gy [950 rad], cesium irradiator) C57BL/6J recipient mice. Donor and host cells were distinguished by allelic expression of CD45 (Ly5.1+ host cells and Ly5.2+ donor cells). Eight weeks after transplantation, mice were killed. Cells from bone marrow, spleen, and thymus were immunostained and analyzed by flow cytometry to assess the different hematopoietic cell lineages.
Establishment of fetal liver stroma
Total cells from 13.5 days after coitus murine fetal liver (106/mL) were seeded in long-term culture medium (Myelocult M5300; Stem Cell Technologies) supplemented with 10–6 M hydrocortisone; half of the medium was changed on a weekly basis. After 4 weeks, cells were treated with trypsin and further cultured and expanded in Dulbecco modified Eagle medium (DMEM; GibcoBRL, Life Technologies, Cergy-Pontoise, France) supplemented with 20% FCS and 50 μM β-mercaptoethanol. The murine adult bone marrow cell line MS5 22 was cultured in α-MEM (GibcoBRL) supplemented with 10% FCS.
Cobblestone area-forming cell (CAFC) assays
For the cobblestone area-forming cell (CAFC) assay, stromal cells (2500 cells per well) were seeded in 96-well plates precoated with 0.1% (wt/vol) gelatin in long-term culture medium (Myelocult M5300; Stem Cell Technologies). The following day, CD45+ sorted murine fetal liver cells were seeded at various concentrations on the adherent layers in long-term culture medium. The number of cells ranged from 5 to 180 cells per well. Cultures were incubated for 5 weeks at 33°C in 5% CO2, with weekly half-medium changes. Wells containing CAFCs were scored after 5 weeks of coculture. CAFC frequency was estimated by plotting, on a logarithmic scale, the proportion of negative wells versus the number of cells per well. Poisson distribution of the CAFCs was verified, and the frequency of CAFCs in the starting CD45+ cell population was measured as being the inverse of the number of seeded cells corresponding to 37% negative wells.
Lentiviral vectors
TRIPΔU3-EF1α-Pitx2c-IRES-EGFP lentiviral vector was constructed from the original vector23 by inserting the entire coding sequence of Pitx2c between BamHI and XhoI restriction sites upstream of the internal ribosomal entry site (IRES) EGFP cassette. The murine Pitx2c coding sequence was amplified by PCR using the plasmid containing Pitx2c cDNA21 as template and the following forward and reverse primers: fwd 5′-CCGGGATCCCGCAGCGGTTCCCTC-3′ and rev 5′-CCGACGTCGACTCACACCGGCCGATCGACTGCA-3′. The PCR-generated fragments contained BamHI and SalI sites at their 5′ and 3′ ends (bold characters), respectively, and a silent mutation in the coding sequence was inserted in the reverse primer (G→A underlined) to allow digestion by BamHI and SalI and directional subcloning into the TRIPΔU3-EF1α-IRES-GFP vector, resulting in TRIPΔU3-EF1α-Pitx2c-IRES-GFP (Figure 5A).
The 19-nt siRNA sequence was chosen to be common to all Pitx2 mRNA isoforms and verified by basic local alignment search tool24 (BLAST) to ensure that it did not have significant sequence homology with other murine genomic sequences or with other genes particularly of the Pitx family. The strategy for constructing the Pitx2 short hairpin RNAi expression vector was to generate by PCR a DNA fragment with a 19-nt inverted repeat motif at both ends (underlined in the primer sequences) separated by a spacer of 12 nt and containing a BglII site at the 5′ end and a HindIII site at the 3′ end using the following primers: fwd 5′-TGCCGAGATCTCCAGCAGCACTCCAGCTTCGGCTGCGGCCGCTC-3′ and rev 5′-ATAGTCAAGCTTGAAAAAAGCAGCACTCCAGCTTCGGGAGCGGCCGCAG-3′, which hybridize to each other only by the 12-nt spacer sequence. The annealing temperature was 55°C, and 5 cycles of amplification were performed. The PCR fragment was then digested by BglII and HindIII and inserted into the same sites of the pH1 plasmid under the transcriptional control of the H1 promoter, resulting in the pH1 Pitx2RNAi plasmid. The final Pitx2 RNAi lentiviral vector was obtained by cloning the digested BamHI/XhoI fragment of the pH1 Pitx2RNAi (containing the H1 promoter followed by the Pitx2 RNAi sequence) between BamHI/XhoI sites of the TRIPΔU3 vector, resulting in the TRIPΔU3-H1Pitx2RNAi vector (Figure 5B). The amplified DNA sequences inserted within the constructs were sequenced in both directions. Vector particles were produced by transient calcium phosphate cotransfection of 293T cells, as previously described.23
Transduction of fetal liver stromal cells
Fetal liver cells were plated at 106/mL in long-term culture medium in 25 cm2 flasks precoated with 0.1% (wt/vol) gelatin. After a 4-hour incubation at 37°C in 5% CO2, lentiviral vector particles were added at a concentration of 2500 ng/mL of viral p24 (HIV-1 capsid protein) (for TRIPΔU3-EF1α-Pitx2c-IRES-EGFP and control TRIPΔU3-EF1α-IRES-EGFP vector). An identical amount of lentiviral vector particles was added 24 hours later, and the incubation was prolonged for a total of 72 hours. Cells were then washed, and fresh medium was added. The culture was then continued to establish stromal cell layers as described above.
Long-term culture assays of human CD34+ cord blood cells on the MS5 stromal cell line
Cord blood samples were collected with the informed consent of the mothers, according to approved institutional guidelines, in accordance with the Declaration of Helsinki. CD34+ cells (more than 80% purity) were purified from mononuclear cells by immunomagnetic bead selection (Miltenyi Biotec). Long-term culture-initiating cell (LTC-IC) assays were performed for 5 weeks in limiting dilution conditions in long-term culture medium consisting of α-MEM supplemented with 12.5% FCS, 12.5% horse serum, and 10–4 M β-mercaptoethanol as described25 on MS5 cell monolayers either untransduced or transduced with TRIPΔU3-H1Pitx2RNAi or control TRIPΔU3-EF1α-EGFP vectors (using doses of vectors corresponding to 500 to 1000 ng of viral p24 per milliliter).
Statistical analysis
Significance of the difference between groups was evaluated by analysis of variance, followed by a 2-tailed Student t test.
Results
Pitx2 gene expression in developing liver
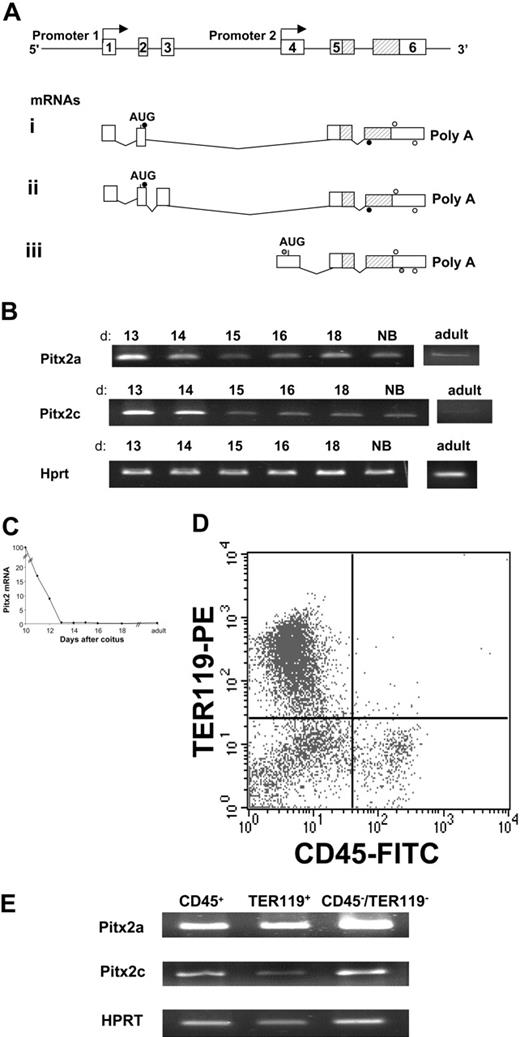
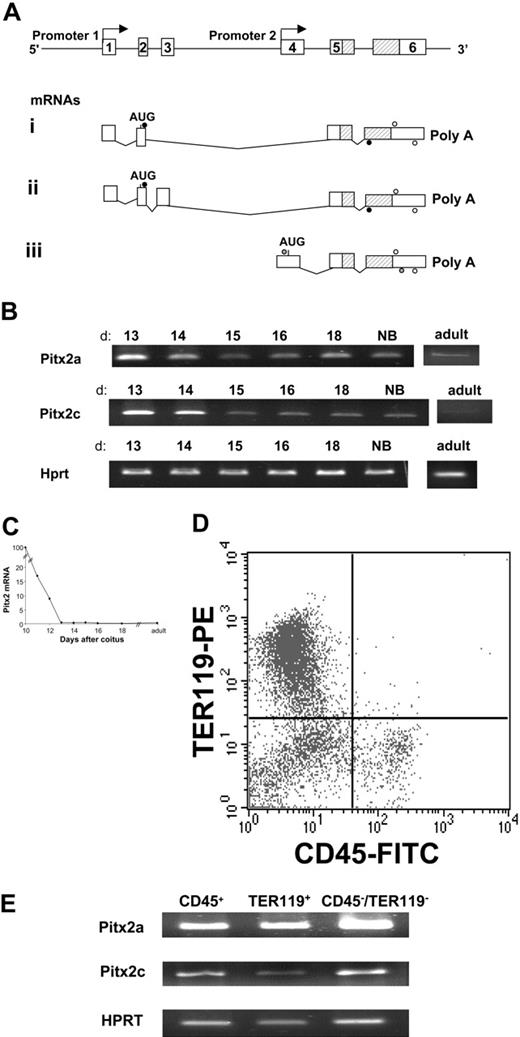
Because Pitx2 gene expression has been shown to be highly regulated in adult bone marrow,21 we decided to study its expression in fetal liver, the major site of hematopoiesis in the embryo after 12.5 days after coitus. We studied Pitx2 gene expression by RT-PCR analysis at different stages of normal fetal liver development. Total RNA was extracted from fetal livers at 10.5 to 18 days after coitus as well as from neonatal and normal adult mouse livers. Pitx2a-, Pitx2b-, and Pitx2c-specific primers were used for amplification by RT-PCR (Figure 1A). Pitx2 mRNAs a and c were expressed from 10.5 days after coitus to birth and in the adult liver (Figure 1B), and Pitx2b mRNA was inconsistently detected. Total Pitx2 mRNA expression, analyzed by quantitative RT-PCR (Figure 1C), decreased rapidly from 10 to 13 days after coitus and then remained stable at a low level until birth and during adult life. To distinguish the fetal liver cell populations that expressed the 3 Pitx2 isoforms, we studied their mRNA levels in fetal hematopoietic cells sorted on the basis of the cell surface expression of 2 hematopoietic markers (Figure 1D): CD45, which is expressed on all hematopoietic cells except erythroblasts, and the TER119 surface marker, which is expressed by erythroblasts. The remaining CD45–/TER119– cells represented the nonhematopoietic cell population. At 13.5 days after coitus, all 3 sorted cell populations expressed Pitx2a and Pitx2c mRNAs (Figure 1E), but more careful analysis, performed later by quantitative RT-PCR, showed that the Pitx2 mRNAs were mostly expressed in CD45–/TER119– cells: Pitx2 gene expression was only 20% in CD45+ cells and 1% in TER119+ cells of that of the double-negative cell population. This explains, by a dilution phenomenon, the dramatic decline in time of Pitx2 mRNAs in total fetal liver (Figure 1C) and especially between days 10 and 13 of gestation, days during which erythroblasts (TER119+) become the more prominent cell population of the fetal liver. These data showed that the Pitx2a and Pitx2c mRNA isoforms are expressed both in hematopoietic and nonhematopoietic cells in normal fetal liver and, thus, we studied these different cell populations in Pitx2-null embryos.
Phenotypic analysis of the fetal liver cell populations
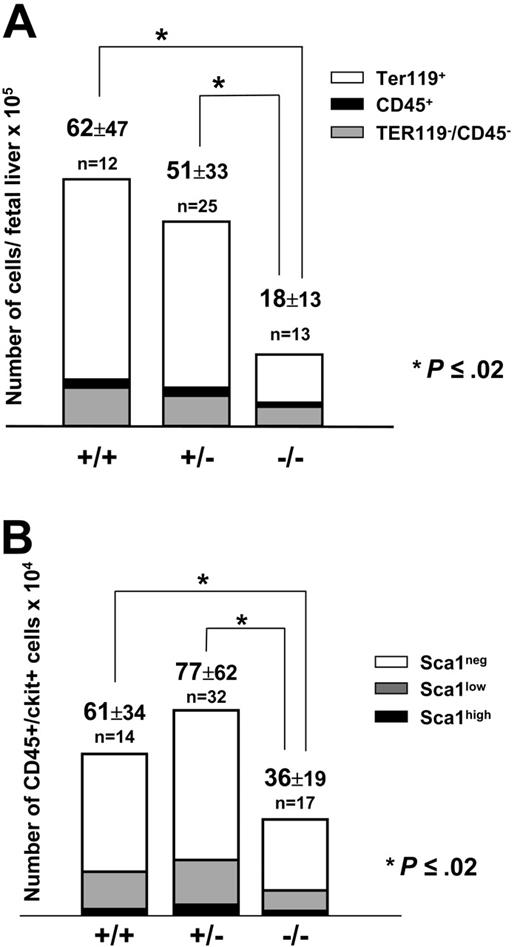
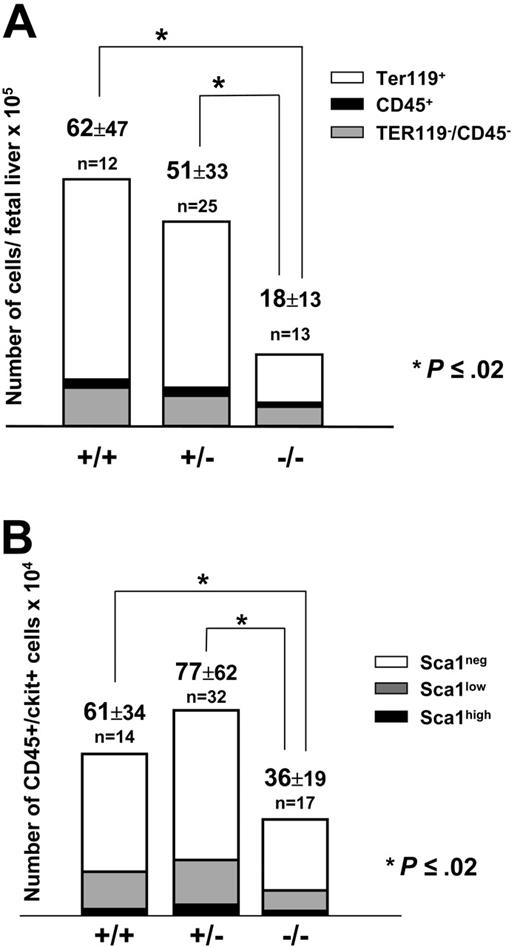
At 13.5 days after coitus, Pitx2–/– embryos had considerably smaller livers compared with their normal littermates (data not shown). Total fetal liver cell counts were 18 × 105 ± 13 × 105 (n = 13) for Pitx2–/– embryos versus 62 × 105 ± 47 × 105 (n = 12; P < .02) for Pitx2+/+ embryos (Figure 2A). Cell counts of Pitx2+/– fetal livers (51 × 105 ± 33 × 105; n = 25) were not statistically different from Pitx2+/+ fetal livers (Figure 2A). We performed a phenotypic analysis of embryonic fetal liver cells based on their expression of the CD45 and TER119 hematopoietic markers. Quantification by FACS analysis of the 3 cell populations showed that they were all reduced in absolute numbers in Pitx2–/– compared with Pitx2+/+ or Pitx2+/– fetal livers (Table 1; Figure 2A). The CD45+ and CD45–/TER119– cell populations were reduced by half, whereas there was a 4-fold reduction of the TER119+ erythroid cell population.
Structure of the Pitx2 gene and its mRNAs and Pitx2 gene expression in normal murine fetal liver. (A) Genomic organization of the Pitx2 gene. Exon sequences are represented by white boxes and are numbered. The position of the homeodomain coding sequences is indicated (hatched bars). Pitx2a and Pitx2b mRNAs are transcribed using promoter 1 and generated by alternative splicing. Pitx2c mRNA is transcribed using internal promoter 2. For RT-PCR, Pitx2a and Pitx2b primers (•) amplify 274-bp and 412-bp products, respectively. Pitx2c primers (gray circles) amplify a 535 bp product and, for real-time RT-PCR. Pitx2 primers (○) amplify a 233-bp product common to all Pitx2 mRNA isoforms. (B) RT-PCR analysis of Pitx2a and Pitx2c mRNA isoforms during normal fetal liver development from 13 days after coitus to birth (NB) and adult life. d indicates days after coitus. (C) Evolution of Pitx2 mRNA accumulation during normal fetal liver development, from 10 days after coitus to adult life, studied by quantitative real-time RT-PCR amplifying all isoforms of Pitx2 mRNA (see panel A). Results are expressed as the percentage of the Pitx2 mRNA level detected in the fetal liver at 10 days after coitus. (D) FACS analysis of normal 13.5 days after coitus fetal liver cells stained by anti-CD45– FITC (panhematopoietic except erythroblasts) and anti-TER119–PE (late erythroid differentiation marker) antibodies. (E) RT-PCR analysis of Pitx2a and Pitx2c isoform expression in the 3 populations seen in panel D after cell sorting.
Structure of the Pitx2 gene and its mRNAs and Pitx2 gene expression in normal murine fetal liver. (A) Genomic organization of the Pitx2 gene. Exon sequences are represented by white boxes and are numbered. The position of the homeodomain coding sequences is indicated (hatched bars). Pitx2a and Pitx2b mRNAs are transcribed using promoter 1 and generated by alternative splicing. Pitx2c mRNA is transcribed using internal promoter 2. For RT-PCR, Pitx2a and Pitx2b primers (•) amplify 274-bp and 412-bp products, respectively. Pitx2c primers (gray circles) amplify a 535 bp product and, for real-time RT-PCR. Pitx2 primers (○) amplify a 233-bp product common to all Pitx2 mRNA isoforms. (B) RT-PCR analysis of Pitx2a and Pitx2c mRNA isoforms during normal fetal liver development from 13 days after coitus to birth (NB) and adult life. d indicates days after coitus. (C) Evolution of Pitx2 mRNA accumulation during normal fetal liver development, from 10 days after coitus to adult life, studied by quantitative real-time RT-PCR amplifying all isoforms of Pitx2 mRNA (see panel A). Results are expressed as the percentage of the Pitx2 mRNA level detected in the fetal liver at 10 days after coitus. (D) FACS analysis of normal 13.5 days after coitus fetal liver cells stained by anti-CD45– FITC (panhematopoietic except erythroblasts) and anti-TER119–PE (late erythroid differentiation marker) antibodies. (E) RT-PCR analysis of Pitx2a and Pitx2c isoform expression in the 3 populations seen in panel D after cell sorting.
We then quantified the hematopoietic progenitors present in the fetal liver based on the expression of both the CD45 and c-Kit surface markers and distinguished 3 subpopulations of progenitors based on the expression of the Sca1 marker: CD45+/c-Kit+/Sca1neg, CD45+/c-Kit+/Sca1low, and CD45+/c-Kit+/Sca1high cells. These cell populations corresponded respectively to monopotent, bipotent, and multipotent progenitors.26 As shown in Figure 2B, the global progenitor cell population was reduced by half in Pitx2–/– fetal livers compared with Pitx2+/+ or Pitx2+/– fetal livers. The Sca1neg, Sca1low, and Sca1high progenitor cell populations were also reduced to the same extent in Pitx2–/– fetal livers. This reduction was statistically significant for Sca1neg and Sca1low progenitor cell populations (P ≤ .02 and P < .001, respectively). However, more likely due to the small size of this population, statistical significance was not reached for this same 2-fold reduction of the Sca1high immature progenitor cell population (P ≤ .003 between Pitx2+/– and Pitx2–/– cell populations but P ≤ .08 between Pitx2+/+ and Pitx2–/– cell populations).
In conclusion, the absence of Pitx2 expression resulted in a quantitative deficit of immature and mature hematopoietic cells in the fetal liver.
Numeration and phenotypic analysis of fetal liver cells at 13.5 days after coitus.(A) Histograms show the absolute number of cells from the total liver. Total cell number (× 105 ± SEM) is indicated at the top of the histograms for Pitx2+/+, Pitx2+/–, and Pitx2–/– fetal livers. Each bar of the histogram is divided in 3 parts, according to the content in each sample of the 3 different cell populations: TER119+ erythroid cells (white), CD45+ hematopoietic cells (black), and CD45–/TER119– nonhematopoietic cells (gray). (B) Histograms show the absolute number of progenitor cells in normal and mutant fetal livers. Total CD45+/c-Kit+ cell number (× 104 ± SEM) is indicated at the top of the histograms for Pitx2+/+, Pitx2+/–, and Pitx2–/– fetal livers. Each bar of the histogram is divided in 3 parts, according to the content in each sample of the 3 different cell populations analyzed: Sca1high (black), Sca1low (gray), and Sca1neg (white). For both panels, the results are shown as the means of n experiments. *Statistically significant differences (P = .02 by Student t test) between total populations.
Numeration and phenotypic analysis of fetal liver cells at 13.5 days after coitus.(A) Histograms show the absolute number of cells from the total liver. Total cell number (× 105 ± SEM) is indicated at the top of the histograms for Pitx2+/+, Pitx2+/–, and Pitx2–/– fetal livers. Each bar of the histogram is divided in 3 parts, according to the content in each sample of the 3 different cell populations: TER119+ erythroid cells (white), CD45+ hematopoietic cells (black), and CD45–/TER119– nonhematopoietic cells (gray). (B) Histograms show the absolute number of progenitor cells in normal and mutant fetal livers. Total CD45+/c-Kit+ cell number (× 104 ± SEM) is indicated at the top of the histograms for Pitx2+/+, Pitx2+/–, and Pitx2–/– fetal livers. Each bar of the histogram is divided in 3 parts, according to the content in each sample of the 3 different cell populations analyzed: Sca1high (black), Sca1low (gray), and Sca1neg (white). For both panels, the results are shown as the means of n experiments. *Statistically significant differences (P = .02 by Student t test) between total populations.
Hematopoietic potential of Pitx2–/– immature hematopoietic cells
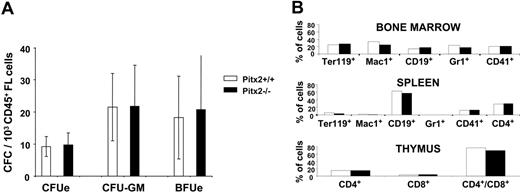
To precisely define the nature of the progenitors present in the fetal livers of Pitx2–/– embryos, we studied their hematopoietic potential in vitro and in vivo. We first performed CFC assays using CD45+ sorted hematopoietic cells of 13.5 days after coitus Pitx2+/+ and Pitx2–/– fetal livers. For identical numbers of CD45+ cells seeded, the number, size, morphology, and types of the colonies obtained were identical whatever the Pitx2 genotype (+/+ or –/–) of the fetal liver cells (Figure 3A). This result indicated that, while the absolute number of CFCs is 2-fold reduced in the Pitx2–/– compared with Pitx2+/+ fetal liver, the clonogenic potential of Pitx2–/– CFCs was normal. The quantitative defect of hematopoietic progenitors could also be accounted for by a reduced size of the hematopoietic stem cell (HSC) pool or by impaired differentiation of HSCs into clonogenic progenitors. To examine HSC function, we performed hematopoietic reconstitution using lethally irradiated mouse recipients. Two doses (1 × 106 or 2 × 105) of total fetal liver cells of each genotype were transplanted into lethally irradiated Ly5.1 C57BL/6J recipient mice. After 2 months, mice receiving transplants were killed and bone marrow cells were analyzed by flow cytometry for the level of donor versus recipient hematopoietic cell reconstitution. Whatever the dose or genotype (Pitx2+/+ or Pitx2–/–) of engrafted fetal liver cells, 100% of the hematopoietic cells in all of the mice receiving transplants were of donor origin based on their Ly5.1/Ly5.2 phenotype (data not shown). Furthermore, Pitx2–/– HSCs had the same capacity as Pitx2+/+ cells to give rise to all of the hematopoietic lineages in the bone marrow, thymus, and spleen of recipient mice as determined by FACS analysis (Figure 3B). These results demonstrate that the Pitx2–/– HSCs have a normal potential for hematopoietic reconstitution and differentiation in normal proportions for the different lineages. Secondary transplantations of Pitx2–/– and Pitx2+/+ bone marrow cells gave rise to the same number of CFU-S12d (colony-forming unit spleen of day 12) (data not shown), further indicating that Pitx2–/– HSCs had a normal proliferative potential.
In summary, Pitx2–/– fetal livers were severely hypoplastic and showed reduced numbers of hematopoietic cells at various stages of differentiation, but these hematopoietic cells retained normal intrinsic hematopoietic potential in vitro and in vivo. These results suggested a nonautonomous cell defect in Pitx2–/– hematopoietic fetal liver cells and, thus, we studied the hematopoietic supportive capacity of the microenvironment in Pitx2–/– and Pitx2+/+ fetal livers.
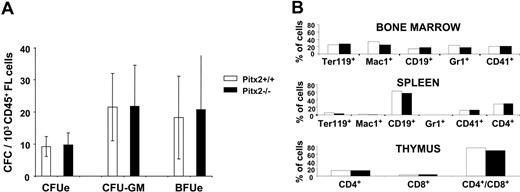
Analysis of hematopoiesis in Pitx2+/+ and Pitx2–/– fetal liver at 13.5 days after coitus. (A) Histograms show the number of CFCs per 1 × 103 CD45+ Pitx2+/+ (□) and Pitx2–/– (▪) fetal liver cells at 13.5 days after coitus. Results represent the mean ± SEM of 4 separate experiments. (B) Multilineage differentiation of donor-derived hematopoietic cells in the bone marrow, thymus, and spleen cells of mice receiving transplants with Pitx2+/+ (□) and Pitx2–/– (▪) fetal liver cells. Differentiation marker expression was studied using antibodies specific for mouse antigens TER119 (erythroid cells), Mac1 (CD11-b, macrophages), CD19 (B cells), Gr1 (Ly-6G, granulocytes), CD41 (megakaryocytic cells), and CD4 and CD8 (T cells).
Analysis of hematopoiesis in Pitx2+/+ and Pitx2–/– fetal liver at 13.5 days after coitus. (A) Histograms show the number of CFCs per 1 × 103 CD45+ Pitx2+/+ (□) and Pitx2–/– (▪) fetal liver cells at 13.5 days after coitus. Results represent the mean ± SEM of 4 separate experiments. (B) Multilineage differentiation of donor-derived hematopoietic cells in the bone marrow, thymus, and spleen cells of mice receiving transplants with Pitx2+/+ (□) and Pitx2–/– (▪) fetal liver cells. Differentiation marker expression was studied using antibodies specific for mouse antigens TER119 (erythroid cells), Mac1 (CD11-b, macrophages), CD19 (B cells), Gr1 (Ly-6G, granulocytes), CD41 (megakaryocytic cells), and CD4 and CD8 (T cells).
Deficient capacity of Pitx2–/– fetal liver stromal cells to support hematopoiesis
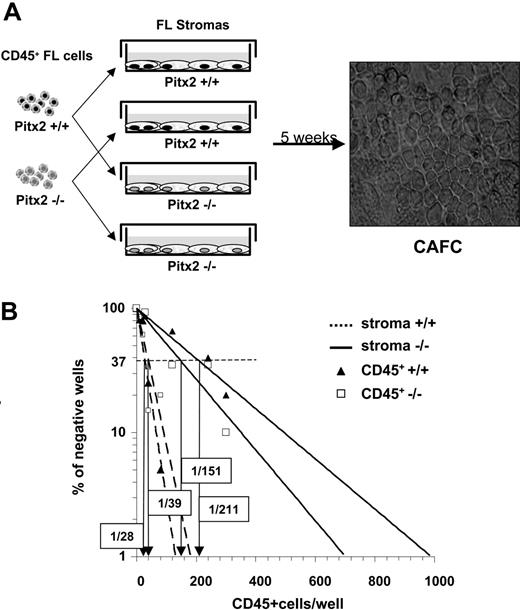
To evaluate the hematopoietic supportive ability of the Pitx2–/– fetal liver, we established primary stromal cell cultures using adherent cells of fetal livers from 13.5 days after coitus embryos of Pitx2–/– and Pitx2+/+ genotypes. As expected from the PCR analysis described in Figure 1D, Pitx2+/+ primary stromal cells expressed a and c isoforms of Pitx2 mRNAs (data not shown). No striking difference was observed during the establishment of the stromal cell layers using cells from either genotype. We then performed quantitative CAFC assays by establishing long-term cocultures between various numbers of CD45+ fetal liver cells of both Pitx2–/– and Pitx2+/+ genotypes and fetal liver stromal cell layers of the 2 Pitx2 genotypes (Figure 4A). CAFCs were enumerated after 5 weeks of coculture, and the frequency of CAFCs was calculated as indicated in “Materials and methods.” The frequency of CAFCs using Pitx2+/+ or Pitx2–/– CD45+ cells was identical when Pitx2+/+ stromal cells were used (Figure 4B; Table 2). However, for CD45+ cells of both genotypes, the frequency obtained after culture on Pitx2–/– stromal cells was more than 4-fold decreased compared with that obtained with Pitx2+/+ stromal cells. Thus, the Pitx2–/– stromal cells exhibited defective hematopoietic supportive capacity.
Pitx2 is directly involved in the hematopoietic supportive ability of primary fetal liver stromal cells
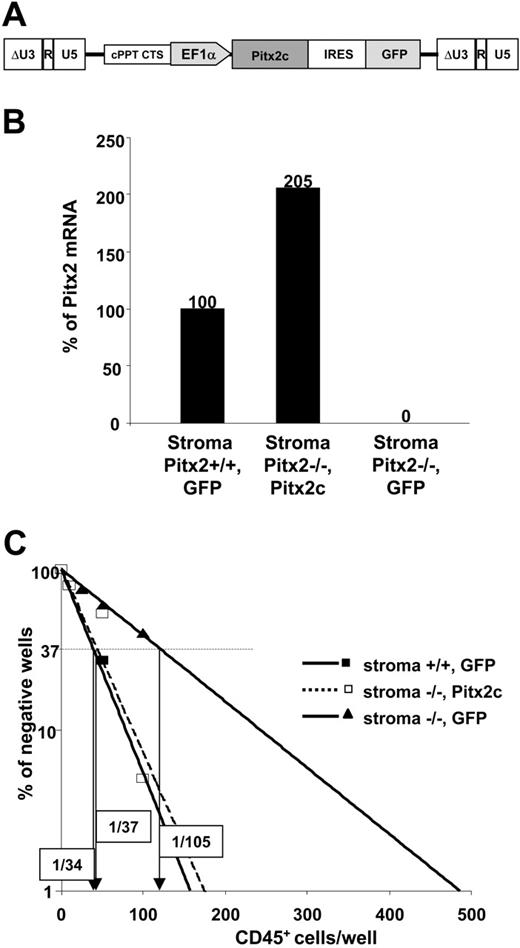
To demonstrate the direct role of Pitx2 in the hematopoietic supportive ability of primary fetal liver stromal cells, we transduced Pitx2–/– fetal liver cells with lentiviral vectors expressing either Pitx2c (Figure 5A) or EGFP mRNAs. Stromal cell layers were established from the transduced cells and designated as (Pitx2–/–, Pitx2c) and (Pitx2–/–, GFP), respectively.
Pitx2 mRNA expression in the different established stromal cell layers was studied by real-time PCR analysis using 2 primers that amplified all Pitx2 mRNA isoforms (Figure 1A). As expected, no Pitx2 mRNA was found in (Pitx2–/–, GFP) stromal cells, while the level of Pitx2 mRNA expression in (Pitx2–/–, Pitx2c) stromal cells was 2-fold higher than in Pitx2+/+ stromal cells (Figure 5B). We then performed quantitative CAFC assays using long-term cocultures between Pitx2+/+ CD45+ fetal liver cells and these (Pitx2–/–, GFP) and (Pitx2–/–, Pitx2c) fetal liver stromal cells. As shown in Figure 5C, expression of Pitx2c resulted in a total reversion of the hematopoietic supportive ability of the (Pitx2–/–, Pitx2c) stromal cells compared with (Pitx2–/–, GFP) stromal cells.
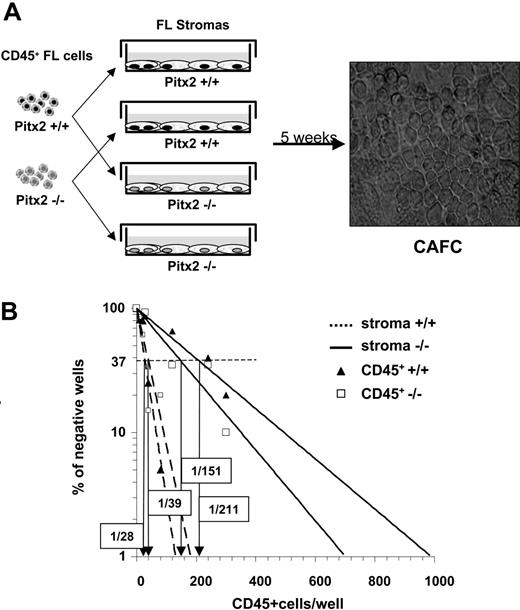
Comparative study of the hematopoietic supportive ability of Pitx2–/– and Pitx2+/+ fetal liver stromal cells. (A) Schematic diagram illustrating CAFC assays using cocultures between preestablished fetal liver stromal cells and CD45+ hematopoietic fetal liver cells and microscopic appearance of CAFCs when they are enumerated after 5 weeks of coculture. Phase-contrast image was obtained using a Nikon Eclipse TE2000S inverted microscope (Nikon, Champigny sur Marne, France) and a Nikon Plan Fluor 20×/0.50 numeric aperture objective, for a total magnification of 200×. Image was captured using a Nikon DXM1200F digital camera and Nikon EclipseNet software. (B) Results of 1 of 3 representative (Table 2, experiment 3) quantitative CAFC experiments generated using Pitx2–/– (solid lines) and Pitx2+/+ (broken lines) fetal liver stromal cells after coculture with Pitx2+/+ (▴) and Pitx2–/– (□) CD45+ hematopoietic cells, respectively. The frequency of wells negative for the presence of CAFCs is plotted against the number of CD45+ cells seeded. The CAFC frequency (indicated in boxes) is defined as the inverse of the number of seeded cells that corresponds to 37% negative wells (Poisson statistics27 ).
Comparative study of the hematopoietic supportive ability of Pitx2–/– and Pitx2+/+ fetal liver stromal cells. (A) Schematic diagram illustrating CAFC assays using cocultures between preestablished fetal liver stromal cells and CD45+ hematopoietic fetal liver cells and microscopic appearance of CAFCs when they are enumerated after 5 weeks of coculture. Phase-contrast image was obtained using a Nikon Eclipse TE2000S inverted microscope (Nikon, Champigny sur Marne, France) and a Nikon Plan Fluor 20×/0.50 numeric aperture objective, for a total magnification of 200×. Image was captured using a Nikon DXM1200F digital camera and Nikon EclipseNet software. (B) Results of 1 of 3 representative (Table 2, experiment 3) quantitative CAFC experiments generated using Pitx2–/– (solid lines) and Pitx2+/+ (broken lines) fetal liver stromal cells after coculture with Pitx2+/+ (▴) and Pitx2–/– (□) CD45+ hematopoietic cells, respectively. The frequency of wells negative for the presence of CAFCs is plotted against the number of CD45+ cells seeded. The CAFC frequency (indicated in boxes) is defined as the inverse of the number of seeded cells that corresponds to 37% negative wells (Poisson statistics27 ).
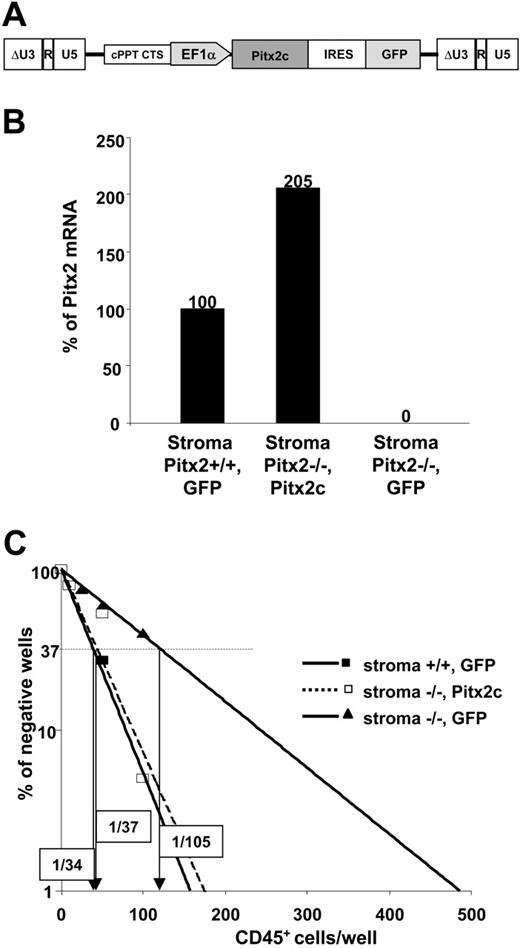
Comparative study of the hematopoietic supportive ability of primary fetal liver stromal cells expressing or not Pitx2 mRNAs. (A) Schematic representation of the functional elements of the TRIPΔU3-EF1α-Pitx2c-IRES-GFP vector. (B) Quantitative RT-PCR was performed on (Pitx2–/–, Pitx2c), (Pitx2–/–, GFP), and control (Pitx2+/+, GFP) stromal cells. Histograms show the results expressed as the percentage of Pitx2 mRNA expression obtained in the control stromal cells. The Hprt housekeeping gene was used as internal standard to normalize the results for cDNA input. All samples were run in duplicate and gave identical results. (C) Results of 1 of 2 representative quantitative CAFC assays performed by coculture of Pitx2+/+ CD45+ fetal liver hematopoietic cells using the different stromal cells described in panel A: Pitx2–/–, GFP (solid lines, ▴); Pitx2–/–, Pitx2c (broken lines, □); and Pitx2+/+, GFP (solid lines, ▪). The frequency of wells negative for the presence of CAFCs is plotted against the number of CD45+ cells seeded. The CAFC frequency, indicated in boxes for each condition, is defined as the inverse of the number of seeded cells that corresponds to 37% negative wells (Poisson statistics27 ).
Comparative study of the hematopoietic supportive ability of primary fetal liver stromal cells expressing or not Pitx2 mRNAs. (A) Schematic representation of the functional elements of the TRIPΔU3-EF1α-Pitx2c-IRES-GFP vector. (B) Quantitative RT-PCR was performed on (Pitx2–/–, Pitx2c), (Pitx2–/–, GFP), and control (Pitx2+/+, GFP) stromal cells. Histograms show the results expressed as the percentage of Pitx2 mRNA expression obtained in the control stromal cells. The Hprt housekeeping gene was used as internal standard to normalize the results for cDNA input. All samples were run in duplicate and gave identical results. (C) Results of 1 of 2 representative quantitative CAFC assays performed by coculture of Pitx2+/+ CD45+ fetal liver hematopoietic cells using the different stromal cells described in panel A: Pitx2–/–, GFP (solid lines, ▴); Pitx2–/–, Pitx2c (broken lines, □); and Pitx2+/+, GFP (solid lines, ▪). The frequency of wells negative for the presence of CAFCs is plotted against the number of CD45+ cells seeded. The CAFC frequency, indicated in boxes for each condition, is defined as the inverse of the number of seeded cells that corresponds to 37% negative wells (Poisson statistics27 ).
These results indicated that the quantitative defect of fetal liver hematopoiesis in Pitx2–/– embryos might be explained by a failure of the hematopoietic supportive capacity of the fetal liver rather than a cell-autonomous defect of hematopoietic cells. Therefore, Pitx2 is an important molecular component of the murine fetal liver stroma involved in the hematopoietic supportive capacity of this organ during development.
Pitx2 is also involved in the hematopoietic supportive ability of MS5 stromal cells for primitive human hematopoietic cells
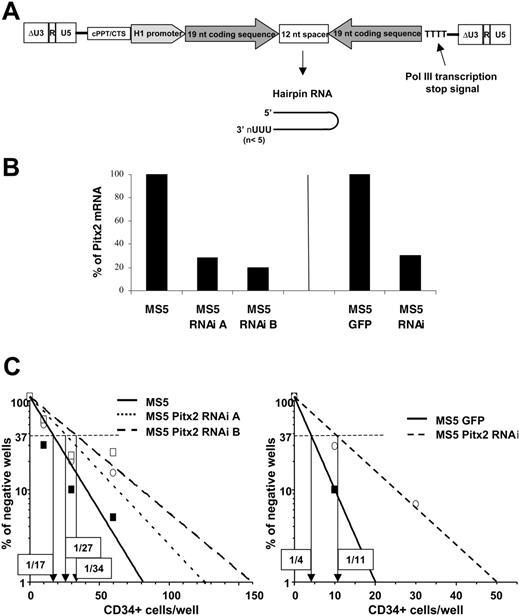
Having demonstrated the important role of Pitx2 in the hematopoietic supportive capacity of murine fetal liver stroma, we examined whether Pitx2 also has a role in the hematopoietic support of adult bone marrow stroma and if this is true for human as well as for murine hematopoiesis. Thus, we transduced the MS5 stromal cell line, which is known for its supportive ability of both murine and human hematopoiesis, with a lentiviral vector expressing an RNAi directed against Pitx2 mRNAs (Figure 6A) to establish 2 different batches of MS5 cells designated as Pitx2 RNAi A and B. Using real-time PCR, the expression level of Pitx2 was found to be decreased to 28% and 20% of that of MS5 control cells in MS5 Pitx2 RNAi batches A and B, respectively (Figure 6B). Quantitative LTC-IC assays using human CD34+ cord blood cells cultured on the MS5 Pitx2 RNAi A and B cells showed that the frequency of LTC-ICs obtained was decreased up to 2-fold when the Pitx2 mRNA level decreased to 20% of the level observed in control cells (Figure 6C).
These results indicate that the role of Pitx2, as an important molecular component of the hematopoietic microenvironment, extends to the human hematopoietic supportive capacity of the adult bone marrow stromal cell line MS5.
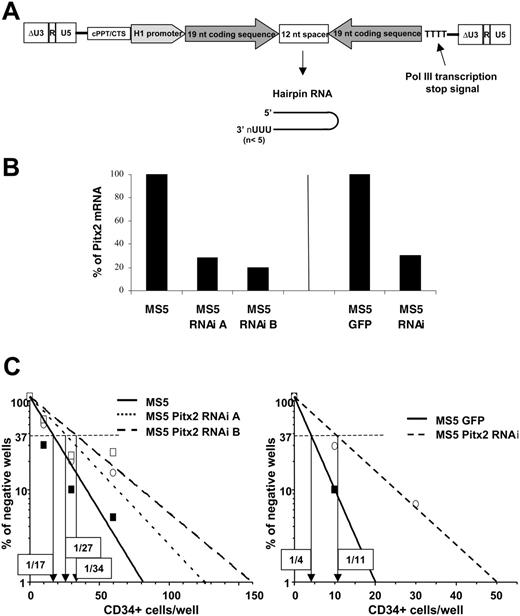
Comparative study of the human hematopoietic supportive ability of control MS5 cells and MS5 cells transduced with vectors expressing RNAi directed against Pitx2 mRNA. (A) Schematic representation of the TRIP ΔU3-H1Pitx2RNAi vector. (B) Quantitative RT-PCR was performed using RNA isolated from control MS5 or MS5 GFP cells and MS5 Pitx2RNAi cells in 3 independent transduction experiments. Histograms show the results expressed as the percentage of Pitx2 mRNA expression in control cells, MS5 (left), and MS5 GFP (right). The Hprt housekeeping gene was used as internal standard to normalize cDNA input. All samples were run in duplicate and gave identical results. (C) Results of representative quantitative LTC-IC assays performed by coculture of human CD34+ cord blood cells using the different MS5 cells described in panel B. The frequency of wells negative for the presence of LTC-ICs is plotted against the number of CD34+ cells seeded, and the LTC-IC frequency, indicated in boxes for each condition, is defined as the inverse of the number of seeded cells that corresponds to 37% negative wells (Poisson statistics27 ).
Comparative study of the human hematopoietic supportive ability of control MS5 cells and MS5 cells transduced with vectors expressing RNAi directed against Pitx2 mRNA. (A) Schematic representation of the TRIP ΔU3-H1Pitx2RNAi vector. (B) Quantitative RT-PCR was performed using RNA isolated from control MS5 or MS5 GFP cells and MS5 Pitx2RNAi cells in 3 independent transduction experiments. Histograms show the results expressed as the percentage of Pitx2 mRNA expression in control cells, MS5 (left), and MS5 GFP (right). The Hprt housekeeping gene was used as internal standard to normalize cDNA input. All samples were run in duplicate and gave identical results. (C) Results of representative quantitative LTC-IC assays performed by coculture of human CD34+ cord blood cells using the different MS5 cells described in panel B. The frequency of wells negative for the presence of LTC-ICs is plotted against the number of CD34+ cells seeded, and the LTC-IC frequency, indicated in boxes for each condition, is defined as the inverse of the number of seeded cells that corresponds to 37% negative wells (Poisson statistics27 ).
Discussion
Here we show that Pitx2, a member of the bicoid family of homeodomain genes, is required for optimal proliferation of definitive hematopoietic cells during fetal liver organogenesis. Pitx2-null embryos have hypoplastic livers with a quantitative defect in nonhematopoietic and hematopoietic cells, particularly affecting the number of erythroblasts. Nevertheless, the quantitative defect of hematopoietic cells seems to be cell nonautonomous because all of the assays of hematopoietic progenitor and stem cell function that we have performed in vitro and in vivo, although not conclusive concerning the size of the HSC pool, gave normal results. Moreover, in a forthcoming paper by Zhang et al,28 further evidence is provided supporting the notion that the defect in hematopoietic fetal liver cells may result from a cell-nonautonomous mechanism. Indeed, they show that chimera mice obtained using Pitx2–/– ES cells have a bone marrow hematopoietic chimerism identical to that of the coat color. Moreover, the percentage of Pitx2–/– hematopoietic cells of all lineages remained stable during 14 months in the chimeric mice, suggesting that the self-renewal ability of Pitx2–/– HSCs seems normal. They also show results of fetal liver cell transplantation that achieved long-term multilineage hematopoietic reconstitution in lethally irradiated mice. However, all of these results do not formally exclude a slight defect in the self-renewal ability of Pitx2–/– HSCs in the fetal liver that could have been demonstrated by serial transplantation experiments in competitive conditions or after hematopoietic stress, as was shown in the case of p21 knock-out hematopoietic cells.29 The absence of a cell-autonomous defect in Pitx2–/– hematopoietic cells, if confirmed, could be explained by a possible redundancy with other homeodomain proteins expressed in these cells.
Nevertheless, we demonstrated a defect in the ability of Pitx2–/– stromal cells to support hematopoiesis, as revealed by quantitative CAFC assays showing a reduced number of CAFCs obtained with Pitx2–/– versus Pitx2+/+ stromal cell layers. Furthermore, using a Pitx2 overexpression vector, we showed that Pitx2 is directly involved in the hematopoietic supportive ability of primary fetal liver stromal cells. Finally, we showed that the role played by Pitx2 in the adult bone marrow stromal cell line MS5 is also crucial for optimal support of human hematopoiesis.
A few known transcription factors have been implicated as playing a role in providing a normal fetal liver hematopoietic supportive microenvironment. These include Xbp1,30 c-Myb,31 and Hlx.32 The fetal livers of knock-out embryos for each of these 3 genes show defects in fetal liver hematopoiesis similar to that seen in Pitx2 knock-out embryos (ie, hypoplastic fetal livers) and reduced numbers of hematopoietic cells. The decrease in hematopoietic cell number is attributed in these cases to a defect in the hematopoietic microenvironment (defect of survival, proliferation or differentiation of hepatoblasts, or defect of cytokine production by stromal cells). Recent results in the literature are consistent with the notion that Pitx2 plays a role in the stroma. A functional genomics approach has shown that Pitx2 mRNA is one of the mRNAs that is preferentially expressed by a stem cell supportive stromal cell line AFT024 compared with nonsupportive cell line 2018.33 The microenvironment supports and regulates the growth, self-renewal, and differentiation of hematopoietic cells. The hematopoietic microenvironment of fetal liver consists of a mixed population of cells, designated as stromal cells, that contains endothelial cells,34,35 macrophages, hepatoblasts,36 vascular smooth muscle cells (VSMCs),37 and primitive biliary epithelial cells.38 Numerous signals for the regulation of hematopoiesis are provided by the microenvironment. These signals are generated by multicomponent molecular networks consisting of adhesion molecules (integrins, selectins, sialomucins, and members of the immunoglobulin superfamily), extracellular matrix molecules, and cytokines (soluble, membrane-bound, or bound to the extracellular matrix).
A number of nonexclusive hypotheses can be proposed for the abnormal function of Pitx2–/– stroma. It could be due to a deficit in the expression of growth factors essential for hematopoiesis, as has been described in c-Myb knock-out fetal liver stroma, which underexpresses SCF,31 as well as in IL-6 knock-out fetal liver stroma.39 There could also be a defect in the expression or function of cell adhesion molecules implicated in the hematopoietic supportive capacity of stromal cells. For example, VCAM-1 interacts with VLA-4 (α4β1), an integrin expressed on HSCs, CAFCs, LTC-ICs, and CFCs.40-42 In addition, a high level of expression of the stem cell antigen Sca1 is correlated with hematopoietic supportive activity of a murine bone marrow stromal cell line.43 Pitx2 could play a role in the regulation of such molecular components of the microenvironment that are important for the support of hematopoiesis. Another hypothesis is that Pitx2 could play a role in the cellular biology of the microenvironment by regulating the proliferation of a particular type of stromal cell essential for hematopoietic supportive activity. Recently, it was reported that Pitx2 is induced by the Wnt/Dvl/β-catenin pathway and is required for the effective cell type–specific proliferation of a cell population in the developing heart by directly activating specific growth-regulating genes such as cyclin D1, cyclin D2, and Myc.44,45 Moreover, β-catenin regulates cell proliferation in developing murine and chicken liver.46-48 Conversely, Pitx2 can also inhibit cell proliferation.49
However, no difference was observed in the expression, studied by FACS analysis, of several surface markers known to be expressed by hematopoietic cell supportive cell lines (Sca1, Thy 1, CD44, integrin α 5 and 6, β 1, and VCAM-1)50 (data not shown). Again, we could not observe any difference in the proliferation of cells during the establishment of Pitx2–/– versus Pitx2+/+ fetal liver stroma. This was confirmed by the quantitative study, by real-time quantitative PCR analysis, of the expression level of cyclins D1 and D2 and c-Myc that gave identical results for the 2 types of stroma (data not shown).
In conclusion, our results highlight the importance of Pitx2 in the hematopoietic supportive function of stromal cells. The study of downstream target genes of this transcription factor constitutes a novel approach to identify new genes involved in the regulation of hematopoiesis by the microenvironment.
Prepublished online as Blood First Edition Paper, September 29, 2005; DOI 10.1182/blood-2005-02-0529.
Supported in part by grants from INSERM and the United States Public Health Service (USPHS) National Institutes of Health (HL56920, HL63357, TWO2296). A.K. was the recipient of successive fellowships from the French Ministère de la Recherche et des Nouvelles Technologies, the Fondation de la Recherche Médicale (FRM), and the Société Française d'Hématologie (SFH). P.-E.M. was the recipient of a fellowship from the Association Française contre les Myopathies (AFM).
A.K. performed most of the experimental work and analysis of data and wrote the paper; J.C. and C.K. contributed to experimental work and analyzed data; P.-E.M. and P.J.G. contributed vital reagents; N.N. and B.I. contributed to experimental work; G.U. provided analytical tools; B.G.F. provided vital reagents, initiated the work, and edited the paper; and A.D.-K. designed research, performed experiments, analyzed data, and wrote the paper.
J.C. and C.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank P. Charbord for helpful advice and discussions. The authors are grateful to C. Dez and the staff of the animal facilities of the IM3 in Creteil and of the INSERM U506 in Villejuif for the care of our mice. A.D.-K. warmly acknowledges M. Cailleret for his excellent technical contribution at the beginning of this project. We are indebted to Dr Y. Rouquet and the midwives of the “Clinique des Noriets” (Vitry, France) for providing cord blood samples. We sincerely thank P. H. Roméo and Sylvie Gisselbrecht for critically reading the manuscript and all the members of the “TOP team” for their support during this work.