Abstract
The granulocyte colony-stimulating factor receptor (G-CSF-R) transmits signals for proliferation and differentiation of myeloid progenitor cells. Here we report on the identification of a rare single nucleotide polymorphism within its intracellular domain (G-CSF-R_Glu785Lys). Screening a cohort of 116 patients with primary myelodysplastic syndromes (MDS), de novo acute myeloid leukemia (AML) (84 patients), as well as 232 age- and sex-matched controls revealed a highly significant association of the G-CSF-R_785Lys allele with the development of high-risk MDS as defined by more than 5% bone marrow blasts (9.7% versus 0.9% in controls; P = .001; odds ratio [OR], 12.5; 95% confidence interval [CI], 2.4-58.9) or an International Prognostic Scoring System score of intermediate-2 or high (13.0% versus 0.9%; P < .001; OR, 14.0; 95% CI, 3.4-85.0). Functional analysis by retroviral transfer of G-CSF-R_785Lys into myeloid progenitor cells of G-CSF-R–deficient mice showed a significantly diminished colony-formation capacity after G-CSF stimulation as compared with cells transduced with the wild-type receptor. These results suggest that lifelong altered G-CSF response by the G-CSF-R_785Lys may render individuals susceptible to development of high-risk MDS.
Introduction
The myelodysplastic syndromes (MDS) are clonal stem-cell disorders characterized by dysplastic and ineffective hematopoiesis and a variable risk of transformation to acute myeloid leukemia (AML). Disease outcome ranges from an indolent course that may span years to a rapid leukemic progression within several months.1,2 The etiology of MDS is unknown in most patients, although the risk of the development of MDS is increased among individuals who have been exposed to benzene, halogenated organics, metals, and petrol compounds or have received radiation or chemotherapy for a primary, most often malignant disease.2-4 MDS emerging after known exposure to such reagents or chemotherapy is defined as “secondary” or “therapy-related” MDS (t-MDS). A genetic predisposition has been implicated in the etiology of MDS5 because polymorphisms in metabolic activation or detoxification enzymes such as nicotinamide adenine dinucleotide phosphate (NAD(P)H): quinone oxidoreductase and glutathione S-transferase–M1 and –T1 have been associated with increased susceptibility to (t-) MDS.6-8 The concept of predisposition is also supported by the observation of defective DNA mismatch repair in patients with t-MDS9 and significant mitochondrial genomic instability in those with primary MDS.10-13 Finally, an inherited predisposition to MDS has long been evident from studies of constitutional genetic disorders such as Shwachman-Diamond syndrome, Fanconi anemia,14 and neurofibromatosis type 1.15
The granulocyte colony-stimulating factor receptor (G-CSF-R) transmits signals essential for proliferation and differentiation of myeloid progenitor cells.16,17 Like other members of the hematopoietin superfamily, the G-CSF-R lacks intrinsic tyrosine kinase activity, but upon binding of its ligand G-CSF and receptor oligomerization, cytoplasmic tyrosine kinases as well as other intracellular proteins like members of the Janus kinase and Src family, members of the signal transducer and activator of transcription (STAT) family, phosphatidylinositol-3 kinase, and components of the mitogen-activated protein kinase (MAPK) pathway are activated.16,17 Somatic mutations in its cytoplasmic domain resulting in premature stop codons and C-terminal truncation of the G-CSF-R have been found in patients with severe congenital neutropenia (SCN) who subsequently developed AML.18-20 These observations led to the assumption that mutations in the cytoplasmic domain of the G-CSF-R may also play a role in the development of other myeloid malignancies.
Here we report on a novel single nucleotide polymorphism (SNP) located within the cytoplasmic domain of the G-CSF-R (Human Genome Organization [HUGO] gene symbol: CSF3R) at position 2591 (G to A transition, G>A), which results in a missense mutation by exchanging glutamic acid to lysine at amino acid position 785 (G-CSF-R_Glu785Lys). Screening a cohort of primary MDS patients, de novo AML patients, as well as age- and sex-matched controls identified this mutation as a rare SNP, which is associated with the development of high-risk MDS. Functional analysis by retroviral transfer of G-CSF-R_785Lys into myeloid progenitor cells of G-CSF-R–deficient (Gcsfr-/-) mice revealed a significantly diminished colony-formation capacity after G-CSF stimulation as compared with cells expressing the wild-type receptor.
Patients, materials, and methods
Patients
This study included 121 primary MDS cases (refractory anemia [RA] and RA with ring sideroblasts [RARS], n = 49; RA with excess of blasts [RAEB], n = 59; RA with excess of blasts in transformation [RAEBt], n = 13) and 5 secondary AMLs evolving from MDSs; 101 cases were diagnosed at the University Hospital Graz and its affiliated hospitals and 25 at the Hanusch Hospital Vienna. Diagnosis of MDS was made using peripheral blood and bone marrow smears according to the criteria of the French-American-British (FAB) Cooperative Group classification.21 Patients with chronic myelomonocytic leukemia, which is increasingly considered to be a separate hematologic entity that is related more closely to the myeloproliferative syndromes,22,23 and patients with t-MDS were excluded from this study. In addition, there was no indication of a familial history of MDS in our patients. For each patient, 2 sex- and age-matched control subjects with normal peripheral blood counts and no history of a malignant hematologic disease were enrolled. In addition, 84 patients with de novo AML were analyzed. All subjects were of white racial background, and informed consent was obtained from all individuals. The institutional review board of the Medical University of Graz approved the study. DNA was isolated from peripheral blood or a bone marrow aspirate after red cell lysis, a buccal swab, or from diagnostic bone marrow slides according to standard procedures,24 or using a DNA extraction kit (Qiagen, Hilden, Germany). In 10 cases (5 RA, 3 RAEB, 2 RAEBt) we were not able to isolate sufficient amounts of DNA, so a total of 116 MDS patients with 232 age- and sex-matched controls and 84 patients with de novo AML were included in the study.
Genotyping of the 2591G>A polymorphism of the G-CSF-R gene
Analysis of the polymorphic site at nucleotide position 2591 of the G-CSF-R gene was done by capillary-based single-strand conformation sensitive gel electrophoresis (SSCSGE).25,26 A 277 base pair fragment including the polymorphic site was amplified by polymerase chain reaction (PCR) using the HotStar Taq polymerase kit (Qiagen) and fluorescent primers G-CSF-R-F1 5′-6-FAM-CAGGTCCTTTATGGGCAGC-3′ and G-CSF-R-R1 5′-HEX-CCAGGAAGCCCTAGAAGCT-3′. The PCR conditions were as follows: preheating at 95°C for 15 minutes; followed by 35 cycles of 94°C for 90 seconds, 60°C for 2 minute, and 72°C for 2 minute; and a final extension at 72°C for 10 minutes. For subsequent denaturation, 0.5 μL 0.3 M NaOH and 10 μL HIDI formamide (Applied Biosystems, Scoresby, Australia) were added to 1 μL of the PCR products (either undiluted or diluted with deionized water depending on efficiency of amplification) and 1 μL of the commercially available DNA length standard Tamra 500 (Applied Biosystems; manufactured by Roche, Branchburg, NJ). The mixture was incubated at 95°C for 2 minutes, placed on ice, and subjected to capillary-based SSCSGE, which was performed on an ABI 310 Genetic Analyzer (Applied Biosystems). The samples were electroinjected at 15 kV for 10 seconds to a 42 cm long capillary (diameter, 50 μm; Applied Biosystems) filled with GeneScan polymer (Applied Biosystems) diluted with Tris-borate-EDTA (Tris–borate–ethylenediaminetetraacetic acid) (TBE) buffer containing 10% glycerol to a final concentration of 5%. Electrophoresis was performed at 13 kV and 30°C for 25 minutes. Filter set C, allowing signal detection at wavelengths 532, 543, 557, and 584 nm, was used in all runs. The results were collected using the Data Collection program (Applied Biosystems) and analyzed using GeneScan software. To obtain reproducible results all electropherograms were calibrated by fixing the positions of peaks produced by the DNA length standard and inclusion of samples with known genotypes.
SSCSGE-based genotyping was verified by direct DNA sequencing for all samples displaying the 2591G>A mutation and 20% of randomly chosen normal samples. After amplification using the PCR conditions stated and nonfluorescent primers G-CSF-R-F1 and G-CSF-R-R1, PCR products were processed and sequenced on both strands on an ABI Prism 310 Genetic Analyzer using the Big Dye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer's protocol.
Production of retroviral vector
The G-CSF-R 2591G>A mutation was introduced with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using primers G-CSF-R_785Lys-F1 5′-CCAAGCCAGAAGGACGACTGTGT-3′ and G-CSF-R_785Lys-R1 5′-ACACAGTCG-TCCTTCTGGCTTGG-3′ and the plasmid pBABE-G-CSF-R 27 as template, resulting in pBABE-G-CSF-R_785Lys. PhoenixE packaging cells (courtesy of G. Nolan, Stanford, CA) were transfected with pBABE-G-CSF-R or pBABE-G-CSF-R_785Lys with CaPO4 precipitation. Supernatants containing high-titer, helper-free recombinant virus were harvested from about 80% confluent producer cells cultured for 16 to 20 hours in Dulbecco modified Eagle medium (DMEM; GIBCO-BRL, Breda, The Netherlands) supplemented with 10% fetal calf serum (FCS), penicillin (100 IU/mL), and streptomycin (100 ng/mL). To determine the virus titers, the virus particles were pelleted by ultracentrifugation at 104 000g (29 000 rpm) for 3 hours (XL-90; Beckman, Mijdrecht, The Netherlands), and RNA was extracted with phenol (pH 4.0) and spot blotted on nitrocellulose filters. This blot was hybridized with a BABE-specific probe (SV40 fragment, BamHI-HinDIII digest).
Cell culture and retroviral gene transfer
Hematopoietic cells were harvested from the femurs and tibiae of FVB G-CSF-R–deficient (Gcsfr-/-) mice as described.28,29 Primitive hematopoietic progenitor cells were harvested as previously described30 and prestimulated for 2 days at a final concentration of 1 × 106/mL in CellGro SCGM (Cellgenix Technologie Transfer, Freiburg, Germany) supplemented with a cytokine cocktail composed of murine interleukin-3 (mIL-3; 10 ng/mL), human (h) FLT3-ligand, human thrombopoietin (hTPO; 10 ng/mL), and murine stem cell factor (mSCF; 100 ng/mL). Retroviral infection with pBABE-G-CSF-R and pBABE-G-CSF-R_785Lys virus was performed as described.28,30
Flow cytometric analysis of G-CSF-R expression
To determine G-CSF-R expression on transduced bone marrow cells, cells were labeled for 30 minutes at 4°C with biotinylated antibody recognizing an N-terminal extracellular epitope of the human G-CSF-R (LMM741; Pharmingen, San Diego, CA) and subsequently incubated for 30 minutes at 4°C with phycoerythrin-conjugated streptavidin (SA-PE; DAKO Diagnostics, Glostrup, Denmark). Cells were subjected to flow cytometric analysis on a FACScan (Becton Dickinson, Sunnyvale, CA).
Colony assay
Bone marrow cells were plated at densities of 1 × 105/mL per dish in triplicate in methylcellulose medium (Methocult M3231; StemCell Technologies, Vancouver, BC) supplemented with G-CSF (100 ng/mL) and 2.5 μg/mL puromycin (Sigma, Zwijndrecht, The Netherlands). On day 6 colonies were counted and examined morphologically using Giemsa-stained cytospin preparations.
Statistics
Statistical analysis was done using SPSS 11.0 for Windows (Chicago, IL). Numeric values were analyzed by the Student t test. The distribution of genotypes in MDS and AML populations compared with the control group was assessed for significance using the Fisher exact test. Odds ratios (ORs) and 95% confidence intervals were calculated by logistic regression analysis. Threshold for significance was P below .05.
Results
Median age of patients with MDS was 73 years (range, 32 to 92 years), and 62 of 116 (53.4%) were male. Controls were age-(± 1 year) and sex-matched to the case subjects. Patients with de novo AML were predominantly women (56%) and between 18 and 84 years of age, with a median of 58 years. Genotype distribution did not deviate from the Hardy-Weinberg equilibrium in patients or controls. In contrast to the low frequency in age- and sex-matched hematologically healthy control subjects (0.9%; 2 of 232; Table 1), the heterozygous G-CSF-R_785Glu/Lys genotype was found in 6.9% (8 of 116) of patients with MDS (P = .003; OR, 8.5; 95% confidence interval [95% CI], 1.8-40.7). Because MDSs represent a heterogeneous group of disorders with a diverse clinical outcome, we classified our MDS patients according to the most important morphologic criterion on survival and transformation to AML— namely, the percentage of blasts in the bone marrow and the International Prognostic Scoring System (IPSS) for MDS. Classifying MDS patients by the percentage of bone marrow blasts into a low-risk group (less than 5% bone-marrow blasts; patients with RA and RARS, n = 44) and a high-risk group of patients with RAEB, RAEBt, and secondary AML following MDS (more than 5% bone-marrow blasts; n = 72) revealed a significantly higher prevalence of the G-CSF-R_785Glu/Lys genotype in high-risk MDS patients than in control subjects (9.7% versus 0.9%; P = .001; OR, 12.5; 95% CI, 2.4-58.9) but not in the low-risk MDS group (Table 1). Classifying MDS patients by the IPSS was possible for 105 patients. In 11 MDS patients, information on cytogenetic abnormalities was not available to allow proper classification. Using the IPSS, MDS patients were divided into 2 groups: an IPSS low-risk group with patients displaying a low or intermediate-1 score (n = 51) and an IPSS high-risk group with patients displaying an intermediate-2 or high score (n = 54). Again, the prevalence of the G-CSF-R_785Glu/Lys genotype was significantly higher in the IPSS high-risk group than in age- and sex-matched controls (13.0% versus 0.9%; P < .001; OR, 14.0; 95% CI, 3.4-85.0) but not in the IPSS low-risk group (Table 1). The clinical characteristics of MDS patients with the G-CSF-R_785Glu/Lys genotype are given in Table 2. In contrast to high-risk MDS patients, the prevalence of the G-CSF-R_785Glu/Lys genotype did not differ significantly in patients with de novo AML as compared with healthy controls (2.4% versus 0.9%; P = .288).
Because MDSs represent clonal hematopoietic stem cell disorders and genotyping of peripheral blood or bone marrow cells could thus display an acquired mutation inherent to the MDS clone indistinguishable from an inherited polymorphism, we extracted DNA from epithelial cells of buccal swabs (n = 8) or postmortem skin biopsies (n = 2) in all MDS and de novo AML patients with a G-CSF-R_785Glu/Lys genotype. The G-CSF-R_785Glu/Lys genotype was detected in epithelial cells in all of these individuals, indicating that this is indeed a constitutional polymorphism.
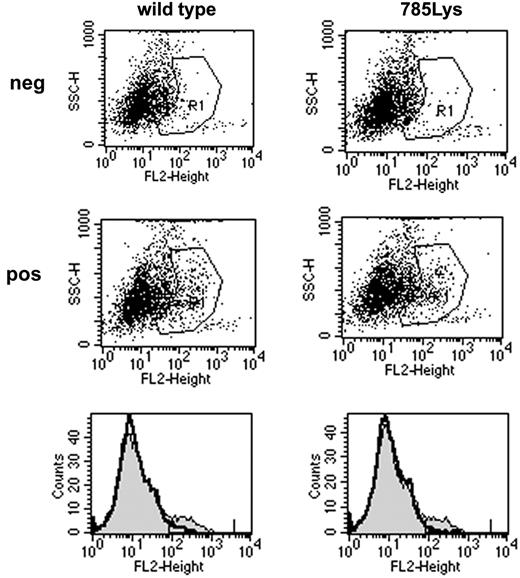
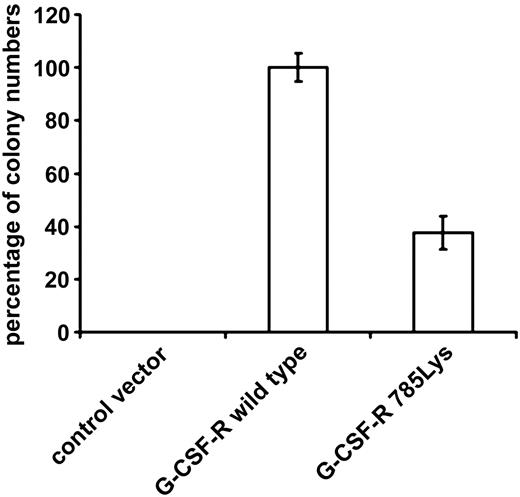
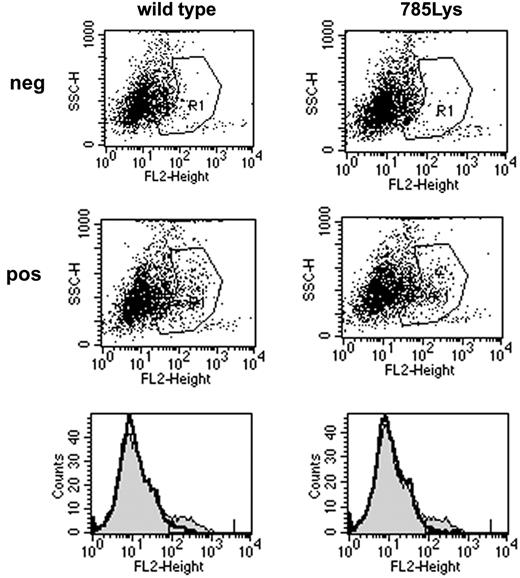
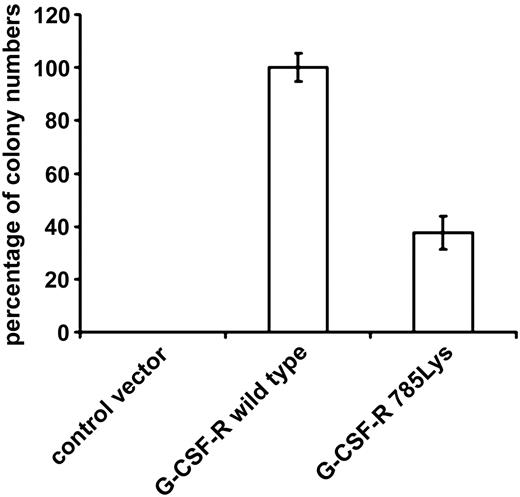
To study the functional relevance of the substitution of glutamic acid by lysine on position 785 of the G-CSF-R in primary hematopoietic cells, we introduced G-CSF-R_785Lys, G-CSF-R wild-type, or BABE control vector into Gcsfr-/- murine bone marrow cells. Flow cytometric analysis of CSF-R_785Lys and G-CSF-R wild-type–transduced Gcsfr-/- bone marrow cells revealed that transduction efficiencies and cell surface expression of the 2 receptor forms were equal (Figure 1). As predicted, no colonies were formed by Gcsfr-/- cells transduced with BABE vector (Figure 2). Cells expressing G-CSF-R_785Lys displayed a significantly decreased G-CSF–induced colony formation capacity as compared with cells transduced with the wild-type G-CSF-R (Figure 2). In contrast, granulocyte/macrophage-CSF–induced colony formation did not differ significantly (data not shown). Mean number of cells per colony and morphologic features of colony cells (mainly maturing myeloid cells at progressive stages of neutrophil differentiation) were also comparable, indicating that G-CSF-R_785Lys is able to transduce signals for differentiation.
Cell surface expression of wild-type G-CSF-R and G-CSF-R_785Lys. Flow cytometric analysis of G-CSF-R expression on Gcsfr-/- bone marrow cells retrovirally transduced with wild-type G-CSF-R or G-CSF-R_785Lys revealed no differences in transduction efficiency (8.02% versus 7.35%) and receptor surface expression (mean fluorescence intensity, 246.8 versus 231.4). The second row of dot plots and thin line in histograms indicate cells stained with biotinylated G-CSF-R antibodies and SA-PE; the first row and bold line in histograms indicate cells stained with SA-PE only.
Cell surface expression of wild-type G-CSF-R and G-CSF-R_785Lys. Flow cytometric analysis of G-CSF-R expression on Gcsfr-/- bone marrow cells retrovirally transduced with wild-type G-CSF-R or G-CSF-R_785Lys revealed no differences in transduction efficiency (8.02% versus 7.35%) and receptor surface expression (mean fluorescence intensity, 246.8 versus 231.4). The second row of dot plots and thin line in histograms indicate cells stained with biotinylated G-CSF-R antibodies and SA-PE; the first row and bold line in histograms indicate cells stained with SA-PE only.
G-CSF–induced colony formation of Gcsfr-/- bone marrow cells transduced with either wild-type G-CSF-R or G-CSF-R_785Lys. Colonies were grown in the presence of 100 ng/mL G-CSF. Percentages of colony numbers as compared with wild-type G-CSF-R from triplicate colony dishes of 4 independent experiments are shown (mean ± SE). Differences between wild-type G-CSF-R and G-CSF-R_785Lys are highly significant (Student t test, P < .001).
G-CSF–induced colony formation of Gcsfr-/- bone marrow cells transduced with either wild-type G-CSF-R or G-CSF-R_785Lys. Colonies were grown in the presence of 100 ng/mL G-CSF. Percentages of colony numbers as compared with wild-type G-CSF-R from triplicate colony dishes of 4 independent experiments are shown (mean ± SE). Differences between wild-type G-CSF-R and G-CSF-R_785Lys are highly significant (Student t test, P < .001).
Discussion
Here we reported on the association of the rare G-CSF-R_Glu785Lys SNP with high-risk MDS and demonstrated its functional consequences. The frequency of the heterozygous G-CSF-R_785Glu/Lys genotype was found to be more than 10 times higher in patients with high-risk MDS than in age- and sex-matched controls. Because we did not find an increased prevalence of this genotype in low-risk MDS patients characterized by a low or intermediate-1 IPSS score or less than 5% BM blasts as well as in de novo AML patients, which is in accordance with Bernard et al, who did not identify this genotype in 40 de novo AML patients,31 these findings suggest a specific role of this SNP in the pathogenesis of high-risk MDS. Due to the low frequency of the G-CSF-R_785Lys allele, we did not identify any patient or control individual with a homozygous G-CSF-R_785Lys/Lys genotype in our cohorts. Interestingly, although in the currently available literature only few patients with MDS were screened for mutations in the intracellular domain of the G-CSF-R,32,33 Carapeti et al34 reported a patient with secondary AML following high-risk MDS who displayed the homozygous G-CSF-R_785Lys/Lys genotype. Because DNA from nonhematopoietic cells from this patient was not studied, it remained unclear whether this homozygous base change represented a polymorphism involving both alleles or was due to an acquired somatic mutation.
Functional consequences of this SNP became evident by expression of G-CSF-R_785Lys in myeloid progenitor cells of Gcsfr-/- mice. In comparison with wild-type G-CSF-R, retroviral transfer of G-CSF-R_785Lys resulted in significantly diminished colony formation of these cells after G-CSF stimulation. In contrast to transduced primary bone marrow cells of Gcsfr-/- mice, we did not observe diminished proliferative signaling from G-CSF-R_785Lys in murine myeloid 32D cells. This may be due to discrepancies in signaling requirements between cell lines and primary cells.35-37
Although the domain affected by the Glu785Lys polymorphism is completely conserved between human and mouse, it has not yet been linked with any of the known signaling pathways of the G-CSF-R. Because differentiation of cells was not affected by the G-CSF-R_785Lys, we performed a comprehensive analysis on so far known signaling pathways involved in mediating proliferation and survival after G-CSF-R activation via the carboxy-terminal region of the G-CSF-R. However, we did not find alterations in extracellular signal-regulated kinase-1 (ERK1), ERK2, p38MAPK, Jun N-terminal kinase (JNK), and STAT5 activation by the G-CSF-R_785Lys in 32D cells (data not shown). These results indicate the involvement of a yet-undefined pathway affected by the G-CSF-R_785Lys.
Increasing evidence links intracellular routing and processing of activated receptor complexes to signaling function.38-40 For hematopoietin receptors, it has recently been demonstrated that the carboxy-terminal part of the erythropoietin receptor (EPO-R) is rapidly ubiquitinated and targeted for proteasomal degradation after activation of the EPO-R/Janus kinase-2 (JAK2) complex.41 Erkeland et al reported the binding of WSB-1 and WSB-2 (WD40-repeat–containing protein with a suppressor of cytokine signaling [SOCS] box) to amino acids 769 to 813 of the G-CSF-R, a region that encompasses the Glu785Lys polymorphism. Binding of WSB-1 and -2 is presumably involved in intracellular trafficking of the G-CSF-R.42 Therefore, disturbances in intracellular processing of G-CSF-R_785Lys may result in incomplete G-CSF-R signaling, causing diminished colony formation after G-CSF stimulation.
Hematopoietic growth factor receptor abnormalities may contribute to hematologic malignancies by circumventing normal growth factor control or altering the balance of proliferation and differentiation. For instance, one of the most common somatic mutations found in AML and MDS is located within the FLT3 gene encoding a transmembrane tyrosine kinase growth factor receptor that is selectively expressed on hematopoietic cells.43-46 Among hematopoietin factor receptors that lack intrinsic tyrosine kinase activity, mutations within the G-CSF-R have been linked to leukemogenesis,47 because in patients with SCN who subsequently developed AML or MDS, somatic nonsense mutations within a critical glutamine-rich stretch of the cytoplasmic domain of the G-CSF-R were found.18-20 In contrast to such acquired mutations leading to ligand-dependent and/or -independent growth as well as survival advantages in affected cells, the inherited G-CSF-R_785Lys allele resulted in diminished G-CSF response in all cells, implicating a completely different mechanism involved in conferring susceptibility to high-risk MDS. Because full activation of the G-CSF-R signaling cascade is not only necessary for adequate granulopoiesis48,49 but also for directing primitive hematopoietic progenitor cells into the common myeloid lineage,50 lifelong improper G-CSF response via G-CSF-R_785Lys during generation of myeloid progenitors may render these cells susceptible to the development of high-risk MDS. This notion is supported by several findings. First, in most MDS patients the neoplastic events occur at the level of a committed myeloid progenitor cell.51-53 Second, a critical role of alterations in the physiologically well-balanced and subtly regulated signaling cascade after G-CSF binding to its receptor is also apparent from the clinical finding of a higher incidence of therapy-induced MDS and AML after G-CSF administration during high-dose chemotherapy for childhood acute lymphoblastic leukemia in a randomized trial.54 Third, hyporesponsiveness to G-CSF and other hematopoietic growth factors is a well-described finding in bone marrow progenitor cells of MDS patients.33,55,56
In summary, we have described a significant association of the hyporesponsive G-CSF-R_785Lys with high-risk MDS. These data suggest that abnormal responses to hematopoietic growth factors may play an early role in the multistep pathogenesis of MDS.
Prepublished online as Blood First Edition Paper, January 11, 2005; DOI 10.1182/blood-2004-06-2094.
Supported in part by Leukämiehilfe Steiermark and by the Dutch Cancer Society (KWF-kankerbestrijding).
A.W. and S.J.E. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.