Abstract
Juvenile myelomonocytic leukemia (JMML) is a lethal disease of young children characterized by hypersensitivity of hematopoietic progenitors to granulocyte-macrophage colony-stimulating factor (GM-CSF). Mutations in PTPN11, which encodes the protein tyrosine phosphatase Shp-2, are common in JMML. We hypothesized that PTPN11 mutations induce hypersensitivity of hematopoietic progenitors to GM-CSF and confer increased GM-CSF–stimulated phospho–extracellular signal-regulated kinase (Erk) levels. To test this hypothesis, the wild-type (WT) and 3 mutant Ptpn11 cDNAs (E76K, D61V, and D61Y) were transduced into murine bone marrow cells to examine GM-CSF–stimulated granulocyte-macrophage colony-forming unit (CFU-GM) growth, macrophage progenitor proliferation, and activation of the Ras signaling pathway. Expression of the Shp-2 mutants induced progenitor cell hypersensitivity to GM-CSF compared with cells transduced with vector alone or WT Shp-2. Macrophage progenitors expressing the Shp-2 mutants displayed both basal and GM-CSF–stimulated hyperproliferation compared with cells transduced with vector alone or WT Shp-2. Consistently, macrophage progenitors transduced with the Shp-2 mutants demonstrated constitutively elevated phospho-Erk levels and sustained activation of phospho-Erk following GM-CSF stimulation compared with vector alone or WT Shp-2. These data support the hypothesis that PTPN11 mutations induce hematopoietic progenitor hypersensitivity to GM-CSF due to hyperactivation of the Ras signaling axis and provide a basis for the GM-CSF signaling pathway as a target for rational drug design in JMML.
Introduction
Juvenile myelomonocytic leukemia (JMML), a childhood leukemia, is a lethal disease of young children characterized by spontaneous growth of peripheral blood hematopoietic progenitors and hypersensitivity of hematopoietic progenitors to the cytokine granulocyte-macrophage colony-stimulating factor (GM-CSF).1,2 Activating mutations of the N-RAS and K-RAS genes and loss of heterozygosity of the tumor suppressor gene NF1 (in individuals with the congenital disorder Neurofibromatosis type 1) have long been recognized as pathogenic in this disease.3-7 However, recently, somatic mutations within PTPN11, which encodes the protein tyrosine phosphatase Shp-2, have also been found to occur commonly in JMML,8 as well as in acute myeloid leukemia, myelodysplastic syndrome, and acute lymphoid leukemia.8-10 These findings were elicited based on the initial observation that individuals with the congenital disorder known as Noonan syndrome, 50% of which bear germline PTPN11 mutations,11,12 have an increased incidence of JMML.13,14
Shp-2 is a nonreceptor protein tyrosine phosphatase that contains 2 Src homology 2 domains (N-SH2 and C-SH2) at the amino terminus and an enzymatic phosphatase domain at the carboxy terminus of the protein.15 Shp-2 has been demonstrated repeatedly to play a positive role in growth factor signaling to Ras,16-21 suggesting that normal Shp-2 function is required for Ras-dependent functions, such as cellular proliferation and survival. Germline PTPN11 mutations observed in Noonan syndrome as well as somatic PTPN11 mutations observed in leukemias commonly involve residues within the N-SH2 domain or within the phosphatase domain.8,11 The affected amino acids disrupt noncovalent interactions between the N-SH2 and phosphatase domains necessary to maintain Shp-2 in a closed, inactive conformation.22 The current hypothesis is that mutant Shp-2 proteins preferentially remain in the open conformation with constitutively active Shp-2 phosphatase activity, thus inducing the observed disease states. A mouse model bearing a common PTPN11 germline mutation found in Noonan syndrome changing amino acid 61 from aspartate to glycine11 faithfully mimics the human phenotype of Noonan syndrome, including facial anomalies, short stature, heart malformations, and myelomonocytic hyperplasia23 ; however, the effect of somatic PTPN11 mutations on primary hematopoietic cell growth and GM-CSF–stimulated signal transduction has not been examined. Additionally, the mechanism of how the leukemia-associated PTPN11 mutations affect the hematopoietic phenotype and whether these mutations function in a similar or disparate fashion is unknown. Therefore, we examined the effect of 3 somatic PTPN11 mutations (E76K, D61V, and D61Y) found in children with JMML on GM-CSF–stimulated hematopoietic progenitor growth, macrophage progenitor proliferation, and signal transduction.
Materials and methods
Retroviral vectors and retroviral transduction
The wild-type (WT) Ptpn11 cDNA was mutated at nucleotides 226 (G > A), 182 (A > T), or 181 (G > T) using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to yield amino acid changes E76K, D61V, or D61Y, respectively. Each cDNA was subcloned into the murine stem-cell virus (MSCV)–based bicistronic retroviral vector, pMIEG3, in tandem with enhanced green fluorescent protein cDNA (EGFP). Following mutatgenesis, each full-length cDNA was sequenced to verify the desired mutation and to rule out aberrantly introduced mutations. Ecotropic retroviral supernatants (pMIEG3, pMIEG3-WT, pMIEG3-E76K, pMIEG3-D61V, and pMIEG3-D61Y) were prepared using Eco-Phoenix packaging cells by the Indiana University Vector Production Facility (Indianapolis, IN). Low-density bone marrow mononuclear cells from C57/Bl6 mice (Jackson Laboratories, Bar Harbor, ME) were purified using a Ficoll gradient. Cells were prestimulated at a concentration of 2 × 106 cells/mL in Iscove modified Dulbecco medium (IMDM), 20% bovine growth serum (Hyclone, Logan, UT), 2 mM glutamine, 1% penicillin/streptomycin, stem-cell factor (SCF), granulocyte colony-stimulating factor (G-CSF), and thrombopoietin (Tpo) all at 100 ng/mL (growth factors from Peprotech, Rocky Hill, NJ) for 24 hours. Cells were then transferred to retroviral supernatant on plates coated with fibronectin fragments (Retronectin; Takara, Madison, WI) with SCF, G-CSF, and Tpo, all at 100 ng/mL for an additional 48 hours. Cells were placed back in the prestimulation media for 24 hours prior to sorting for EGFP+ cells using fluorescence activated cell sorting (FACS).
Granulocyte-macrophage progenitor (granulocyte-macrophage colony-forming unit) analysis
Transduced, EGFP+ cells were plated at a concentration of 8000 cells/mL in 0.9% methylcellulose-based media containing IMDM, 2 mM glutamine, 1% penicillin/streptomycin, 10-5 β-mercaptoethanol, 30% fetal bovine serum (FBS), and increasing concentrations of GM-CSF (0, 0.01, 0.1, 1, and 10 ng/mL). Cultures were incubated in a humidified incubator at 37°C in 5% CO2 and colonies were counted on day 7. Assays were conducted on 3 independent occasions and plated in duplicate in each experiment. The data are represented as a percentage of maximal colony formation, which is calculated by dividing the number of colonies at each dose by the number of colonies at the maximal GM-CSF concentration (10 ng/mL).
Macrophage progenitor differentiation and analysis
Sorted, EGFP+ cells were plated at a concentration of 10 000 cells/mL in IMDM, 20% bovine growth serum, 2 mM glutamine, 1% penicillin/streptomycin, and 50 ng/mL macrophage-colony stimulating factor (M-CSF) for 4 days at 37°C in 5% CO2 for differentiation to macrophage progenitors. Cells were collected using cell dissociation buffer (Invitrogen, Grand Island, NY) and stained with anti–Mac-1 conjugated to allophycocyanin (APC; BD Pharmingen, San Diego, CA), anti-F4/80 conjugated to phycoerythrin (PE; Serotec, Oxford, United Kingdom), or anti–M-CSF receptor (2-4A5; Santa Cruz Biotechnologies, Santa Cruz, CA) secondarily stained with PE-conjugated F(ab′)2 fragment goat anti–rat immunoglobulin G (IgG; Beckman Coulter, Fullerton, CA) and analyzed using FACS.
Western blot analysis
Sorted, EGFP+ cells were differentiated to macrophage progenitors, and protein extracts were prepared as previously described,17 electrophoresed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and probed with anti–Shp-2 (C-18; Santa Cruz Biotechnologies), anti-GM-CSFα receptor (M-20; Santa Cruz Biotechnologies), or anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Biodesign International, Saco, ME). For signal transduction studies, macrophage progenitor cultures were serum and growth factor deprived for 24 hours. Cells either remained unstimulated (for baseline measurements) or were stimulated with 50 ng/mL GM-CSF for 5, 10, 30, or 60 minutes followed by protein extract preparation, as described,17 and probed with anti–extracellular signal-regulated kinase (Erk) or anti–phospho-Erk (Cell Signaling, Beverly, MA). All signals were detected by enhanced chemiluminescence and quantitated using ImageJ (National Institutes of Health, Bethesda, MD).
Thymidine incorporation assays
Sorted, EGFP+ cells were differentiated to macrophage progenitors as described. Cultures were serum and growth factor deprived for 24 hours followed by either treatment with GM-CSF (50 ng/mL), M-CSF (100 ng/mL), or no stimulation (baseline) for 24 hours. Cultures were pulsed with 0.037 MBq (1.0 μCi) of [3H]-thymidine (Amersham, Piscataway, NJ) for 6 hours followed by harvesting. Thymidine incorporation was measured using a Beckman Coulter LS 6500 Scintillation Counter (Beckman Coulter, Fullerton, CA). For MIEG3, WT, E76K, and D61V, assays were conducted on 3 independent occasions with each condition plated in triplicate. For D61Y, assays were conducted on 2 independent occasions with each condition plated in triplicate.
Statistical analysis
For the granulocyte-macrophage colony-forming unit (CFU-GM) and thymidine incorporation assays, comparisons were conducted using randomeffects analysis of variance (ANOVA) to deal with repeated measurements within each independent assay. The P value for pairwise comparisons was adjusted using the Tukey method.24 For comparisons of phospho-Erk band intensities, a 2-tailed, unpaired Student t test was used.
Results
Transduction of murine bone marrow cells and ectopic Shp-2 expression
The WT Ptpn11 cDNA was mutated at nucleotides 226 (G > A), 182 (A > T), or 181 (G > T) to yield amino acid changes E76K, D61V, or D61Y, respectively. Bone marrow low density mononuclear cells from 8- to 12-week-old C57/Bl6 mice were subjected to fibronectin-assisted retroviral transduction with vector alone (pMIEG3), WT Shp-2 (pMIEG3-WT), or each of the mutants (pMIEG3-E76K, -D61V, or -D61Y), and transduced cells were enriched using FACS (Figure 1A). We next examined the level of Shp-2 expression in each of the transduced populations. As seen in Figure 1B, expression of Shp-2 was higher in the cells transduced with WT and mutant Shp-2 compared with cells transduced with vector alone. When Shp-2 expression levels were normalized to the constitutively expressed GAPDH protein using densitometry, no difference in the level of Shp-2 was observed in cells expressing exogenously introduced WT or mutant Shp-2 (Figure 1B).
Transduction of murine bone marrow cells and ectopic Shp-2 expression. (A) Low-density bone marrow mononuclear cells from C57/Bl6 mice were transduced with ecotropic retroviral supernatants (pMIEG3, pMIEG3-WT, pMIEG3-E76K, pMIEG3-D61V, and pMIEG3-D61Y) and tranduced cells were enriched by sorting for EGFP expression. Representative FACS showing the percentage of EGFP+ cells for collection of transduced cells is demonstrated. (B, left) Western blot analysis demonstrated that the exogenously introduced Shp-2 proteins are equally expressed in each group of transduced cells. Shp-2 band intensities normalized to GAPDH are shown graphically (right). Representative Western blot of 2 independent experiments with similar results.
Transduction of murine bone marrow cells and ectopic Shp-2 expression. (A) Low-density bone marrow mononuclear cells from C57/Bl6 mice were transduced with ecotropic retroviral supernatants (pMIEG3, pMIEG3-WT, pMIEG3-E76K, pMIEG3-D61V, and pMIEG3-D61Y) and tranduced cells were enriched by sorting for EGFP expression. Representative FACS showing the percentage of EGFP+ cells for collection of transduced cells is demonstrated. (B, left) Western blot analysis demonstrated that the exogenously introduced Shp-2 proteins are equally expressed in each group of transduced cells. Shp-2 band intensities normalized to GAPDH are shown graphically (right). Representative Western blot of 2 independent experiments with similar results.
PTPN11 mutants E76K, D61V, and D61Y induce hematopoietic progenitor hypersensitivity to GM-CSF
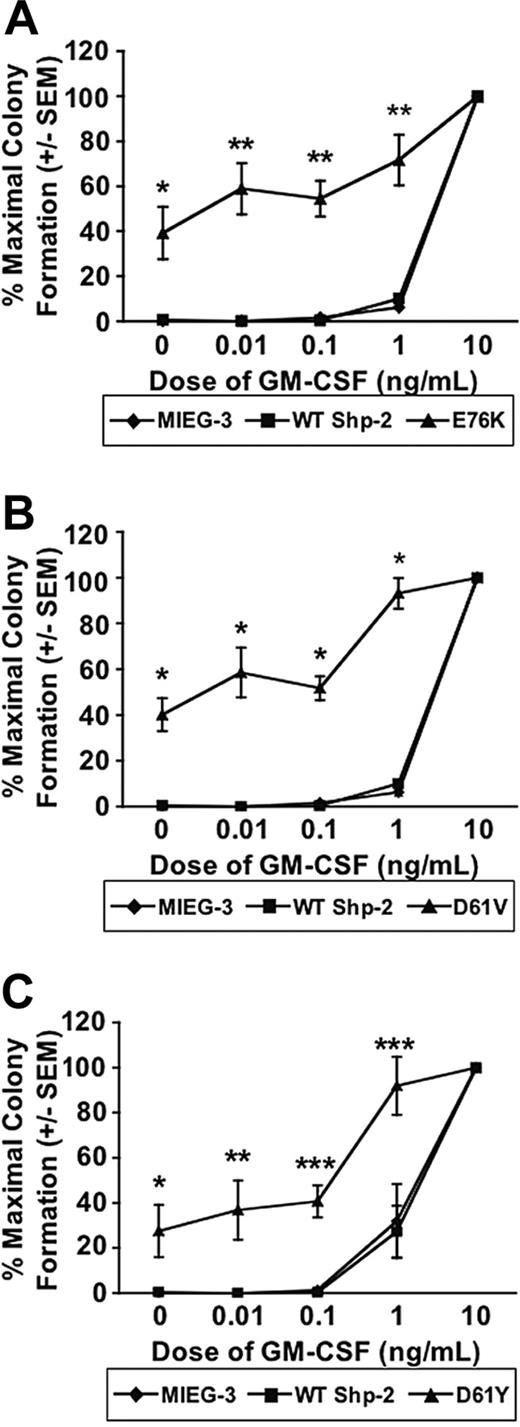
To examine the effect of the mutant Shp-2 molecules on hematopoietic progenitor growth in response to GM-CSF, sorted EGFP+ cells were collected and plated in a CFU-GM assay with increasing concentrations of GM-CSF (0-10 ng/mL). Similar to the GM-CSF hypersensitivity noted in JMML patients,1 hematopoietic progenitors transduced with each of the Shp-2 mutants also demonstrated significant hypersensitivity to GM-CSF compared with cells transduced with the vector alone or with WT Shp-2 (Figure 2). Given the similar level of ectopic WT Shp-2 and mutant Shp-2 (E76K, D61V, and D61Y) expression following transduction (Figure 1B), the induction of GM-CSF hypersensitivity appears to be specific to the mutant Shp-2 protein rather than to an overall increase in Shp-2 expression. Importantly, cells bearing each of the 3 Shp-2 mutants demonstrated a similar degree of hypersensitivity to GM-CSF. These results suggest that functionally, the 3 Shp-2 mutants that we examined behave in a similar fashion.
PTPN11 mutants E76K, D61V, and D61Y induce hematopoietic progenitor hypersensitivity to GM-CSF. Transduced EGFP+ cells were plated in CFU-GM progenitor assays in duplicate in 3 independent experiments. The data are represented as a percentage of maximal colony formation, which is calculated by dividing the number of colonies at each dose by the number of colonies at the maximal GM-CSF concentration (10 ng/mL). Error bars indicate standard error of the mean. Statistical analysis was conducted using random-effects ANOVA to deal with repeated measurements from duplicate plates. (A) E76K vs MIEG3 and WT; *P = .0014 for E76K vs MIEG3 and *P = .0015 for E76K vs WT; **P < .0001 for E76K vs MIEG3 or WT. (B) D61V vs MIEG3 and WT; *P < .0001 for D61V vs MIEG3 or WT. (C) D61Y vs MIEG3 and WT; *P = .0179 for D61Y vs MIEG3 and *P = .02 for D61Y vs WT; **P = .0058 for D61Y vs MIEG3 or WT; ***P < .0001 for D61Y vs MIEG3 or WT.
PTPN11 mutants E76K, D61V, and D61Y induce hematopoietic progenitor hypersensitivity to GM-CSF. Transduced EGFP+ cells were plated in CFU-GM progenitor assays in duplicate in 3 independent experiments. The data are represented as a percentage of maximal colony formation, which is calculated by dividing the number of colonies at each dose by the number of colonies at the maximal GM-CSF concentration (10 ng/mL). Error bars indicate standard error of the mean. Statistical analysis was conducted using random-effects ANOVA to deal with repeated measurements from duplicate plates. (A) E76K vs MIEG3 and WT; *P = .0014 for E76K vs MIEG3 and *P = .0015 for E76K vs WT; **P < .0001 for E76K vs MIEG3 or WT. (B) D61V vs MIEG3 and WT; *P < .0001 for D61V vs MIEG3 or WT. (C) D61Y vs MIEG3 and WT; *P = .0179 for D61Y vs MIEG3 and *P = .02 for D61Y vs WT; **P = .0058 for D61Y vs MIEG3 or WT; ***P < .0001 for D61Y vs MIEG3 or WT.
PTPN11 mutants E76K, D61V, and D61Y induce macrophage progenitor hyperproliferation in response to GM-CSF but not to M-CSF
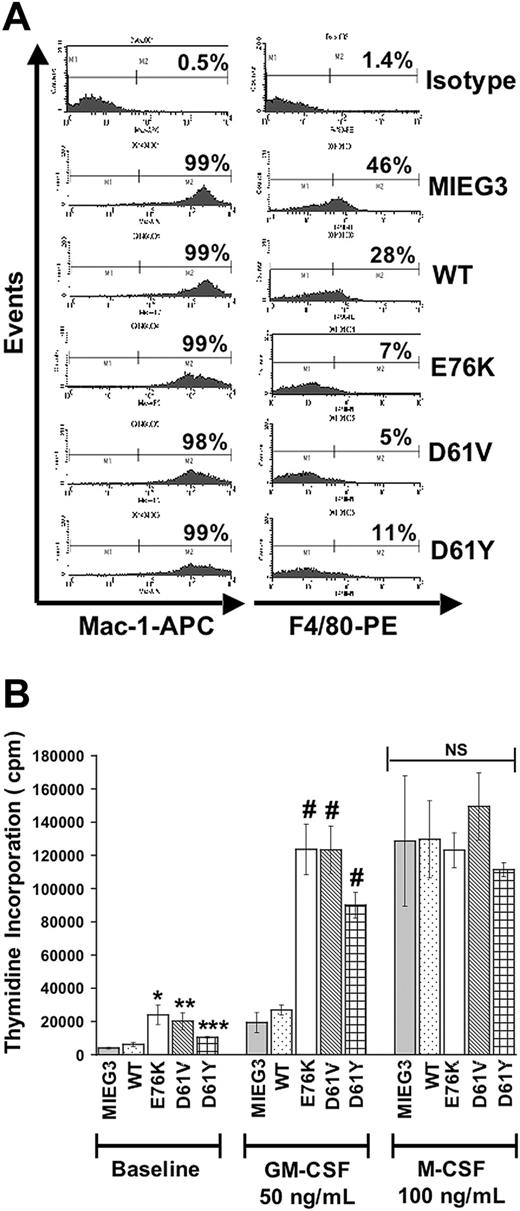
We next sought to investigate altered cellular and signaling parameters induced by each of the 3 Shp-2 mutants. Given the observed human hematopoietic phenotype of myelomonocytic hyperplasia and leukemia within patients bearing PTPN11 mutations, we examined baseline and growth factor–stimulated proliferation of macrophage progenitors induced to differentiate in the presence of M-CSF. Sorted EGFP+ cells were cultured in media containing 50 ng/mL M-CSF for 4 days. Following 4 days of culture, we found that all cultures consisted of 95% or more cells positive for Mac-1, a marker specific for myelomonocytic cells (Figure 3A). We also examined F4/80, a surface marker specific for macrophages. We found that cells expressing the Shp-2 mutants expressed lower levels of F4/80 compared with cells transduced with vector alone or with WT Shp-2, suggesting that cells expressing the mutant Shp-2 molecules do not efficiently undergo maturation within the macrophage lineage. Interestingly, in this regard, overexpression of WT Shp-2 also appeared to inhibit F4/80 expression, but to a lesser extent than that observed in cells expressing the mutant Shp-2 molecules. Taken together, these results suggest that overexpression of WT Shp-2 or the presence of activating Shp-2 mutants interferes with normal macrophage maturation.
PTPN11 mutants E76K, D61V, and D61Y induce macrophage progenitor hyperproliferation in response to GM-CSF but not to M-CSF. (A) Sorted EGFP+ cells subjected to macrophage progenitor differentiation conditions were stained with anti–Mac-1 or with anti-F4/80 and analyzed using FACS. Representative FACS analysis showing the percentage of Mac-1+ or F4/80+ cells is depicted; 2 independent experiments conducted with consistent results. (B) Macrophage progenitor cultures were serum- and growth factor–deprived for 24 hours and then stimulated with GM-CSF or M-CSF. Data shown as ± SEM. For pMIEG3, WT, E76K, and D61V, 3 independent experiments with each condition in triplicate were conducted; for D61Y, 2 independent experiments with each condition in triplicate were conducted. Statistical analysis was conducted using random-effects ANOVA to deal with repeated measurements from triplicate conditions; *P < .0001 for E76K vs MIEG3 or WT; **P = .0002 for D61V vs MIEG3 and **P = .0014 for D61V vs WT; ***P = .019 for D61Y vs MIEG3 and ***P = .074 for D61Y vs WT; #P < .0001 comparing E76K, D61V, or D61Y to MIEG3 or WT following GM-CSF treatment. No statistical difference was observed between cultures following stimulation with M-CSF.
PTPN11 mutants E76K, D61V, and D61Y induce macrophage progenitor hyperproliferation in response to GM-CSF but not to M-CSF. (A) Sorted EGFP+ cells subjected to macrophage progenitor differentiation conditions were stained with anti–Mac-1 or with anti-F4/80 and analyzed using FACS. Representative FACS analysis showing the percentage of Mac-1+ or F4/80+ cells is depicted; 2 independent experiments conducted with consistent results. (B) Macrophage progenitor cultures were serum- and growth factor–deprived for 24 hours and then stimulated with GM-CSF or M-CSF. Data shown as ± SEM. For pMIEG3, WT, E76K, and D61V, 3 independent experiments with each condition in triplicate were conducted; for D61Y, 2 independent experiments with each condition in triplicate were conducted. Statistical analysis was conducted using random-effects ANOVA to deal with repeated measurements from triplicate conditions; *P < .0001 for E76K vs MIEG3 or WT; **P = .0002 for D61V vs MIEG3 and **P = .0014 for D61V vs WT; ***P = .019 for D61Y vs MIEG3 and ***P = .074 for D61Y vs WT; #P < .0001 comparing E76K, D61V, or D61Y to MIEG3 or WT following GM-CSF treatment. No statistical difference was observed between cultures following stimulation with M-CSF.
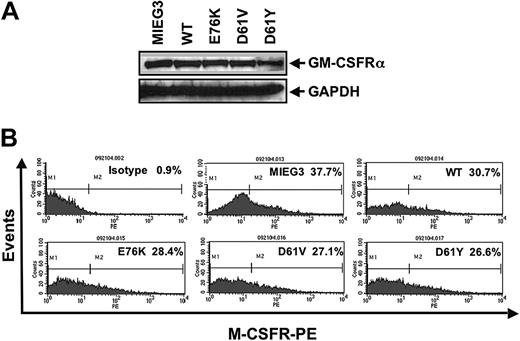
To determine if the expression of activating Shp-2 mutants in macrophage progenitors induced hypersensitivity to GM-CSF as well as to M-CSF, we compared the proliferative capacity of each of the transduced macrophage progenitors using a thymidine incorporation assay. As shown in Figure 3B, macrophage progenitors transduced with each of the Shp-2 mutants demonstrated modest but significant hyperproliferation in the absence of cytokine stimulation at baseline and dramatic hyperproliferation in response to GM-CSF stimulation compared with cells transduced with the vector alone or with WT Shp-2. In contrast, each of the macrophage progenitor cultures proliferated similarly in response to M-CSF. Given that we observed a defect in macrophage maturation in cells transduced with the Shp-2 mutants (Figure 3A), we examined GM-CSF receptor (GM-CSFRα) and M-CSF receptor (M-CSFR) expression in each of the macrophage progenitor cultures to address the possibility of variable receptor levels accounting for the different responses to GM-CSF and M-CSF stimulation, respectively. As shown in Figure 4A, expression of GM-CSFRα in each of the macrophage progenitor cultures was similar. A slight decrease in the expression of M-CSFR was observed in cultures expressing WT Shp-2 and each of the activating Shp-2 mutants (Figure 4B). These findings suggest that the activating Shp-2 mutants preferentially confer hypersensitivity to the GM-CSF signaling pathway, although a modest involvement of the M-CSF signaling pathway cannot be completely ruled out.
Macrophage progenitors derived from MIEG3-, WT-, E76K-, D61V-, and D61Y-transduced cells express similar levels of the GM-CSF and M-CSF receptors. Given the variable response observed in macrophage proliferation in response to GM-CSF and M-CSF, receptor levels for each of these cytokines was examined. (A) Protein extracts prepared from macrophage progenitor cultures were evaluated for the unique α chain of the GM-CSF receptor. (B) Macrophage progenitors were collected, incubated with anti–M-CSFR, and secondarily stained with PE-conjugated F(ab′)2 fragment goat anti–rat IgG followed by FACS analysis to determine the percentage of M-CSFR+ cells.
Macrophage progenitors derived from MIEG3-, WT-, E76K-, D61V-, and D61Y-transduced cells express similar levels of the GM-CSF and M-CSF receptors. Given the variable response observed in macrophage proliferation in response to GM-CSF and M-CSF, receptor levels for each of these cytokines was examined. (A) Protein extracts prepared from macrophage progenitor cultures were evaluated for the unique α chain of the GM-CSF receptor. (B) Macrophage progenitors were collected, incubated with anti–M-CSFR, and secondarily stained with PE-conjugated F(ab′)2 fragment goat anti–rat IgG followed by FACS analysis to determine the percentage of M-CSFR+ cells.
PTPN11 mutants E76K, D61V, and D61Y confer constitutively elevated and prolonged GM-CSF–stimulated phospho-Erk levels in macrophage progenitors
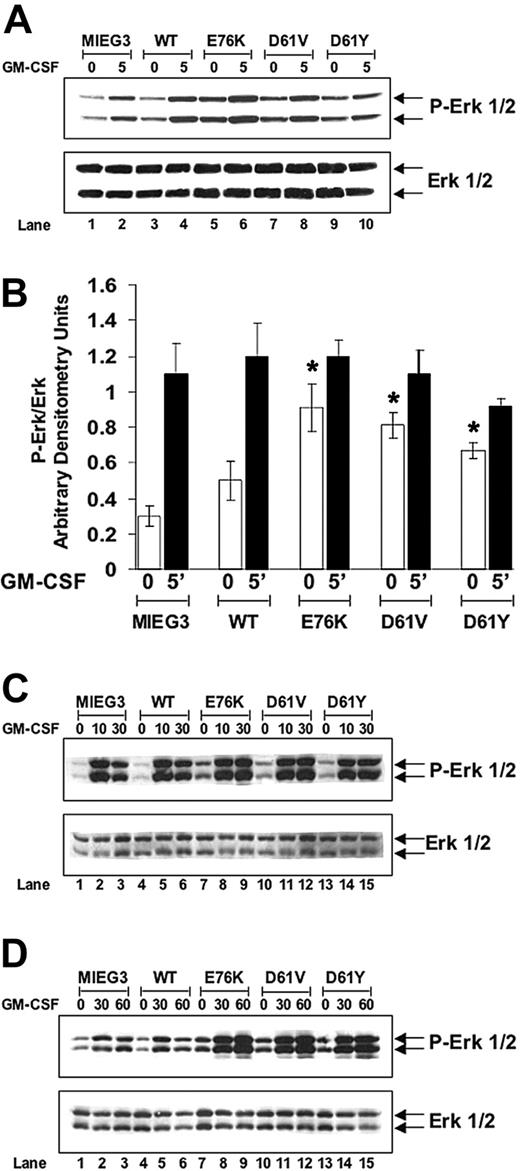
Activating RAS mutations and loss of function mutations in the tumor suppressor gene NF1 have implicated up-regulation of the Ras signaling pathway in the pathogenesis of JMML. Shp-2 is known to be a positive mediator of ligand-stimulated Ras activation and previous studies have demonstrated that the positive effect of Shp-2 on Ras activation is at least partially dependent on its phosphatase function.16,17,19-21 Therefore, we hypothesized that GM-CSF stimulation of cells bearing gain-of-function Shp-2 mutants would yield elevated activation of downstream effectors of Ras, including Erk. To test this hypothesis, sorted EGFP+ cells were cultured in macrophage differentiation media, as described, and were serum deprived for 24 hours followed by stimulation with GM-CSF for various amounts of time. We observed constitutively elevated levels of phospho-Erk at baseline in cultures transduced with each of the Shp-2 mutants compared with those transduced with the vector alone or with WT Shp-2 (compare lanes 5, 7, and 9 to lanes 1 and 3 in Figure 5A). Densitometry data from multiple independent experiments were compiled demonstrating that the baseline levels of phospho-Erk were significantly elevated in cultures transduced with the Shp-2 mutants compared with those transduced with WT Shp-2 or with the vector alone (Figure 5B). Following 5 minutes of GM-CSF stimulation, no significant difference between the level of phospho-Erk was observed between any of the macrophage progenitor cultures (Figure 5B). However, when examining the kinetics of GM-CSF–stimulated Erk activation, we observed that the level of phospho-Erk started to decline by 30 minutes after stimulation in the cells transduced with vector alone or with WT Shp-2 in contrast to that observed in cells transduced with the Shp-2 mutants (compare lanes 9, 12, and 15 to lanes 3 and 6 in Figure 5C). Further studies demonstrated sustained elevated phospho-Erk levels in cells transduced with each of the Shp-2 mutants as long as 60 minutes after GM-CSF stimulation (compare lanes 9, 12, and 15 to lanes 3 and 6 in Figure 5D).
PTPN11 mutants E76K, D61V, and D61Y confer constitutively elevated and prolonged GM-CSF–stimulated phospho-Erk levels in macrophage progenitors. (A) Representative Western blot showing 0- and 5-minute time points of GM-CSF stimulation of macrophage progenitor cultures; repeated in 3 independent experiments for MIEG3, WT, E76K, and D61V; repeated in 2 independent experiments for E61Y. (B) Band intensities were quantitated using densitometry, phospho-Erk band intensities were normalized to Erk band intensities, and data were compiled from all experiments and demonstrated graphically. Data shown as ± SEM; *P < .05 for E76K, D61V, and D61Y compared with MIEG3 at baseline. No statistical difference between MIEG3 and WT at baseline. (C) Western blot analysis demonstrating phospho-Erk levels following GM-CSF stimulation for 10 and 30 minutes demonstrating prolonged activation of Erk in macrophage progenitors transduced with each of the Shp-2 mutants. (D) A second independent experiment showing prolonged activation of Erk in macrophage progenitors transduced with each of the Shp-2 mutants for as long as 60 minutes following GM-CSF stimulation.
PTPN11 mutants E76K, D61V, and D61Y confer constitutively elevated and prolonged GM-CSF–stimulated phospho-Erk levels in macrophage progenitors. (A) Representative Western blot showing 0- and 5-minute time points of GM-CSF stimulation of macrophage progenitor cultures; repeated in 3 independent experiments for MIEG3, WT, E76K, and D61V; repeated in 2 independent experiments for E61Y. (B) Band intensities were quantitated using densitometry, phospho-Erk band intensities were normalized to Erk band intensities, and data were compiled from all experiments and demonstrated graphically. Data shown as ± SEM; *P < .05 for E76K, D61V, and D61Y compared with MIEG3 at baseline. No statistical difference between MIEG3 and WT at baseline. (C) Western blot analysis demonstrating phospho-Erk levels following GM-CSF stimulation for 10 and 30 minutes demonstrating prolonged activation of Erk in macrophage progenitors transduced with each of the Shp-2 mutants. (D) A second independent experiment showing prolonged activation of Erk in macrophage progenitors transduced with each of the Shp-2 mutants for as long as 60 minutes following GM-CSF stimulation.
Discussion
JMML is a leukemia of young children with limited treatment options. Currently, the only therapy available producing a potential cure is allogeneic stem cell transplantation; however, leukemia recurrence remains unacceptably high at approximately 35% following this rigorous treatment.25 Hyperactivated Ras has clearly been shown to be a central molecular participant in the etiology of JMML either due to activating mutations in K-RAS and N-RAS6,7 or due to loss of function mutations in NF1,3-5 a guanosine triphosphatase (GTPase)–activating protein that accelerates the conversion of active Ras-GTP to inactive Ras–guanosine diphosphate (GDP).26 Additionally, gain-of-function mutations in PTPN11, a phosphatase shown in multiple studies to act upstream of and to positively mediate the activation of Ras,16,19-21 provide an additional means of Ras hyperactivation in JMML.
We have examined the effect of 3 somatic PTPN11 mutants observed from leukemic cells from children with JMML on GM-CSF–stimulated hematopoietic progenitor growth, macrophage progenitor proliferation, and macrophage progenitor signaling. Our data support the hypothesis that Shp-2 gain-of-function mutants induce hypersensitivity to GM-CSF, similar to the classically described phenotype of hematopoietic progenitors from patients with JMML. By examining GM-CSF–stimulated macrophage progenitor proliferation, we found that GM-CSF functions to increase the growth of hematopoietic progenitors bearing Shp-2 mutations in part by increasing cell proliferation. We also found that GM-CSF had a much more significant effect on macrophage progenitor proliferation than did M-CSF, supporting the hypothesis that Shp-2 gain-of-function mutants preferentially confer hypersensitivity to the GM-CSF signaling pathway. These studies also revealed that hematopoietic cells expressing each of these Shp-2 mutants displayed a delay of macrophage maturation in response to M-CSF based on F4/80 expression. Diminished macrophage differentiation in the context of JMML seems counterintuitive based on the observation that colonies grown in CFU-C assays from the peripheral blood or bone marrow of patients with JMML are composed almost exclusively of monocytes and macrophages (in contrast to colonies derived from healthy individuals, which also contain granulocytic and erythroid elements); however, aberrantly reduced differentiation may account for the increased numbers of immature myeloid cells observed in the tissue and peripheral blood of patients with JMML.27,28 Although this finding needs to be examined further by conducting an in-depth kinetic study of macrophage differentiation using cells expressing each of the Shp-2 mutants, this result implies that an additional characteristic of leukemic cells from patients with JMML may be an arrest or a slowing of macrophage progenitor maturation.
At the molecular level, we examined the effect of each of these Shp-2 mutants on GM-CSF–stimulated phospho-Erk levels, a downstream effector of Ras. We found that each of the Shp-2 mutants produced constitutively elevated phospho-Erk levels following serum and growth factor deprivation as well as sustained levels of phospho-Erk following GM-CSF stimulation. This finding of sustained GM-CSF–induced phospho-Erk levels is consistent with the findings of Zhang et al,29 who found prolonged GM-CSF–stimulated mitogen-activated protein kinase (MAPK) levels in Nf1-/- fetal liver cells as well as with the findings of Tartaglia et al,8 who observed a similar result in epidermal growth factor (EGF)–stimulated fibroblasts expressing Shp-2 mutant proteins. In contrast, Araki et al did not observe increased levels of phospho-Erk in murine embryonic fibroblasts derived from mice bearing either the heterozygous or homozygous Ptpn11 germline mutation D61G in response to EGF.23 It is possible that examination of phospho-Erk levels at time points longer than 20 minutes after EGF stimulation would have revealed differences in the sustainability of phospho-Erk levels between WT and heterozygous and/or homozygous D61G murine embryonic fibroblasts. However, another possibility is that Shp-2 gain-of-function–induced hyperactivation of the Ras signaling cascade may be both cell type and growth factor specific. This possibility is supported by our observation that cells expressing Shp-2 gain-of-function proteins are exquisitely hypersensitive to GM-CSF, but not to M-CSF, as evaluated by proliferation. Taken together, these functional and biochemical studies support the hypothesis that Shp-2 mutants induce hypersensitivity of hematopoietic cells to GM-CSF via hyperactivation of Ras and effector molecules and substantiate the GM-CSF pathway as a tractable target for improved therapies in JMML.
This study also demonstrated that 2 of the most common PTPN11 mutations found in patients with JMML, E76K and D61Y (each constituting approximately 8% of the patients with JMML examined by Tartaglia et al8 ), as well as a less common mutation, D61V, act similarly in inducing the functional abnormality of hematopoietic cell hypersensitivity to GM-CSF. This finding suggests that these mutant Shp-2 molecules recruit similar substrates that integrate at the molecular level to induce hyperactivation of the Ras signaling axis in response to GM-CSF stimulation. As the vast majority of somatic PTPN11 mutations found in leukemic cells reside within the N-terminal–SH2 domain of Shp-2, it is likely that additional PTPN11 mutations will act in a similar fashion. However, a global characterization of the substrates recruited by various activating Shp-2 mutations will allow for improved rational drug design for this devastating disease of childhood.
The historical observation that hematopoietic progenitors from patients with JMML display hypersensitivity to GM-CSF in vitro provides clues to potential interventions at the molecular level that may curtail leukemic cell growth in vivo. Indeed, WT mice reconstituted with hematopoietic cells from Nf1-/- animals develop a myeloproliferative disease similar to JMML,29,30 which is ameliorated by transplanting these cells into mice lacking GM-CSF expression.31 Although the participation of Shp-2 in multiple hematopoietic cytokine and growth factor pathways has been examined,32-36 surprisingly, little is known about the role of Shp-2 in GM-CSF–stimulated signal transduction in hematopoietic cells. Studies have shown that GM-CSF stimulation results in phosphorylation and recruitment of Shp-2 to tyrosines 577, 612, and 695 of the GM-CSF receptor in conjunction with activation of the Ras-mediated MAPK pathway.37 An additional well-studied out-come of GM-CSF receptor stimulation is the activation of JAK2 and the transcription factor STAT538 as well as of the myeloid-specific Src kinases Lyn and Hck.39 The future goals of this field will be directed at dissecting out the interaction of the Shp-2 gain-of-function mutants with these well-described effectors of GM-CSF stimulation using genetic and biochemical models in an effort to define molecular targets that can be blocked pharmacologically to inhibit GM-CSF–stimulated Ras hyperactivation, to specifically target leukemic cell growth, and to provide improved therapies for JMML in children.
Prepublished online as Blood First Edition Paper, January 11, 2005; DOI 10.1182/blood-2004-10-4002.
Supported by the Indiana 21st Century Research and Technology Fund (R.J.C.) and National Institutes of Health grant RO1 HL075816 (R.K.). R.J.C. is a recipient of a March of Dimes Basil O'Connor Starter ScholarAward (5-FY03-136).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Chris Starkey and Lisa Duffy in the Indiana University Vector Production Facility; Susan Rice, Denessa Luckett and Lizz J. Gasyna in the Indiana University Flow Cytometry Facility; Arliene K. Britt for assistance in manuscript preparation; and Wade Clapp for critical review of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal