Abstract
MYH9-related disorders are autosomal dominant syndromes, variably affecting platelet formation, hearing, and kidney function, and result from mutations in the human nonmuscle myosin-IIA heavy chain gene. To understand the mechanisms by which mutations in the rod region disrupt nonmuscle myosin-IIA function, we examined the in vitro behavior of 4 common mutant forms of the rod (R1165C, D1424N, E1841K, and R1933Stop) compared with wild type. We used negative-stain electron microscopy to analyze paracrystal morphology, a model system for the assembly of individual myosin-II molecules into bipolar filaments. Wild-type tail fragments formed ordered paracrystal arrays, whereas mutants formed aberrant aggregates. In mixing experiments, the mutants act dominantly to interfere with the proper assembly of wild type. Using circular dichroism, we find that 2 mutants affect the α-helical coiled-coil structure of individual molecules, and 2 mutants disrupt the lateral associations among individual molecules necessary to form higher-order assemblies, helping explain the dominant effects of these mutants. These results demonstrate that the most common mutations in MYH9, lesions in the rod, cause defects in nonmuscle myosin-IIA assembly. Further, the application of these methods to biochemically characterize rod mutations could be extended to other myosins responsible for disease.
Introduction
The autosomal dominant, giant-platelet disorders—May-Hegglin anomaly (MHA), Fechtner (FTNS), Sebastian (SBS), Epstein (EPS), and Alport-like syndromes with macrothrombocytopenias—result from mutations in the human MYH9 gene, which encodes nonmuscle myosin-IIA heavy chain.1-5 Virtually all patients exhibit platelet macrocytosis, thrombocytopenia, and leukocyte inclusions. Some individuals also exhibit high-tone sensorineural deafness, cataracts, and nephritis to varying degrees. Additional genetic or environmental factors may contribute to the variable penetrance of the nonhematologic clinical manifestations.6,7 A single family with predominantly hearing loss has been reported.8 Together, these illnesses represent a spectrum of disorders collectively termed MYH9-related disorders.6,7,9
Of the 82 mutations identified in MYH9-related disorder families, only 21 amino acids, out of a possible 1961, are mutated.3,7,10 Identical MYH9 mutations occur in unrelated families and can arise spontaneously, suggesting that these disorders result from highly specific disruption of the MYH9 structure and/or function. Mutations at other sites may occur and remain undetected owing to (1) a completely recessive alteration in myosin-II function, (2) a weakly penetrant dominant effect that escapes clinical detection, or (3) severe dysfunction that is incompatible with life.
In humans, the 3 nonmuscle myosin-II heavy chains (IIA, IIB, and IIC) are encoded by distinct genes (MYH9, MYH10, and MYH14, respectively).11 Only nonmuscle myosin-IIA is expressed in platelets,12,13 and no mutations in nonmuscle myosin-IIB or myosin-IIC have been reported. Nonmuscle myosin-II motor proteins are heterohexamers of polypeptides that include 2 heavy chains (approximately 220 kDa) and 2 pairs of light chains (17 and 15 kDa) (Figure 1A). Forty-three percent of the heavy chain consists of an N-terminal head responsible for motor function and a neck that plays a role in light-chain binding, regulation, and chemomechanical force production. The remainder of the heavy chain forms a coiled-coil dimer that characterizes the rodlike, native myosin-II tail.
Schematic of a native nonmuscle myosin-IIA molecule, its assembly steps, and the purified tail fragments used in this study. (A) A schematic of nonmuscle myosin-IIA shows 2 heavy chains (white and black globular and coiled domains) and the essential and regulatory light chains (ELC and RLC are indicted by arrows). An arrow marks the junction (P836) between the motor head domain and the rod. The arrowhead marks the junction between subfragment 2 (S2) and light meromysoin (LMM) of the rod. The bounded arrow indicates the region (amino acids [aa] 1102-1960) of the wild-type rod used in this study, and an asterisk signifies the relative position of the 4 mutations studied (R1165C, D1424N, E1841K, R1933Stop, reading from left to right). (B) Assembly steps of nonmuscle myosin-IIA. The rod of individual heavy chains fold into an α-helical structure that dimerizes with another heavy chain, forming a coiled coil. A cross-section through a coiled coil, appearing below the diagram, shows the relationship of amino acids in the characteristic heptad repeat. To form higher order assemblies, such as contractile filaments, dimers must laterally associate with one another in proper register. Periodic charged regions along the rod play important roles during this alignment. The α-helix and coiled-coil diagrams are reprinted from Figures 3-6 and 3-9 of Lodish et al14 with permission. (C) Here, 5 μg each recombinantly purified protein were subjected to electrophoresis on an acrylamide gel and Coomassie stained (lane 1 indicates wild type; lane 2, R1165C; lane 3, D1424N; lane 4, E1841K; and lane 5, R1933Stop). Two to 3 minor bands, probably breakdown products, can be seen in each lane.
Schematic of a native nonmuscle myosin-IIA molecule, its assembly steps, and the purified tail fragments used in this study. (A) A schematic of nonmuscle myosin-IIA shows 2 heavy chains (white and black globular and coiled domains) and the essential and regulatory light chains (ELC and RLC are indicted by arrows). An arrow marks the junction (P836) between the motor head domain and the rod. The arrowhead marks the junction between subfragment 2 (S2) and light meromysoin (LMM) of the rod. The bounded arrow indicates the region (amino acids [aa] 1102-1960) of the wild-type rod used in this study, and an asterisk signifies the relative position of the 4 mutations studied (R1165C, D1424N, E1841K, R1933Stop, reading from left to right). (B) Assembly steps of nonmuscle myosin-IIA. The rod of individual heavy chains fold into an α-helical structure that dimerizes with another heavy chain, forming a coiled coil. A cross-section through a coiled coil, appearing below the diagram, shows the relationship of amino acids in the characteristic heptad repeat. To form higher order assemblies, such as contractile filaments, dimers must laterally associate with one another in proper register. Periodic charged regions along the rod play important roles during this alignment. The α-helix and coiled-coil diagrams are reprinted from Figures 3-6 and 3-9 of Lodish et al14 with permission. (C) Here, 5 μg each recombinantly purified protein were subjected to electrophoresis on an acrylamide gel and Coomassie stained (lane 1 indicates wild type; lane 2, R1165C; lane 3, D1424N; lane 4, E1841K; and lane 5, R1933Stop). Two to 3 minor bands, probably breakdown products, can be seen in each lane.
To understand how rod mutations dominantly impair platelet formation and function, it is important to understand nonmuscle myosin-II assembly. To participate in contractility, nonmuscle myosin-IIs must successfully assemble into functional bipolar filaments. This depends on structural considerations that can be conceptually divided into at least 2 discrete steps (Figure 1B). First, individual heavy chains must dimerize by correctly folding into an α-helical coiled-coil structure. Dimerization occurs when 2 α-helices, each formed through hydrogen bonding between polypeptide backbone amide hydrogens and carbonyl oxygens, wrap around one another. The α-helices of coiled-coil proteins are characterized by a heptad repeat pattern of residues (a, b, c, d, e, f, g) with similar chemical properties, allowing for a buried hydrophobic seam (a, d residues) to form along the coiled-coil interface.15,16 Residues in the e and g positions play electrostatic roles to help hold the coiled coil together.
The second assembly step involves lateral associations of heavy-chain dimers, in proper register and orientation, to form functional bipolar filaments (Figure 1B). The α-helical coiled-coil rod is divided into 2 regions by a short stretch of residues, or hinge. A head-distal, light meromysoin (LMM) fragment self-associates to form the core of the bipolar filament and a head-proximal fragment, subfragment 2 (S2), which allows the myosin-II head to swing out from the bipolar filament backbone. Sequences near the carboxy-terminus of LMM (ie, the globular tail piece) and the periodic distribution of positively and negatively charged zones along the length of LMM, primarily at the b, c, and f heptad positions in 28-residue periods (ie, 4 heptad repeats), provide electrostatic interactions that specify self-assembly and alignment of dimers to form a filament.17-20 In principle, defects in either step, coiled-coil dimer formation or assembly of myosin-II dimers in proper register, could result in the inability to form functional nonmuscle myosin-IIA bipolar filaments.
The size and shape of myosin-II bipolar filaments depend on their origin. Nonmuscle myosin-IIA from human platelets forms small bipolar filaments (approximately 320 nm long, 10 to 11 nm wide in the center of the bar zone, and with each filament consisting of approximately 28 individual myosin-II molecules)21 that are comparable in size to filaments formed by myosin-IIs from other nonmuscle sources.22-24 Regardless of their source or size, bipolar filaments are characterized by self-assembly into multistart helical arrays with heads at either end, a helical repeat of approximately 43 nm, and an axial translation of 14.3 nm between layers of the heads. Self-assembly is reversible and salt-dependent. Filaments form at physiologic or low-salt conditions and are sedimentable at 100 000g. In contrast, in high salt they dissolve into individual myosin-II molecules that are soluble. As a consequence, by determining the solubility over a range of salt concentrations, one can generate solubility curves to provide a quantitative analysis of the fraction of myosin-II in higher-order assemblies (filaments, paracrystals, or aggregates).25-28
Self-assembly of myosin-II into bipolar filaments requires full-length (or nearly full-length) myosin, even though assembly is specified and mediated by sequences in the myosin-II tail. Nevertheless, appropriate myosin-II tail fragments readily assemble into large paracrystaline arrays showing the same 43 nm and 14.3 nm repeats and low-salt dependence as filaments, indicating that the molecular interactions that drive assembly of bipolar filaments also specify paracrystal assembly.29 Presumably, steric constraints, provided by the bulky myosin-II heads, prevent the formation of paracrystals by native (intact) myosin-II molecules. Because native, full-length myosin-IIs are difficult or impossible to make in recombinant systems, analysis of paracrystal formation is an accepted and proven model system for the assembly of myosin-IIs.29 Paracrystal assembly by myosin-II fragments produced in Escherichia coli or in Spodoptera frugiperda–9 (SF-9) cells has been used to characterize the portions of the tail required for assembly27,28,30 and the influence of naturally occurring splice-form variants of the smooth-muscle myosin-II tail on assembly.29
The biochemical mechanisms by which mutations in MYH9 disrupt myosin-II function are incompletely understood. Recently, Hu et al31 used recombinant approaches to examine the effects of mutations in the MYH9 head (N93K and R702C) and found both mutations impaired enzymatic activity, significantly reducing the mutant protein's ability to translocate actin filaments in vitro compared with wild type. Interestingly, about 75% of families with MYH9-related disorders have mutations in the rod of MYH9, and 88% of these mutations occur at only 4 residues (1165, 1424, 1841, and 1933), fewer than 1% of possible residues, suggesting that these mutations disrupt the rod structure in very specific ways. Several groups have suggested these mutations probably interfere with some aspect of assembly,1,10,32 and one paper suggests that, for the D1424N mutation, haploinsufficiency is the cause.33
We chose to study 4 of the most common rod mutations, R1165C, D1424N, E1841K, and R1933Stop, which have been identified in 5, 11, 20, and 13 independent families, respectively, and are responsible for 59% of all MYH9 families (F.D., Sufong Li, Núria Pujol-Moix, et al, manuscript submitted, October 2004). We find that all mutants form dimers, but the dimers do not form wild-type myosin-II assemblies and, indeed, interfere with the capability of wild-type protein to assemble. Thus, we can biochemically explain the dominant behavior of these mutations. Two mutants have altered coiled-coil structures and perturb lateral associations required for assembly, whereas the other 2 mutants only perturb lateral associations. Together, these results establish the biochemical mechanisms by which rod mutations disrupt nonmuscle myosin-IIA function to cause MYH9-related disorders.
Materials and methods
Tail-fragment constructs
A wild-type MYH9 tail construct was cloned in 3 separate pieces from first-strand cDNA synthesis with the use of random primers and mRNA from an Epstein-Barr virus (EBV)–transformed lymphoblastoid cell line established from an individual without an MYH9 mutation. Piece 1 (bp 3304-4038 of GenBank accession no. NM002473.2) was amplified by polymerase chain reaction (PCR) with a 5′ BamH1 site and an endogenous 3′ EcoR1 site; piece 2 (bp 4008-5171) was amplified by PCR to contain endogenous 5′ EcoR1 and 3′ SacII restriction sites; and piece 3 (base pair [bp] 5045-5883) was amplified by PCR with the endogenous 5′ SacII and a 3′ HindIII restriction sites. Each was separately cloned into the pSTBlue-1 vector (Novagen brand; EMD Biosciences, Madison, WI) and sequenced. Site-directed mutagenesis was then performed by means of QuikChange (Stratagene, La Jolla, CA) to make the R1165C, D1424N, and E1841K mutations in the appropriate pieces. For the R1933Stop mutation, a separate piece 3 was amplified from cDNA encompassing bp 4038-5797 followed by an inframe stop codon. Piece 2 was digested and ligated into the vector containing piece 3 with the use of EcoR1 and SacII. Next, piece 1 was digested and added to the vector containing both piece 2 and piece 3 with the use of BamH1 and EcoR1. This resulted in a final assembled tail region (bp 3304-5883, aa 1102-1961) in pSTBlue-1 containing the wild type or the respective mutations. Each was directly subcloned into the pMW172 bacterial expression vector26,34 by digesting with BamH1 and HindIII. Positive clones were verified by complete sequencing of the assembled MYH9 fragment.
Purification of wild-type and mutant tail-fragment proteins
Wild-type and mutant plasmids were transformed into BL-21 cells, and purification was performed as described previously.26 For salt-dependent solubility curves and electron microscopy (EM) studies, the BioRad DC Protein Assay (Bio-Rad Laboratories, Mississauga, ON, Canada) was used to estimate the concentration of each mutant tail fragment with the use of bovine serum albumin as a standard.
Salt-dependent solubility assays
First, 100 μg protein (wild type and each mutant tail) was dialyzed into buffer (10 mM imidazole-Cl, pH 6.8) containing various salt concentrations between 50 mM and 500 mM NaCl. Two 10-μL aliquots were removed; the total protein concentration was measured and averaged. The remainder was centrifuged at 100 000g for 50 minutes. Two 10-μL aliquots of the supernatant were removed; their protein content determined and averaged, and then the percentage of soluble protein was calculated.
Negative staining of paracrystals for electron microscopy
Tail proteins (1 mg/mL) were dialyzed from a high-salt buffer (10 mM imidazole-Cl, pH 6.8; 600 mM NaCl, and 1 mM EDTA [ethylenediaminetetraacetic acid]) into a low-salt buffer (the same as for high-salt buffer but with 10 mM NaCl), adjusted to 0.5 mg/mL, and then prepared for EM as described elsewhere.35,36 A Phillips 301 microscope, calibrated by means of a diffraction grating no. 10 002 of 2160 lines per mm (Ernest F. Fullam, Latham, NY), was used to analyze grids. Micrographs were obtained at a magnification of 31 000× or 41 000× using a Philips 301 electron microscope (Philips Electron Optics, Eindhoven, the Netherlands). After development of 8.3 cm × 10.2 cm film, negatives were scanned and imported into Adobe Photoshop (Adobe, San Jose, CA). For mixing experiments, equal amounts (100 μg) of wild-type tail fragment and each mutant were mixed and treated as described for each individual tail fragment. Periodicity was determined by counting the total number of periods, to within half a period, over a set length.
Circular dichroism analysis and melting temperature determination
Wild-type and mutant tail fragments were dialyzed into 20 mM NaPO4, pH 7.0, and 600 mM NaCl, adjusted to approximately 0.2 μM total protein, and then degassed. Circular dichroism (CD) was measured with an Aviv circular dichroism spectrometer model 202 (Proterion, Piscataway, NJ), at 10°C by stepping in 1-nm increments from 250 nm to 200 nm and integrating for 10 seconds at each wavelength. Thermal melts began at 10°C and moved to 90°C by 1° steps, allowing 1 minute for temperature equilibration before a 30-second measurement at 222 nm. Data conversion and analysis were performed by means of Kaleidagraph (Synergy Software, Reading, PA). TCEP (Tris(2-carboxyethyl)phophine-Cl [tris(hydroxymethyl)aminomethane(2-carboxyethyl)phophine-Cl]) was purchased from Sigma (St Louis, MO).
Results
Full-length myosin-II heavy chain, for biochemical analysis, is very difficult to obtain in sufficient quantities from recombinant systems.29 Therefore, we cloned sequences encoding 76% of the wild-type rod (amino acid [aa] 1102-1960), which spans approximately 100 amino acids of S2 and the entire LMM (hereafter referred to as “tail fragment”) (Figure 1A). We used site-directed mutagenesis to create 4 of the most prevalent MYH9 rod mutations. All tail fragments were soluble in E coli and when purified had the expected electrophoretic mobility of approximately 90 kDa (Figure 1C). Our ability to purify each using standard LMM purification protocols that entail low-salt–dependent precipitation, high-salt–dependent solubility, and ethanol precipitation26,37 strongly suggests that all tail fragments formed expected α-helical coiled-coil dimers.
Rod mutations cause defects in assembly and morphology
To investigate the effect of rod mutations on nonmuscle myosin-II assembly, we used electron microscopy (EM) to analyze paracrystals of tail fragments formed at low salt concentrations (Figure 2).29 The wild-type tail fragment (Figure 2A) formed large, well-ordered paracrystals with uniform periodicity (14.3 ± 0.4 nm for n = 11 paracrystals). This value is identical to the periodicity for the LMM (approximately 14.3) of other myosin-IIs38 and corresponds to the helical periodicity of full-length myosin-II packed into bipolar filaments (see “Introduction”).
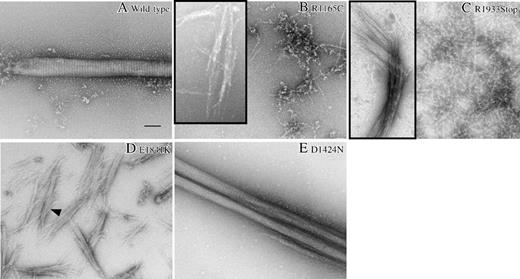
Negative staining and electron microscopy of MYH9 mutant tail fragments. Negative staining and electron microscopy reveal aberrant paracrystal morphology in MYH9 mutant tail fragments. The insets show extremely rare examples and represent the best structures observed. The scale bar is 100 nm. (A) The wild-type tail fragment formed paracrystals with expected morphology. They were large and well-ordered and had uniform periodicity throughout the paracrystal. (B) The R1165C tail fragment is unable to form paracrystals or any other type of large assembly. Instead, we found chainlike structures in a random array. On extremely rare occasions, we found faint examples of assemblies (shown in the inset), but they lacked size, uniformity, and periodicity. (C) The R1933Stop tail fragment predominantly formed chainlike structures similar to R1165C; however, a few paracrystallike assemblies were observed (inset). These lacked size, uniformity, and periodicity. (D) For E1841K, chainlike structures were often observed; however, we also found paracrystals with very different morphology compared with wild-type. Not only are these structures very short in length, but they appear to fray apart at the ends. Periodicity can be found only in the central region of some paracrystals (arrowhead). (E) The D1424N tail fragment assembled into paracrystals (the best example is shown), which were wild type in size, but had altered periodicity.
Negative staining and electron microscopy of MYH9 mutant tail fragments. Negative staining and electron microscopy reveal aberrant paracrystal morphology in MYH9 mutant tail fragments. The insets show extremely rare examples and represent the best structures observed. The scale bar is 100 nm. (A) The wild-type tail fragment formed paracrystals with expected morphology. They were large and well-ordered and had uniform periodicity throughout the paracrystal. (B) The R1165C tail fragment is unable to form paracrystals or any other type of large assembly. Instead, we found chainlike structures in a random array. On extremely rare occasions, we found faint examples of assemblies (shown in the inset), but they lacked size, uniformity, and periodicity. (C) The R1933Stop tail fragment predominantly formed chainlike structures similar to R1165C; however, a few paracrystallike assemblies were observed (inset). These lacked size, uniformity, and periodicity. (D) For E1841K, chainlike structures were often observed; however, we also found paracrystals with very different morphology compared with wild-type. Not only are these structures very short in length, but they appear to fray apart at the ends. Periodicity can be found only in the central region of some paracrystals (arrowhead). (E) The D1424N tail fragment assembled into paracrystals (the best example is shown), which were wild type in size, but had altered periodicity.
The assemblies formed by 3 of the 4 mutant proteins were very different in their morphology. Each will be discussed here, and in subsequent figures, in the order of severity of the paracrystal aberrations. R1165C (Figure 2B), did not form well-ordered assemblies, only amorphous chains, random lattices, or mesh-works. R1933Stop (Figure 2C) assembled into chainlike lattices; only rarely were there higher-ordered structures (Figure 2C, inset). E1841K (Figure 2D) formed small numbers of paracrystals having very altered morphology. D1424N (Figure 2E) formed large paracrystaline assemblies, but periodicity was typically difficult to observe and increased compared with wild type: in only 4 of 11 paracrystals could periodicity be determined (16.5 ± 0.5 nm for n = 4; the best example is shown). No overlap in the measured periodicities between D1424N and wild type were observed. By t test, this difference is significant (P < .0001).
Since MYH9-related disorders are dominant, we examined the effect of mutant protein on the assembly of wild-type tail fragments. Equal amounts of wild-type and each mutant tail fragment were mixed at high salt and then dialyzed into low salt to induce paracrystal formation (Figure 3). If mutant dimers failed to interact with wild type (ie, had no dominant activity), we would expect to see both wild-type paracrystals and aggregates characteristic of each mutation. If mutant dimers had dominant effects on assembly, then wild-type paracrystals should be observed only rarely, if at all.
Mixing wild-type and mutant tail fragments. Mixing wild-type and mutant tail fragments demonstrates these mutations have a dominant activity. First, 100 μg wild-type and each mutant tail fragment were mixed in high salt and then dialyzed into low salt to induce paracrystal formation. Scale bar = 100 nm. (A) Wild-type tail fragment alone formed large well-ordered paracrystals with uniform periodicity. (B) Mixing R1165C with wild type, we found a large number of chainlike structures. Occasionally, small assemblies were observed (inset); however, they lacked size and organization. (C) Mixing R1933Stop with wild type, we typically found chainlike structures; however, on rare occasions we observed assemblies. These structures were short with little to no sign of periodicity. (D) Mixing E1841K with wild type, we sometimes found chainlike structures but most often found very short paracrystals with frayed ends—similar to E1841K alone. (E) Mixing D1424N with wild type, we routinely found paracrystals with morphology similar to that of wild type. Periodicity was normal; however, the mixed paracrystals appeared narrower than wild-type ones. Note that this image shows 2 or 3 paracrystals lying next to one another.
Mixing wild-type and mutant tail fragments. Mixing wild-type and mutant tail fragments demonstrates these mutations have a dominant activity. First, 100 μg wild-type and each mutant tail fragment were mixed in high salt and then dialyzed into low salt to induce paracrystal formation. Scale bar = 100 nm. (A) Wild-type tail fragment alone formed large well-ordered paracrystals with uniform periodicity. (B) Mixing R1165C with wild type, we found a large number of chainlike structures. Occasionally, small assemblies were observed (inset); however, they lacked size and organization. (C) Mixing R1933Stop with wild type, we typically found chainlike structures; however, on rare occasions we observed assemblies. These structures were short with little to no sign of periodicity. (D) Mixing E1841K with wild type, we sometimes found chainlike structures but most often found very short paracrystals with frayed ends—similar to E1841K alone. (E) Mixing D1424N with wild type, we routinely found paracrystals with morphology similar to that of wild type. Periodicity was normal; however, the mixed paracrystals appeared narrower than wild-type ones. Note that this image shows 2 or 3 paracrystals lying next to one another.
Wild-type tail fragment alone (Figures 2A and 3A) formed large, well-ordered paracrystals with uniform periodicity. When R1165C was mixed with wild type (Figure 3B), chainlike structures very similar to R1165C alone were most frequently observed. Small assemblies were observed but lacked size and organization (Figure 3B, inset). When R1933Stop was mixed with wild type (Figure 3C), chainlike structures, similar to the mutant alone, were predominantly observed. Some small assemblies were found, but their morphology was very different from wild type. When E1841K was mixed with wild type (Figure 3D), we observed small assemblies similar to E1841K alone. These structures were small, had frayed ends, and showed only rare signs of periodicity. When D1424N (Figure 3E) was mixed with wild type, paracrystal morphologies similar to wild type were observed. While these paracrystals had essentially wild-type periodicity (14.2 ± 0.3 nm for n = 6 paracrystals), we note that these paracrystals were generally more narrow than wild type. Overall, the morphology of “mixed” assemblies were very similar to the morphology of the respective mutant protein alone (Figure 2), indicating that each mutant appears to disrupt formation of wild-type paracrystals, thereby functioning with a dominant biochemical effect.
Salt-dependent solubility of wild-type and mutant tail fragments
Next, we tested each mutant protein's solubility as a function of salt concentration in standard sedimentation assays (Figure 4).19,25,28 None of the mutant myosin tail fragments had a major effect on the salt-dependent solubility, indicating that in each case dimers form into sedimentable complexes. Moreover, this demonstrates that even the chainlike structures sedimented, consistent with other studies that show mutant myosin tail proteins with altered paracrystal morphology had salt-dependent solubilities similar to those of their wild-type counterparts.19 Thus, while in principle, reversible, salt-dependent sedimentation can quantitatively probe the ionic interactions that drive myosin assembly, it cannot be used to distinguish among the varied supramolecular complexes that myosin can form. Nevertheless, the curves for R1165C (Figure 4A) and E1841K (Figure 4C) showed minor, yet reproducible, shifts in solubility that we address in “Discussion.”
Salt-dependent assembly curves of tail fragments. Salt-dependent assembly curves of mutant tail fragments show some variability compared with wild type. First, 100 μg each tail fragment were dialyzed into different NaCl concentrations, ranging from 50 mM to 500 mM (x-axis). After dialysis, the percent solubility (y-axis) of each protein at each salt concentration was determined. Solubility curves were generated for each mutant tail (solid line with squares) compared with wild type (dashed line with circles). Lines shown are the averaged values with dots representing the experimental values. The R1165C mutant protein (panel A) had higher solubility through the physiologic salt concentrations and then became similar to wild type. The E1841K mutant protein (panel C) appeared less soluble than wild type throughout most concentrations tested. The D1424N (panel D) and R1933Stop (panel B) mutant proteins had very similar solubility profiles throughout most concentrations.
Salt-dependent assembly curves of tail fragments. Salt-dependent assembly curves of mutant tail fragments show some variability compared with wild type. First, 100 μg each tail fragment were dialyzed into different NaCl concentrations, ranging from 50 mM to 500 mM (x-axis). After dialysis, the percent solubility (y-axis) of each protein at each salt concentration was determined. Solubility curves were generated for each mutant tail (solid line with squares) compared with wild type (dashed line with circles). Lines shown are the averaged values with dots representing the experimental values. The R1165C mutant protein (panel A) had higher solubility through the physiologic salt concentrations and then became similar to wild type. The E1841K mutant protein (panel C) appeared less soluble than wild type throughout most concentrations tested. The D1424N (panel D) and R1933Stop (panel B) mutant proteins had very similar solubility profiles throughout most concentrations.
Determination of coiled-coil structure and melting temperature
Given that the mutants formed dimers, we considered 2 different mechanisms by which each mutant could interfere with myosin-II assembly, as observed in our EM analysis. First, mutants might alter the coiled-coil structure locally. Such structural defects in individual nonmuscle myosin-IIA molecules could result in the inability to pack with one another to form higher-order assemblies. Second, mutants may leave the coiled-coil structure essentially intact, but cause disruptions by either perturbing formation of lateral associations or, if these associations do occur, altering the axial relationship between adjacent dimers in higher-ordered assemblies.
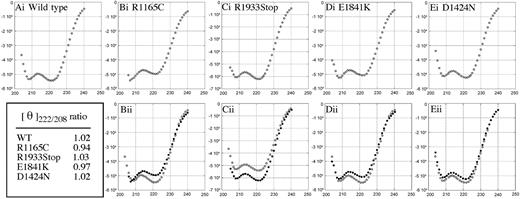
To distinguish between these 2 possibilities, we used CD spectroscopy39 under high-salt conditions where only dimers are present. Proteins with α-helical coiled-coil regions, like the tail of myosin-II, have characteristic minima at 222 nm and 208 nm, the ratio of which (θ222/208) can be used to determine if a protein is coiled coil (θ222/208 = 1.02), versus purely α-helical (θ222/208 = 0.85). If the ratio is below 1.0, then some loss in coiled-coil structure has occurred.40-42 We analyzed the spectra (240 nm to 200 nm) for each tail fragment. The spectrum for wild type (Figure 5A) showed the expected 222 nm and 208 nm minima and had a molar ellipticity ratio (θ222/208) of 1.02 (Figure 5 table). This same spectral pattern was observed for R1933Stop (Figure 5Ci) and D1424N (Figure 5Ei), and both had molar ellipticity ratios similar to wild type, indicating no alteration in the coiled-coil structure. Interestingly, the R1165C (Figure 5Bi) and E1841K (Figure 5Di) mutations showed spectra altered in such a way that the 208 nm minima was greater than the 222 nm (molar ellipticity ratios were 0.94 and 0.97, respectively; Figure 5 table), indicating a partial loss of coiled-coil structure.40,42,43 Overlays of each mutant spectra with wild type highlight these differences (Figure 5 Bii,Cii,Dii,Eii).
CD spectra analysis. CD spectra demonstrate that the R1165C and E1841K mutations decrease the coiled-coil structure of the tail. Spectra from 240 nm to 200 nm (x-axis) were taken for all samples and were plotted versus their mean residue ellipticity (degrees cm2 dmole–1) (y-axis). All show the characteristic pattern (minima at 222 nm and 208 nm) for helical proteins, but not all show the characteristic pattern for coiled-coil proteins (222 nm minima is greater than 208 nm minima). The wild-type tail fragment (panel Ai) showed a spectrum very typical of coiled-coil proteins as did R1933Stop (Ci) and D1424N (Ei). Two mutations R1165C (Bi) and E1841K (Di) have altered CD spectra (208 nm is greater than 222 nm) indicating a partial loss of coiled-coil structure. Overlays of each mutant (dark circles) compared with wild type are also shown: WT versus R1165C (Bii), WT versus R1933Stop (Cii), WT versus E1841K (Dii), and WT versus R1933Stop (Eii). The ratios of molar ellipticity at 222 nm to 208 were calculated and tabulated. For completeness and accuracy, only CD spectra data points with a dynode voltage lower than 600 V are shown.
CD spectra analysis. CD spectra demonstrate that the R1165C and E1841K mutations decrease the coiled-coil structure of the tail. Spectra from 240 nm to 200 nm (x-axis) were taken for all samples and were plotted versus their mean residue ellipticity (degrees cm2 dmole–1) (y-axis). All show the characteristic pattern (minima at 222 nm and 208 nm) for helical proteins, but not all show the characteristic pattern for coiled-coil proteins (222 nm minima is greater than 208 nm minima). The wild-type tail fragment (panel Ai) showed a spectrum very typical of coiled-coil proteins as did R1933Stop (Ci) and D1424N (Ei). Two mutations R1165C (Bi) and E1841K (Di) have altered CD spectra (208 nm is greater than 222 nm) indicating a partial loss of coiled-coil structure. Overlays of each mutant (dark circles) compared with wild type are also shown: WT versus R1165C (Bii), WT versus R1933Stop (Cii), WT versus E1841K (Dii), and WT versus R1933Stop (Eii). The ratios of molar ellipticity at 222 nm to 208 were calculated and tabulated. For completeness and accuracy, only CD spectra data points with a dynode voltage lower than 600 V are shown.
To evaluate whether any mutant affected the global stability of the tail fragment (unlikely, since the mutant tail fragments were purified through cycles of salt-dependent solubility), we determined the transition from a fully folded, native state to a fully denatured state as a function of temperature for each mutant under high-salt conditions (Figure 6). All the mutations had melting temperatures very similar to wild type (45.2°C), except for R1165C (49.9°C), indicating most mutations do not affect the overall stability of the tail (Figure 6). The increase in melting temperature for R1165C suggested a stabilization of the tail fragment. Mutation to a cysteine could cause the formation of a disulfide bond. Experimentally, disulfide bonds within coiled-coil proteins can occur and often dramatically stabilize structure.44 However, owing to the reducing environment of the cytoplasm, in vivo disulfide bonds are expected to be rare. We added TCEP to reduce the cysteines and performed an additional thermal melt. As with most reducing agents, TCEP interferes with CD measurements at high concentrations, so we used concentrations unlikely to completely reduce cysteines. The melting temperature decreased (R1165Cred = 47.9°C), suggesting that a disulfide bond is, at least partially, responsible for the increased stability. We hypothesize that in a cellular environment with fully reduced cysteines, this mutation is unlikely to confer additional stability.
Thermal melt analysis of tail fragments. Thermal melt analysis shows all tail fragments, except R1165C, have stability similar to that of wild type. The melting temperature for each tail fragment was determined by CD monitoring at 222 nm as the temperature was raised from 10°C to 80°C. Shown are the data points for each tail fragment along with the curve used to calculate the melting temperature. Mean residue ellipticity (degrees cm2 dmol–1) was plotted versus temperature (x-axis), and a best-fit equation was generated. The midpoint of the transition was then determined by this equation, and a vertical dashed line was added.
Thermal melt analysis of tail fragments. Thermal melt analysis shows all tail fragments, except R1165C, have stability similar to that of wild type. The melting temperature for each tail fragment was determined by CD monitoring at 222 nm as the temperature was raised from 10°C to 80°C. Shown are the data points for each tail fragment along with the curve used to calculate the melting temperature. Mean residue ellipticity (degrees cm2 dmol–1) was plotted versus temperature (x-axis), and a best-fit equation was generated. The midpoint of the transition was then determined by this equation, and a vertical dashed line was added.
Discussion
Mutations perturb different aspects of myosin-II assembly
Nearly 90% of rod mutations associated with MYH9-related disorders occur at only 4 residues, suggesting these amino acids play special and essential roles in the tail structure and function. We focused on investigating the biochemical properties of 4 of the most common mutations compared with wild type. Nonmuscle myosin-II assembly involves at least 2 steps: dimerization of 2 α-helices to form a coiled-coil rod structure, and lateral associations, in proper register, of the coiled coils to form a functional filament. We find all 4 mutant proteins fail to form wild-type assemblies and interfere with the ability of wild-type tail fragments to assemble. Interestingly, we find mutation-specific alterations that can affect either step of assembly. Our experimental findings for each mutant, their relative position in the heptad repeat, and previously reported clinical manifestations are summarized in Table 1.
R1165C. The R1165C mutation disrupts assembly (assayed by EM), apparently by causing defects in the tail's coiled-coil structure (assayed by CD spectroscopy). Given that the R1165 amino acid lies in the d position of its heptad repeat, along the dimer interface, this is not surprising. It is interesting that an arginine (large, positively charged side chain), rather than a hydrophobic amino acid, lies in the d position in wild type (Figure 1). In patients with MYH9-related disorders, 2 distinct mutations at aa 1165 have been observed: one replaces arginine with cysteine (studied here), the other with leucine. Surprisingly, each “mutant” is expected to support coiled-coil formation better than the wild-type arginine.45 We suggest that the arginine at this position plays a role in interactions other than hydrophobicity: amino acids at some a or d positions could form interhelical salt bridges to help stabilize coiled-coil structures. Such salt bridges were found in the crystal structure of cortexilin I, including arginine residues in the d position forming interhelical salt bridges with glutamate residues that follow.46 The arginine at 1165 in MYH9 is also followed by a glutamate, suggesting that an interhelical salt bridge between R1165 and E1166 may be required for native tail structure. Mutations that remove the positively charged arginine (or the glutamate that follows) would remove this salt bridge and potentially destabilize the coiled coil. The small increase in solubility that we observe as a function of salt concentration is consistent with this interpretation. Our EM studies showed this mutant lacked the ability to form paracrystals, and dominantly perturbed paracrystal formation when mixed with wild type. Overall, our in vitro data suggest that in vivo, this mutation encodes a protein that retains sufficient coiled-coil structure to interact with wild type, but that the mutant dimers inhibit the formation of wild-type filaments.
R1933Stop. The R1933Stop truncation mutation removes a region of the globular tail essential for proper assembly. As this truncation mutation lies outside the heptad repeats, it is not surprising the CD spectrum was similar to that of wild type. By EM, these mutant tails failed to form wild-type assemblies; only chainlike structures were found. When these mutants were mixed with wild type, chainlike structures were predominantly observed, while small, frayed structures were occasionally seen. Four other mutations that perturb the globular tail through frameshift and truncation cause MYH9-related disorders (5774delA, 5779delC, 5825delG, E1945Stop). The inability to assemble, while leaving the coiled-coil region intact, suggests that the last 28 amino acids of the small globular tail are necessary for correct self-assembly into ordered structures. This agrees with studies on other myosin-IIs that show the importance of this region.27 While the precise role of these amino acids is unclear, our results suggest that this portion of the globular tail is required for the proper self-assembly and alignment of individual nonmuscle myosin-IIA molecules into functional filaments in vivo.
E1841K. The E1841K mutation causes local defects in the tail's coiled-coil structure (CD analysis) that disrupt assembly of the malformed E1841K dimers into wild-type structures (EM analysis). Interestingly, defects in coiled-coil structure were not apparent by prediction algorithms.47,48 The E1841K mutation lies in the e position of its heptad repeat, which can contribute to attractive electrostatic interactions between the 2 helices. Substituting a negatively charged glutamate for a positively charged lysine is expected to have dramatic effects on coiled-coil structure.40 The amino acid in the g position of this heptad is another lysine (aa1843); therefore, in the mutant, 2 positive charges are opposite one another. Zhou et al40 showed that lysine residues in the e and g position of the same heptad caused some loss of coiled-coil structure in model polypeptides. To our surprise, the salt-dependent solubility of this mutant protein suggested that the mutant structure is more stable (ie, requires higher salt to solubilize) than wild type. We are forced to conclude that this e position is affecting coiled-coil directions between more than expected, and further analysis will be required to fully understand the mechanisms by which this mutant influences salt-dependent solubility. By EM, E1841K was sometimes capable of forming paracrystallike structures, although morphologically they were very short, had frayed ends, and showed only slight signs of periodicity. Similar structures were observed when mutant was mixed with wild type, demonstrating a dominant activity. Overall, our results suggest this mutation retains sufficient coiled-coil structure to interact with wild type, but its coiled-coil structure is perturbed and prevents the assembly of wild-type filaments in vivo, similar to what happens with the R1165C mutation.
D1424N. By EM, D1424N had the least effect on tail fragment assembly. No defects in coiled-coil structure (CD spectroscopy) were observed. This is consistent with the location of the mutation in the c position of its heptad repeat, an exposed region of the helix generally not important for coiled-coil formation. By EM, D1424N formed paracrystals, but they had altered periodicity. Wild-type tail fragment protein mixed with mutant protein had normal periodicity, but the resulting paracrystals appeared more narrow than wild type alone, indicating some dominant effect of the mutant on structure. In MYH9, this residue lies in the c position of the second heptad in its 28-residue repeat. These charge repeats span the LMM (“Introduction”; Figure 1) and play important roles in correct alignment and register of dimers into paracrystals or filaments.17,18 Typically, the c residues in the second and third heptad of these 28-residue repeats are negatively charged.18 In other studies, alteration of these charged patterns also allowed paracrystals to form; however, individual dimers were not held in proper register and periodicity was altered,19 defects similar to D1424N. Indeed, the D1424N mutation changes this conserved negative charge to an uncharged asparagine. Two other mutations that replace asparagine at aa1424, tyrosine and histidine, have been identified in patients with MYH9-related disorder. These 3 mutations strongly suggest that a key feature of residue 1424 is its negative charge, which plays a crucial role in the correct alignment, or registry, of individual nonmuscle myosin-IIA molecules. We propose that the loss of negative charge at 1424 perturbs the correct alignment of individual molecules that drives assembly of filaments in vivo.
Deutsch et al33 recently reported the D1424N mutation results in an unstable protein causing a approximately 50% decrease in the total amount of nonmuscle myosin-IIA in platelets. They propose that this decrease indicates haploinsufficiency is responsible for May Hegglin/Fechtner syndromes. Our study does not address the total amount of mutant D1424N protein in vivo, but does suggest that any remaining protein could serve to poison wild-type myosin-IIA function. Additionally, Deutsch et al did not look at MYH9 levels in megakaryocytes; therefore, wild-type levels of myosin-IIA (equal amounts of wild-type and mutant protein) might be present in these cells, but the mutant protein fails to segregate into mature platelets as they form.
Myosin-II mutations and human pathology
How perturbation of nonmuscle myosin-IIA function causes macrocytosis, thrombocytopenia, and leukocyte inclusions remains unclear. Megakaryocyte cells, found in bone marrow, fragment to form hundreds of platelets.49,50 Initially, long, tubelike extensions of the megakaryocyte cytoplasm, called proplatelets, form, requiring the microtubule cytoskeleton.49 Proplatelets then bend and bifurcate, through a completely actin-dependent process, to form branched structures from which new processes can extend. Next, proplatelets form constrictions along their length. Soon afterwards, the megakaryocyte cytoplasm retracts, releasing proplatelets, which may fragment further into platelets.49 It is likely that the proplatelet constrictions are functionally analogous to the cleavage furrows that form during cytokinesis, a process known to require myosin-II function. One attractive possibility is that perturbation of nonmuscle myosin-IIA contractility results in fewer, or reduced, constriction events causing aberrant fragmentation and resulting in reduced and enlarged platelets.
Defects in megakaryocyte fragmentation may not fully explain all the hematologic characteristics. MYH9-mutated proteins may also directly perturb platelet functions. Platelets undergo dynamic cell-shape changes, requiring cytoskeletal reorganization, from discoid to spheroid shape on activation.51 Several reports have implicated nonmuscle myosin-IIA contractility via phosphorylation of its regulatory light chain52-54 and functional RhoA,55,56 an upstream activator nonmuscle myosin-II, in these platelet cell shape changes. Therefore, it is likely that dominant MYH9 mutations cause a loss of myosin-II contractility in platelets, which results in a reduced ability to properly change shape and function normally.
Several groups have characterized the leukocyte inclusions from patients with MYH9-related disorders, demonstrating that inclusions are not membrane bound and consist of ribosomes and microfilaments.57-59 Recently, immunofluorescence and EM studies showed the distribution of nonmuscle myosin-IIA in neutrophils is altered in patients with MYH9-related disorders.7,32,60,61 Instead of a homogenous pattern throughout the cytoplasm, bright foci of nonmuscle myosin-IIA staining, corresponding to the sites of inclusions, are observed. Some diffuse cytoplasmic staining persists, suggesting some nonmuscle myosin-IIA is distributed outside these inclusions. Kunishima et al60 reported that wild-type nonmuscle myosin-IIA was present in MYH9-related leukocyte inclusions (at least for 5779delC, 5818delG, and R1933Stop). While inclusions are not observed for all mutations, Seri et al7 reported that the R702C motor head mutation had inclusions, demonstrating that inclusions are not specific to tail mutations. The presence of ribosomes suggests that inclusion formation is either concurrent with, or occurs shortly after, translation.
Our results suggest how MYH9 mutations perturb nonmuscle myosin-IIA function and demonstrate a good correlation between the dominant biochemical effects of the mutant proteins on assembly and the dominant nature of MYH9-related disorders. Our results further suggest that knowing how nonmuscle myosin-IIA function is perturbed cannot fully explain the variability in the clinical observations (Table 1). A more complete understanding of the precise role of nonmuscle myosin-IIA in megakaryocyte fragmentation and platelet cell dynamics will be important in further elucidating the exact mechanism of MYH9-related disorders at the cellular level. In addition, investigations considering other genetic or environmental factors that may affect the hematologic and nonhematologic characteristics should be pursued.
Interestingly, rod mutations in other conventional (or type II) myosins are also known to cause autosomal dominant disorders. Mutations in β-myosin heavy chain (MYH7) cause familial hypertrophic cardiomyopathy (FHC), a leading cause of sudden death in otherwise healthy individuals,62,63 and dilated cardiomyopathy.64,65 Interestingly, 17% of these MYH7 mutations lie in the rod of the β-myosin heavy chain.63,66 Other MYH7 rod mutations are also associated with a skeletal myopathy,67 causing muscle weakness and wasting and hyaline body myopathy68 (HBM), a congenital neuromuscular disorder. To date, a biochemical explanation of how MYH7 rod mutations impair function to cause these disorders is lacking. We believe that this experimental approach can be extended to understand how rod mutations in other myosins alter structure and assembly to cause disease.
On the basis of our findings, and reports by other investigators, we suggest the following model for the mutational perturbation of MYH9 function. When a cell requires nonmuscle myosin-IIA–based contractility, both wild-type and mutant heavy chains are recruited to sites of activity and bipolar filaments assemble. Proper assembly is perturbed because mutant heavy chains dominantly interact with wild-type heavy chains. As a result, insufficient functional wild-type filaments form, resulting in a loss of contractility, causing the cellular dynamics of the affected cell (megakaryocyte and/or platelet) to be impaired.
Prepublished online as Blood First Edition Paper, August 31, 2004; DOI 10.1182/blood-2004-06-2067.
Supported by grants from the National Institute of Health (GM 33830) (D.P.K.) and the National Heart, Lung and Blood Institute (HL 66192) (M.J.K.). M.J.K. is the recipient of a Merit Review Award from the Department of Veteran's Affairs.
M.J.K. and D.P.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank members of the Kiehart and Kelley laboratories and Meg Titus for suggestions and Thomas Ortel and Ruth Montague for critically reading the manuscript. We also thank Preeti Chunga and Terry Oas for assistance and use of the Duke University CD Spectrometer.

![Figure 1. Schematic of a native nonmuscle myosin-IIA molecule, its assembly steps, and the purified tail fragments used in this study. (A) A schematic of nonmuscle myosin-IIA shows 2 heavy chains (white and black globular and coiled domains) and the essential and regulatory light chains (ELC and RLC are indicted by arrows). An arrow marks the junction (P836) between the motor head domain and the rod. The arrowhead marks the junction between subfragment 2 (S2) and light meromysoin (LMM) of the rod. The bounded arrow indicates the region (amino acids [aa] 1102-1960) of the wild-type rod used in this study, and an asterisk signifies the relative position of the 4 mutations studied (R1165C, D1424N, E1841K, R1933Stop, reading from left to right). (B) Assembly steps of nonmuscle myosin-IIA. The rod of individual heavy chains fold into an α-helical structure that dimerizes with another heavy chain, forming a coiled coil. A cross-section through a coiled coil, appearing below the diagram, shows the relationship of amino acids in the characteristic heptad repeat. To form higher order assemblies, such as contractile filaments, dimers must laterally associate with one another in proper register. Periodic charged regions along the rod play important roles during this alignment. The α-helix and coiled-coil diagrams are reprinted from Figures 3-6 and 3-9 of Lodish et al14 with permission. (C) Here, 5 μg each recombinantly purified protein were subjected to electrophoresis on an acrylamide gel and Coomassie stained (lane 1 indicates wild type; lane 2, R1165C; lane 3, D1424N; lane 4, E1841K; and lane 5, R1933Stop). Two to 3 minor bands, probably breakdown products, can be seen in each lane.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/1/10.1182_blood-2004-06-2067/6/m_zh80010571790001.jpeg?Expires=1767785244&Signature=dBJijIbYrQDh3d5xR2qefm5azFOV2mzCGxGEjEXiJKaznMFazslskMvPkDXuLHFGBQQd1dlD8l7Uzz~Qy4IvkzI~6yyyanrvBQzAatGrHxDha5u4wC0rL8YvFQ9xn6amCE6N98HSy8fS5I0O7U62T5uqcVUXTQkM-ik6umekfJds5L7-JZ4~XYWqTtVsWdd1aOlLU-nthDYkByZeWCy9ViVmm1PYe0Hv62Tqg1MJX4htO2vGHallUG9K7FqD2Xq8DwE6G-YbtzaBp5en1eEQ6hEBCdJqH4Ewi-s0ds7llOwQ7y6sPIEtGXCjyh-pJUrXcnjIOZtekmbxIlIPG7ibZQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Figure 1. Schematic of a native nonmuscle myosin-IIA molecule, its assembly steps, and the purified tail fragments used in this study. (A) A schematic of nonmuscle myosin-IIA shows 2 heavy chains (white and black globular and coiled domains) and the essential and regulatory light chains (ELC and RLC are indicted by arrows). An arrow marks the junction (P836) between the motor head domain and the rod. The arrowhead marks the junction between subfragment 2 (S2) and light meromysoin (LMM) of the rod. The bounded arrow indicates the region (amino acids [aa] 1102-1960) of the wild-type rod used in this study, and an asterisk signifies the relative position of the 4 mutations studied (R1165C, D1424N, E1841K, R1933Stop, reading from left to right). (B) Assembly steps of nonmuscle myosin-IIA. The rod of individual heavy chains fold into an α-helical structure that dimerizes with another heavy chain, forming a coiled coil. A cross-section through a coiled coil, appearing below the diagram, shows the relationship of amino acids in the characteristic heptad repeat. To form higher order assemblies, such as contractile filaments, dimers must laterally associate with one another in proper register. Periodic charged regions along the rod play important roles during this alignment. The α-helix and coiled-coil diagrams are reprinted from Figures 3-6 and 3-9 of Lodish et al14 with permission. (C) Here, 5 μg each recombinantly purified protein were subjected to electrophoresis on an acrylamide gel and Coomassie stained (lane 1 indicates wild type; lane 2, R1165C; lane 3, D1424N; lane 4, E1841K; and lane 5, R1933Stop). Two to 3 minor bands, probably breakdown products, can be seen in each lane.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/1/10.1182_blood-2004-06-2067/6/m_zh80010571790001.jpeg?Expires=1767785245&Signature=zGhR8wg1LmU2-ZycwvC98xa3JNKNjdJmzgufEPEXPQoXvDK~pDPfVug8Ppnb1rSWXzrP2tGb-euVhN7~9ZHZaA5C00esZITx9QRUgZWIDsnWgCqXhmml~FT6Z0QWQ2HN113WkBdYqpUhithwyTysIDgaN2YgXxULo6-8jgfv2qHyJsUd~n1kmWC7SI-~9bQ7w~VGSy01A4dyp5R-TA009XY0Pc2q5qRTOwWJi7gMd090pYeg4-DVkPdHs2jDeD8bjwdF3qfdg3tf60DzvKa7bUKwhDt6JbgW2hj29fWT25y~JFWj5uhUlm3mz0RrE8OGhGBGElYCHL0VJooHNDSpuQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




