Abstract
Oncostatin M (OSM) is a multifunctional cytokine that belongs to the interleukin 6 (IL-6) family. As OSM is expressed in adult as well as embryonic hematopoietic tissues, OSM has been considered to play a role in hematopoiesis. To uncover roles of OSM, we have generated mutant mice deficient in the OSM-specific receptor β subunit (OSMR). While OSMR–/– mice were healthy and fertile, hematologic analysis of OSMR–/– mice demonstrated that the numbers of peripheral erythrocytes and platelets were reduced compared with wild-type mice. Consistent with this, progenitors of erythroid and megakaryocyte lineages were reduced in OSMR–/– bone marrow (BM), suggesting that OSM is required for the maintenance of erythroid and megakaryocyte progenitor pools in BM. To investigate whether OSM acts on the hematopoietic progenitors directly or indirectly, we performed BM transplantation experiments. The OSMR–/– mice, engrafted with wild-type BM cells, failed to produce erythrocytic and megakaryocytic progenitors to the levels in wild-type mice, indicating that OSM affects hematopoietic microenvironments. On the other hand, erythrocytic and megakaryocytic progenitors were reduced in the wild-type mice reconstituted with OSMR–/– BM cells. Thus, OSM regulates hematopoiesis in vivo by stimulating stromal cells as well as hematopoietic progenitors, in particular megakaryocytic and erythrocytic progenitors.
Introduction
Oncostatin M (OSM) is a multifunctional cytokine and is a member of the interleukin 6 (IL-6) family that includes IL-6, IL-11, leukemia inhibitory factor (LIF), ciliary neurotrophic factor (CNTF), cardiotrophin-1, and novel neutrophin-1/B-cell–stimulating factor-3.1-3 Among the family members, OSM is the most closely related to LIF structurally, genetically, and functionally.4,5 OSM was initially recognized by its biologic activity to inhibit the proliferation of A375 melanoma cells and its cDNA was originally isolated from phorbol ester–stimulated human histiocytic lymphoma U937 cells.6 It is known that OSM is secreted from activated T cells and monocytes stimulated by cytokines and plays roles in inflammatory reactions.7,8 In humans, OSM and LIF exhibit various overlapping biologic responses such as growth regulation, differentiation, gene expression, and cell survival, while it is also known that OSM displays some unique biologic properties that are not shared with LIF (eg, growth inhibition of A375 melanoma cells,9 growth of AIDS-related Kaposi sarcoma cells in an autocrine manner,10 and up-regulation of α1-proteinase inhibitor in lung-derived epithelial cells).11 These common and unique functions of OSM are now well explained by the receptor structures. Two types of functional OSM receptors are present in humans; the type I OSM receptor is identical to the high-affinity LIF receptor that consists of gp130 and the LIF receptor β subunit12,13 and the type II OSM receptor consists of gp130 and the OSM-specific receptor β subunit (OSMR).14,15 Thus, many overlapping biologic responses between human OSM (hOSM) and hLIF are mediated by the shared type I receptor (ie, the LIF receptor), whereas OSM manifests its specific responses through the type II receptor. OSMR is expressed in a wide variety of cell types including endothelial cells, hepatic cells, lung cells, skin cells, and many tumor cell lines.16-20 Curiously, unlike hOSM, mouse OSM (mOSM) transduces its signals through the OSM-specific receptor consisting of gp130 and OSMR but does not use the type I OSM receptor (ie, the LIF receptor).21
Previously we isolated mOSM cDNA as a signal transducer and activator of transcription 5 (Stat5)–inducible gene in hematopoietic cells by the cDNA subtraction method and found that mOSM is expressed in hematopoietic tissues such as bone marrow (BM) and spleen, suggesting that mOSM may play a role in hematopoiesis.22 OSM is expressed not only in adult tissues but also in embryonic hematopoietic tissues. We found that mOSM is expressed in the embryonic aorta-gonad-mesonephros (AGM) region where definitive hematopoiesis originates and that mOSM stimulates generation of hematopoietic cells and endothelial cells in the primary culture of the AGM cells, possibly acting on their common precursors.23 Moreover, mOSM is also expressed in CD45+ hematopoietic cells in fetal liver and stimulates differentiation of fetal hepatocytes, suggesting that OSM may coordinate development of hematopoiesis and liver.24,25 In adults, while there are many reports that OSM functions as a proinflammatory factor, the role of OSM in steady-state hematopoiesis in vivo remains largely unknown. In this report, we describe the generation of OSMR-deficient mice and the role of OSM in hematopoiesis in vivo.
Materials and methods
Mice
C57/BL6 mice (8-12 weeks) were purchased from Japan SLC (Hamamatsu, Japan). Green fluorescent protein (GFP) mice were from Dr M. Okabe (Keio University, Tokyo, Japan).26
Targeted disruption of OSMR by homologous recombination in embryonic stem cells
The targeting vector was constructed by inserting the LacZ-neomycin phosphotransferase (pgkNeo) cassette for positive selection (a kind gift of Dr Takahashi, Tsukuba University, Tsukuba, Japan) and the MC1–diphtheria toxin (DT-A) cassette for negative selection. The ApaI site was introduced at the 18-bp downstream from ATG to fuse LacZ to ATG in frame. The 5′ 1.8-kb HindIII-ApaI fragment and the 3′ 7.2-kb SacII-BglII fragment were inserted upstream and downstream of the LacZ-pgkNeo cassette, respectively. The DT-A cassette was inserted at the 5′ side. The construct was linearized by BglII and electroporated into CCE embryonic stem (ES) cells. G418-resistant ES cell clones were isolated, expanded, and screened by Southern blot analysis. Genomic DNA was digested with EcoRV, transferred onto a nylon membrane, and prehybridized and hybridized at 45°C in high–sodium dodecyl sulfate (SDS) buffer (Roche Diagnostics, Mannheim, Germany) and probed with a 700-bp EcoRI-BglII fragment (5′ probe) derived from the cloned genomic DNA fragment. The labeling of probe and detection were performed using the Digoxigenin (DIG) system (Roche Diagnostics) according to the manufacturer's protocol. Predicted size of the wild-type DNA fragment detected by 5′ probe is 9.0 kb and that of the mutant DNA is 6.5 kb. These cells were injected into blastocysts. The chimeric mice were mated with C57/BL6 female mice and the heterozygous offspring were interbred to yield wild-type, heterozygous, and mutant mice. Genotyping was performed by polymerase chain reaction (PCR) using specific primers from genomic DNA derived from their tails. PCR reactions were performed in 50 μL with 1 × PCR buffer (Takara, Tokyo, Japan), 0.2 mM each of dNTP (deoxynucleoside triphosphate), 0.5 U Taq polymerase (Takara), and 10 pmol of each primer: 5′-GTAATCAGACCAATGGCTTTCTC-3′,5′-GATCCAACAGAGCAATCATGAAGC-3′, and 5′-GCACATCTGAACTTCAGC-3′. Amplification conditions were an initial denaturation at 94°C for 2 minutes followed by 35 cycles of 94°C for 1 minute, 60°C for 1 minute, and 72°C for 90 seconds. The wild-type mice generated a 364-bp PCR product, whereas the OSMR–/– mice had only a 750-bp fragment. The heterozygous mice had both fragments.
Northern blot analysis
Lung mRNA was prepared from adult lung derived from wild-type, heterozygous, and mutant mice with the FastTrack mRNA purification kit (Invitrogen, Carlsbad, CA). Standard Northern blot analysis of 1 μg mRNA was performed. A 502-bp fragment (nucleotides 1166-1667 from GenBank accession no. AB015978) was DIG-labeled by PCR using primers (5′-CAACTGGAGTTCCTGGAAAG-3′ and 5′-AGTGATTTGCCAGGTATAGG-3′) and DIG DNA labeling Mix (Roche Diagnostics) and then used for Northern blotting.
Hematologic analysis
Orbital plexus blood was collected from anesthetized mice. Peripheral blood cells were analyzed by using an automated counter Sysmex K-4500 (TOA Medical Instruments, Kobe, Japan). BM and spleen progenitor cells were assayed using semisolid methylcellulose cultures as previously described.27 In short, 25 000 BM or 100 000 spleen nucleated cells were plated in triplicate in 1-mL methylcellulose culture. Cells were cultured in the presence of combinations of purified recombinant growth factors at the following final concentrations: murine erythropoietin (EPO; Roche Diagnostics) at 4 U/mL, human stem cell factor (SCF; Sigma Chemical, St Louis, MO) at 80 ng/mL, murine IL-3 at 10 ng/mL. After 3 days of incubation in a fully humidified atmosphere at 37°C supplemented with 5% CO2, erythroid colony-forming units (CFU-Es) were counted. Granulocyte-macrophage colony-forming cells (GM-CFCs), erythroid burst-forming units (BFU-Es), granulocyte-erythrocyte-megakaryocyte-macrophage CFUs (CFU-GEMMs), and megakaryocyte-erythrocyte CFUs (CFU-MEs) were enumerated after 7 days of incubation. Average counts of triplicate cultures are presented in each experiment. Megakaryocytic progenitors were assessed by liquid culture of BM. Nucleated BM cells (100 000 cells) were incubated in Iscove modified Dulbecco medium (IMDM) containing 1% Nutridoma SP (Roche Diagnostics) and thrombopoietin (TPO; Sigma Chemical) at 25 ng/mL in a 96-well plate at 37°C supplemented with 5% CO2 for 3 days. Mature megakaryocyte was identified by acetylcholinesterase (AchE) staining.28 Plates were fixed with 2.5% glutaraldehyde at room temperature for 20 minutes. After washing with phosphate-buffered saline (PBS; twice), AchE staining solution (0.5 mg acetylthiocholine iodide in 1 mL of 5 mM sodium citrate, 1.5 mM CuSO4, 0.25 mM potassium ferricyanide) was added into the plate. After 1 hour of incubation at 37°C, AchE-positive cells were counted.
Phenylhydrazine and 5-FU administration
A single dose of phenylhydrazine (60 mg/kg body weight) was administered intraperitoneally for 2 consecutive days. Wild-type and mutant mice were killed at 1, 2, 4, and 7 days after the final administration. CFU-Es of 25 000 BM nucleated cells were assessed by semisolid methylcellulose cultures as described in hematologic analysis. Likewise, a single dose of 5-fluorouracil (FU; 150 mg/kg body weight) was administered intravenously. Wild-type and mutant mice were killed at 2, 5, and 8 days after administration. Then, 50 000 (days 2) and 25 000 (days 5 and 8) BM nucleated cells were assessed by semisolid methylcellulose cultures as described in hematologic analysis.
Flow cytometry
Marrow cells were flushed from the femurs with a syringe containing 2 mL alfa-MEM-10% FCS (α-modified minimum essential media–10% fetal calf serum). After depletion of mature red cells by hypotonic lysis (0.38% NH4Cl for 7 minutes on ice), the GPF-positive cells were sorted from mononuclear cells of BM and spleen by FACS Vantage (Becton Dickinson Labware, Bedford, MA) and used for semisolid methylcellulose cultures.
Transplantation assay
Transplantation of cells into busulfan-treated neonatal mice was performed as previously described29 with a slight modification. In short, pregnant recipient mice were intraperitoneally injected with busulfan (Sigma Chemical) at 12.5 mg/kg body weight on pregnancy days 17 and 18. The transplanted cells were prepared from BM of a GPF-expressing wild-type or mutant mouse followed by negative selection for lineage markers such as TER119, Gr-1, CD4, and CD8 (PharMingen, San Diego, CA). Then, 24 to 48 hours after birth, 7 × 105 cells in 25 μL of PBS were injected into each neonatal liver. To measure the chimerism of GFP-positive cells in recipient mice, orbital plexus blood was collected from anesthetized mice to a heparinized capillary tube and centrifuged at 800g for 10 minutes. The white blood cell layer at the interface was collected into PBS, then mature red cells were depleted by hypotonic lysis. The chimerism of GFP-positive cells was measured by fluorescence-activated cell sorter (FACS) analysis of white blood cells.
RT-PCR
Total RNA was prepared from wild-type and mutant BM and spleen by TRIzol reagent (Gibco BRL, Grand Island, NY) according to the manufacturer's instruction. For reverse transcriptase–PCR (RT-PCR) analysis, first-strand cDNA was synthesized from total RNA by the First-Strand cDNA Synthesis Kit (Amersham Biosciences, Piscataway, NJ). Each sample was prepared from 3 mice of the same genotype. Synthesized cDNA samples were used as templates for PCR amplification for OSM, SCF, and GAPDH (glyceraldehyde phosphate dehydrogenase). Amplification conditions were as follows: an initial denaturation at 96°C for 30 seconds followed by 28 to 44 cycles of 96°C for 20 seconds, 52°C for 30 seconds, and 72°C for 90 seconds. PCR primers used in this study were as follows: OSM (5′-CATCCTGAGCATGGCACTGG-3′ and 5′-GCCACAGCTGCCTATCTTGG-3′), SCF (5′-TCTTCCAAATGACTATATGATAACCCTC-3′ and 5′-ATTCCTAAGGGAGCTGGCTGCAACAGGG-3′), and GAPDH as a positive control (5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′).
Results
Generation of OSMR-deficient mice
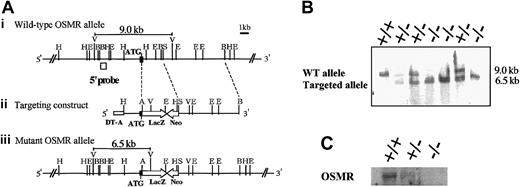
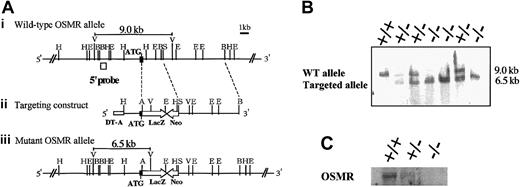
In order to disrupt the OSMR gene, the proximal region of the initiation codon was replaced with the LacZ and neomycin expression cassette, resulting in the deletion of the N-terminal region including the signal sequence (Figure 1A). Neomycin-resistant ES cells were screened by Southern blot using a probe corresponding to the 5′ end of the targeting construct. One of 6 successfully targeted ES clones heterozygous for the OSMR locus was injected into blastocysts to generate chimeric mice. Intercross of heterozygous mice resulted in 3 genotypes of mice (wild-type OSMR+/+, heterozygous OSMR+/–, and homozygous OSMR–/–; Figure 1B), and they were born with a normal mendelian pattern of inheritance. The growth rate of OSMR–/– mice was comparable to wild-type littermates from neonate to adult. These results indicate that OSMR–/– mice have no apparent growth disadvantage. To confirm the disruption of the OSMR locus, we performed Northern blot analysis using RNA prepared from the lung with an OSMR cDNA probe, as it is known that OSMR was highly expressed in lung.30 A single band corresponding to mOSMR mRNA was detected in wild-type and heterozygous mRNA, whereas no such signal was present in homozygous mice (Figure 1C). To further verify the elimination of OSMR function by targeting, we used 2 primary culture systems using fetal liver and the AGM region, which were previously shown to respond to mOSM.23,24 Neither of the culture systems derived from OSMR–/– mice responded to OSM (data not shown), indicating the lack of functional OSMR in the homozygous mice.
Targeting disruption of the mOSMR gene. (Panel A) Targeting strategy of the mOSMR gene. (i) Schematic diagram of the wild-type allele. The start codon is indicated by ATG. The open box represents the location of the probe used to detect the restriction fragment of genomic DNA by Southern blot analysis. Restriction enzyme sites are indicated as ApaI (A), EcoRI (E), EcoRV (V), HindIII (H), BglII (B), and SacII (S). (ii) Gene targeting vector. The vector contains the 5′ and 3′ regions of homology and the cDNAs encoding neomycin transferase (Neo), LacZ, and diphtheria toxin (DT-A). (iii) Diagram of the targeted mutant OSMR allele. (Panel B) Southern blot analysis of the EcoRV-digested genomic DNA extracted from the OSMR+/+, OSMR+/–, and OSMR–/– mice. (Panel C) Northern blot analysis of neonatal lung mRNA (+/+, +/–, and –/–) with mOSMR probe.
Targeting disruption of the mOSMR gene. (Panel A) Targeting strategy of the mOSMR gene. (i) Schematic diagram of the wild-type allele. The start codon is indicated by ATG. The open box represents the location of the probe used to detect the restriction fragment of genomic DNA by Southern blot analysis. Restriction enzyme sites are indicated as ApaI (A), EcoRI (E), EcoRV (V), HindIII (H), BglII (B), and SacII (S). (ii) Gene targeting vector. The vector contains the 5′ and 3′ regions of homology and the cDNAs encoding neomycin transferase (Neo), LacZ, and diphtheria toxin (DT-A). (iii) Diagram of the targeted mutant OSMR allele. (Panel B) Southern blot analysis of the EcoRV-digested genomic DNA extracted from the OSMR+/+, OSMR+/–, and OSMR–/– mice. (Panel C) Northern blot analysis of neonatal lung mRNA (+/+, +/–, and –/–) with mOSMR probe.
OSM was previously shown to stimulate Sertoli cells in vitro, suggesting a role for OSM in reproduction.31 However, as OSMR null mice bred normally, these mice were intercrossed with C57/BL6 repeatedly for at least 8 generations and used for further analysis.
Hematologic analysis
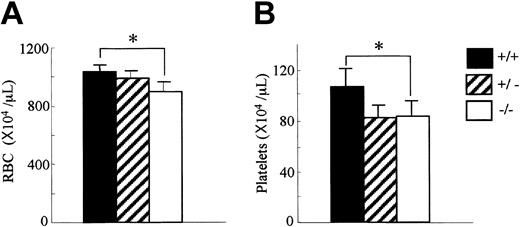
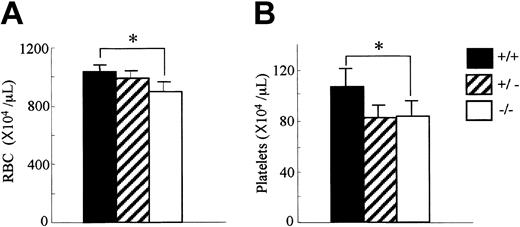
Since OSM is expressed in hematopoietic tissues, we first analyzed the peripheral blood cells in OSMR–/– mice. The number of white blood cells in the peripheral blood was comparable among genotypes (data not shown). In contrast, the red cell number in the peripheral blood of the OSMR–/– mice was reduced compared with the wild-type mice (894 ± 64 × 104/mL in OSMR–/– vs 987 ± 52 × 104/mL in OSMR+/– vs 1028 ± 52 × 104/mL in OSMR+/+ mice; Figure 2A). Consistent with this reduction, the hematocrit of OSMR–/– mice was also reduced. Interestingly, the platelet number in the peripheral blood of OSMR–/– mice was also reduced (82 ± 10 × 104/mL in OSMR–/– vs 78 ± 13 × 104/mL in OSMR+/– vs 106 ± 15 × 104/mL in OSMR+/+ mice; Figure 2B). Although there are reports describing effects of the IL-6–family cytokines such as IL-6, IL-11, and LIF on proliferation and differentiation of megakaryocytes in vitro32-34 and in vivo,35-37 no mice with mutant alleles of either cytokine or its specific receptor have been reported to show thrombocytopenia.38-40 Therefore, mOSM may be the most important IL-6–family cytokine for megakaryopoiesis in vivo.
Hematologic profiles of OSMR+/+, OSMR+/–, and OSMR–/– mice. Orbital plexus blood was collected from anesthetized mice. Peripheral red blood cells (RBCs; panel A) and platelets (B) were analyzed by using automated counter Sysmex K-4500. Numbers show mean cell number ± SD (n = 17 of OSMR+/+ and OSMR–/–,n = 6 of OSMR+/– for RBCs; n = 14 of OSMR+/+ and OSMR–/–,n = 6 of OSMR+/– for platelets). *P < .001 between wild-type and mutant mice.
Hematologic profiles of OSMR+/+, OSMR+/–, and OSMR–/– mice. Orbital plexus blood was collected from anesthetized mice. Peripheral red blood cells (RBCs; panel A) and platelets (B) were analyzed by using automated counter Sysmex K-4500. Numbers show mean cell number ± SD (n = 17 of OSMR+/+ and OSMR–/–,n = 6 of OSMR+/– for RBCs; n = 14 of OSMR+/+ and OSMR–/–,n = 6 of OSMR+/– for platelets). *P < .001 between wild-type and mutant mice.
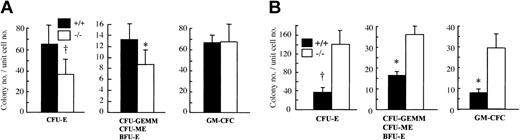
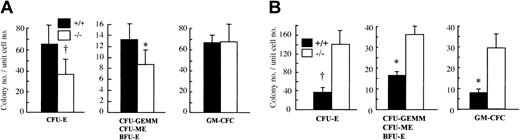
Since OSM is highly expressed in BM,22 we hypothesized that OSM would affect the maintenance of hematopoietic progenitors in BM. To examine this possibility, we counted the number of hematopoietic progenitors in BM by in vitro clonal cultures with multiple cytokine combinations. The number of CFU-Es, the proerythroblasts, was significantly reduced in OSMR–/– BM compared with the wild-type control (Figure 3A). The number of erythrocyte-producing colonies (ie, CFU-GEMMs, CFU-MEs, and BFU-Es) was also significantly reduced in OSMR–/– BM (Figure 3B). In contrast, there was no difference in granulocyte-macrophage colony-forming cells (GM-CFCs) including granulocyte CFUs (CFU-Gs), macrophage CFUs (CFU-Ms), and granulocyte-macrophage CFUs (CFU-GMs) (Figure 3C). These results are consistent with the peripheral blood cells in mutant mice.
Colony-forming activity of hematopoietic progenitor cells from OSMR+/+ and OSMR–/– mice. CFU-Es (A,D), CFU-GEMMs/CFU-MEs/BFU-Es (B,E), and GM-CFCs (C,F) from hematopoietic cells of OSMR+/+ and OSMR–/– mice are shown. The figure shows mean total colony number ± SD (n = 5 to 7) in triplicate cultures containing 25 000 BM cells (A-C) or 100 000 spleen cells (D-F) stimulated with final concentrations of IL-3 at 10 ng/mL, SCF at 80 ng/mL, and EPO at 4 U/mL. *P < .05 between wild-type and mutant mice. †P < .01 between wild-type and mutant mice.
Colony-forming activity of hematopoietic progenitor cells from OSMR+/+ and OSMR–/– mice. CFU-Es (A,D), CFU-GEMMs/CFU-MEs/BFU-Es (B,E), and GM-CFCs (C,F) from hematopoietic cells of OSMR+/+ and OSMR–/– mice are shown. The figure shows mean total colony number ± SD (n = 5 to 7) in triplicate cultures containing 25 000 BM cells (A-C) or 100 000 spleen cells (D-F) stimulated with final concentrations of IL-3 at 10 ng/mL, SCF at 80 ng/mL, and EPO at 4 U/mL. *P < .05 between wild-type and mutant mice. †P < .01 between wild-type and mutant mice.
It is known that the spleen is a second hematopoietic organ and becomes a major site for murine erythropoiesis under conditions that severely damage erythrocytes.41 Since OSMR is expressed in spleen at a relatively high level, we examined hematopoietic progenitors in mutant and wild-type spleen by in vitro clonal cultures. In contrast to BM, the number of CFU-Es in mutant spleen was markedly increased compared with wild-type control (Figure 3D). In addition, the numbers of all types of colonies including CFU-GEMMs, CFU-MEs, BFU-Es, and GM-CFCs were also maintained at higher levels (Figure 3E-F). These results suggested that OSM might have an opposite effect on hematopoiesis in BM and spleen.
Recovery of erythrocytes from chemically induced acute hemolysis
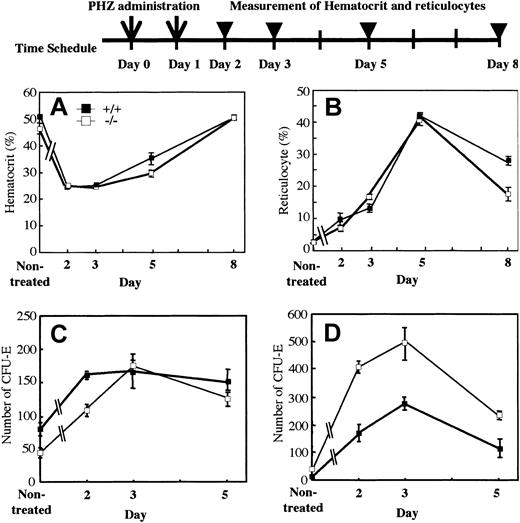
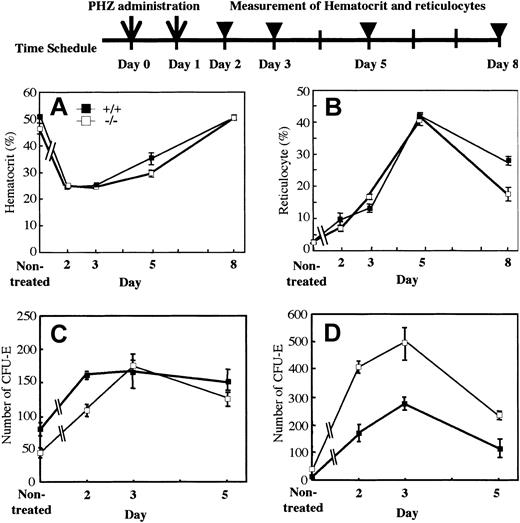
The mutant mice displayed a slightly anemic phenotype and a reduced reservoir of erythrocyte precursors in marrow. To investigate whether OSM itself affects erythropoiesis or not, we assessed wild-type and mutant erythropoiesis in response to acute hemolysis induced by phenylhydrazine (PHZ). We injected PHZ into mutant and wild-type mice intraperitoneally on days 0 and 1. The recovery was monitored by measurement of peripheral red cells, reticulocytes, and CFU-Es in marrow and spleen. The recovery of peripheral red cells as measured by hematocrit was similar in both genotypes (Figure 4A). Reticulocytes are newly generated red blood cells and become mature red cells within 2 days. After administration of PHZ, the number of reticulocytes in wild-type and mutant mice increased sharply with similar kinetics (Figure 4B). Analysis of erythroid progenitor CFU-Es revealed that there was no difference in the initial rate of increase between wild-type and mutant marrow (Figure 4C). Although the number of CFU-Es in mutant spleen was higher than that of wild-type spleen, the rate of increase was not so different, indicating that CFU-E numbers in spleen after PHZ administration reflected the original ratio of CFU-Es between wild-type and mutant mice (Figure 4D). These results strongly suggested that erythroid progenitors in OSMR-deficient mice are capable of responding to acute hemolysis and of differentiating to mature erythrocytes normally.
Kinetics of recovery after acute hemolysis by phenylhydrazine administration. A single dose of phenylhydrazine (60 mg/kg body weight) was administered intraperitoneally on days 0 and 1 (arrows). Wild-type and mutant mice were killed at 1, 2, 4, or 7 days after the final administration (arrowheads). Recovery was monitored by measurement of hematocrit (A) and reticulocytes (B). CFU-Es (mean ± SD) of 25 000 BM nucleated cells (C) and 100 000 spleen nucleated cells (D) from wild-type and mutant mice on days 2, 3, and 5 (n = 3 to 5 for each genotype) were assessed by semisolid methylcellulose cultures.
Kinetics of recovery after acute hemolysis by phenylhydrazine administration. A single dose of phenylhydrazine (60 mg/kg body weight) was administered intraperitoneally on days 0 and 1 (arrows). Wild-type and mutant mice were killed at 1, 2, 4, or 7 days after the final administration (arrowheads). Recovery was monitored by measurement of hematocrit (A) and reticulocytes (B). CFU-Es (mean ± SD) of 25 000 BM nucleated cells (C) and 100 000 spleen nucleated cells (D) from wild-type and mutant mice on days 2, 3, and 5 (n = 3 to 5 for each genotype) were assessed by semisolid methylcellulose cultures.
Since 5-FU selectively kills cycling cells and results in myelosuppression,42 the recovery from such stress needs proliferation and differentiation of dormant stem cells. To assess the recovery of hematopoiesis in BM, we intravenously injected 5-FU into mutant and wild-type mice. The BM progenitors in a femur were examined at 2, 5, and 8 days after 5-FU administration. The kinetics of recovery of BM progenitors in OSMR–/– mice paralleled those of OSMR+/+ mice (data not shown). These results suggested that BM progenitor cells of OSMR–/– mice are able to respond to acute hemolysis and myelosuppression normally.
Analysis of megakaryocyte progenitors
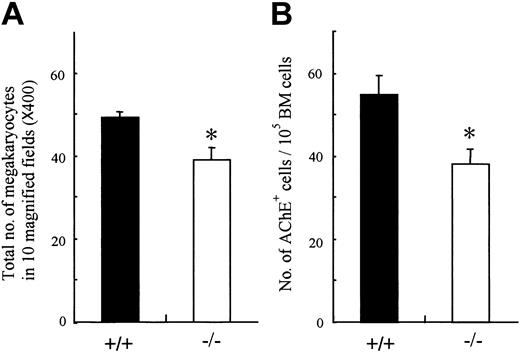
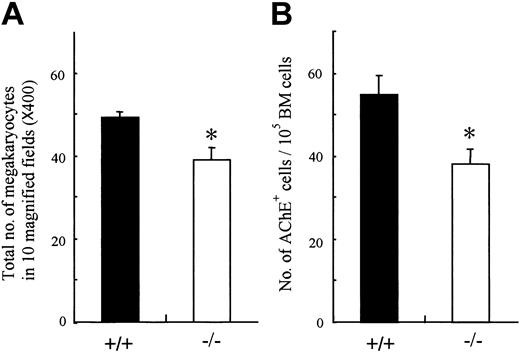
Analysis of peripheral blood cells demonstrated that the platelets as well as erythrocytes were reduced in OSMR–/– mice. Megakaryopoiesis is a regulated process by which multipotential hematopoietic cells commit to megakaryocytes that release platelets after several maturation steps.43,44 Therefore, the number of peripheral platelets depends on the number and maturation process of megakaryocyte progenitors. To determine which stage of platelet production is affected by the lack of OSM function, we examined the number of megakaryocytes in mutant and wild-type BM. The number of megakaryocytes in mutant BM was significantly reduced compared with control (Figure 5A). To compare the numbers of megakaryocyte progenitors in mutant and wild-type BM, we performed liquid culture of BM cells in the presence of TPO and counted cells stained with AchE after 3 days of culture. The number of megakaryocyte progenitors in mutant BM was also reduced (Figure 5B). These data suggested that the reduced peripheral platelet number in mutant mice was in part due to the decrease of megakaryocyte progenitors, CFU-GEMMs and CFU-MEs, in BM, whereas it still remains possible that OSM affects the maturation process of megakaryocytes.
Megakaryocyte and its progenitor from BM of OSMR+/+ and OSMR–/– mice. (A) The sections of femora from OMSR+/+ and OSMR–/– mice were stained with hematoxylin and eosin. (A) The number of mature megakaryocytes in 10 fields (original magnification, × 40) of femoral sections was counted. Error bars indicate mean ± SD, n = 3 of each genotype. (B) Megakaryocytic progenitors were assessed by liquid culture of BM. BM nucleated cells (100 000 cells) were incubated in IMDM containing 1% Nutridoma SP and TPO at 25 ng/mL in 96-well plate at 37°C supplemented with 5% CO2 for 3 days. The number of mature megakaryocytes identified by AchE staining was counted (mean ± SD, n = 7 of each genotype). *P < .05 between wild-type and mutant mice.
Megakaryocyte and its progenitor from BM of OSMR+/+ and OSMR–/– mice. (A) The sections of femora from OMSR+/+ and OSMR–/– mice were stained with hematoxylin and eosin. (A) The number of mature megakaryocytes in 10 fields (original magnification, × 40) of femoral sections was counted. Error bars indicate mean ± SD, n = 3 of each genotype. (B) Megakaryocytic progenitors were assessed by liquid culture of BM. BM nucleated cells (100 000 cells) were incubated in IMDM containing 1% Nutridoma SP and TPO at 25 ng/mL in 96-well plate at 37°C supplemented with 5% CO2 for 3 days. The number of mature megakaryocytes identified by AchE staining was counted (mean ± SD, n = 7 of each genotype). *P < .05 between wild-type and mutant mice.
Transplantation of OSMR+/+ hematopoietic stem cells in OSMR–/– mice
OSMR–/– mice showed impaired hematopoiesis in BM, whereas hematopoiesis in the spleen increased. However, the mechanism of the OSM effects on hematopoiesis remained unclear. To determine whether OSM affects immature hematopoietic progenitors directly or indirectly through acting on their microenvironments such as BM stromal cells, we transplanted wild-type hematopoietic stem cells (HSCs) into OSMR–/– mice. If OSM directly stimulates the hematopoietic progenitors, but not the microenvironment in BM, the engrafted wild-type HSCs in the BM of OSMR–/– mice should proliferate and recover to the normal level of progenitors. To address this question, lineage-negative BM cells from transgenic mice constitutively expressing GFP were injected into the livers of busulfan-treated neonatal mice26,29 (Figure 6A). OSMR–/– GFP-positive BM cells as well as OSMR+/+ GFP-positive BM cells were injected into OSMR–/– neonates. Donor contribution in the recipient mice was examined by flow cytometric analysis of the peripheral blood (Figure 6B). Wild-type GFP-transgenic mice were used as OSMR+/+ controls. Eight and 7 recipients were engrafted with wild-type and mutant BM cells, respectively. Four and 5 of those mice that showed relatively high donor contribution were used for the following analysis (Table 1). Hematologic analysis of these mice revealed that engraftment of wild-type lineage-negative BM cells into OSMR–/– mice failed to increase the numbers of red blood cells and platelets in peripheral blood (Figure 6C-D), suggesting that the hematopoietic microenvironments must be altered in OSMR–/– mice. To verify the hematopoietic potential of the engrafted cells, the donor-derived GFP-positive cells were sorted by FACS and were subjected to semisolid colony assays. The colony-forming potential of GFP-positive OSMR+/+ cells in mutant recipients was not increased, indicating that the hematopoietic potential depends on the environment of the recipient (Figure 6E-F). These results indicated that OSM affected BM hematopoiesis indirectly, possibly acting on the BM stromal cells that constitute the hematopoietic microenvironment. However, the results do not preclude the possibility that OSM affects the hematopoietic cells directly, which has been shown in vitro experiments.45
Transplantation of BM cells in OSMR–/– mice. (A) To investigate whether wild-type hematopoietic cells are able to convert the hematologic profile of mutant mice to the wild type, BM cells were transplanted into busulfan-treated neonatal OSMR–/– mice. To distinguish donor cells from recipients, BM cells from GFP-transgenic mouse were used as input cells. –/– (–/–) indicates OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; –/– (+/+), OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; and +/+, GFP-transgenic OSMR+/+ mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Of fifteen recipient mice, 4 or 5 mice exhibiting relatively high proportion of donor type were used for the following hematologic analyses. (B) Representative FACS profiles of a mouse from each group. (i) –/– (–/–); (ii) –/– (+/+); (iii) +/+; and (iv) non-GFP control. (C-D) Hematologic analyses of the reconstituted OSMR–/– mice and wild-type GFP mice. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (C) and platelets (D) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (E-F) GFP-positive donor cells were sorted from BM (E) and spleen (F) of each engrafted recipient mouse. GFP-transgenic wild-type mouse was used as OSMR+/+ control. The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the sorted donor BM and spleen cells was assessed. The figure represents the mean ± SD [n = 4 for –/– (–/–), n = 4 for –/– (+/+) and n = 3 for +/+]. *P < .05 between wild-type and mutant mice. †P < .01 between wild-type and mutant mice.
Transplantation of BM cells in OSMR–/– mice. (A) To investigate whether wild-type hematopoietic cells are able to convert the hematologic profile of mutant mice to the wild type, BM cells were transplanted into busulfan-treated neonatal OSMR–/– mice. To distinguish donor cells from recipients, BM cells from GFP-transgenic mouse were used as input cells. –/– (–/–) indicates OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; –/– (+/+), OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; and +/+, GFP-transgenic OSMR+/+ mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Of fifteen recipient mice, 4 or 5 mice exhibiting relatively high proportion of donor type were used for the following hematologic analyses. (B) Representative FACS profiles of a mouse from each group. (i) –/– (–/–); (ii) –/– (+/+); (iii) +/+; and (iv) non-GFP control. (C-D) Hematologic analyses of the reconstituted OSMR–/– mice and wild-type GFP mice. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (C) and platelets (D) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (E-F) GFP-positive donor cells were sorted from BM (E) and spleen (F) of each engrafted recipient mouse. GFP-transgenic wild-type mouse was used as OSMR+/+ control. The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the sorted donor BM and spleen cells was assessed. The figure represents the mean ± SD [n = 4 for –/– (–/–), n = 4 for –/– (+/+) and n = 3 for +/+]. *P < .05 between wild-type and mutant mice. †P < .01 between wild-type and mutant mice.
Transplantation of OSMR–/– hematopoietic stem cells in OSMR+/+ mice
In order to test whether OSM directly acts on hematopoietic progenitors, we performed the reciprocal experiment (ie, transplantation of mutant HSCs in OSMR+/+ mice). If OSM also stimulates the hematopoietic progenitors directly, the engrafted OSMR–/– HSCs in the wild-type BM would exhibit impaired hematologic properties similar to the OSMR-deficient mice. To assess the property of OSMR–/– hematopoietic cells in OSMR+/+ mice, the effect of the endogenous OSMR+/+ hematopoietic cells should be excluded. For this reason, evaluation of transplanted cells requires nearly complete chimerism of the transplanted HSCs. Two mice with high donor contribution were used for the following analysis (Table 2). Hematologic analysis revealed that both mice reconstituted with mutant HSCs exhibited a phenotype similar to mutant mice (Figure 7A-B), suggesting that OSM might also affect hematopoietic cells directly in normal hematopoiesis. To verify the hematopoietic potential of the engrafted cells, semisolid colony assay was performed. In this case, BM and spleen cells without sorting were used for the colony assay because of high chimerism. In agreement with the high chimerism of mutant hematopoietic cells, almost all colonies were fluorescent under the fluorescent microscopy, indicating that the engrafted mutant hematopoietic cells contributed to nearly all hematopoietic cells in the recipient mouse. The colony-forming potential of the engrafted mutant cells in BM exhibited the mutant phenotype (ie, reduced numbers of erythrocytic and megakaryocytic progenitors in BM, whereas the GM-CFC number was unchanged [Figure 7C]). These data suggested that OSM might affect the hematopoietic cells directly. Interestingly, hematopoietic progenitors in spleen of the wild-type mice that received OSMR-deficient BM cells exhibited phenotype similar to wild-type mice (Figure 7D).
Transplantation of BM cells in OSMR+/+ mice. OSMR+/+ or OSMR–/– GFP-positive BM cells were transplanted into busulfan-treated neonatal OSMR+/+ mice. +/+ (+/+) indicates OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; +/+ (–/–), OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; and –/–, GFP-transgenic OSMR–/– mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Four +/+ (+/+) mice exhibiting relatively high proportion of donor-type and 2 +/+ (–/–) mice (nos. 1 and 2) exhibiting high chimerism were used for the following hematologic analyses. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (A) and platelets (B) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (C-D) The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the BM (C) and spleen (D) cells were assessed. The figure represents the mean ± SD [n = 4 for +/+ (+/+) and –/–] and each value for +/+ (–/–).
Transplantation of BM cells in OSMR+/+ mice. OSMR+/+ or OSMR–/– GFP-positive BM cells were transplanted into busulfan-treated neonatal OSMR+/+ mice. +/+ (+/+) indicates OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; +/+ (–/–), OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; and –/–, GFP-transgenic OSMR–/– mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Four +/+ (+/+) mice exhibiting relatively high proportion of donor-type and 2 +/+ (–/–) mice (nos. 1 and 2) exhibiting high chimerism were used for the following hematologic analyses. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (A) and platelets (B) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (C-D) The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the BM (C) and spleen (D) cells were assessed. The figure represents the mean ± SD [n = 4 for +/+ (+/+) and –/–] and each value for +/+ (–/–).
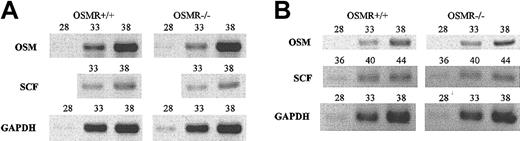
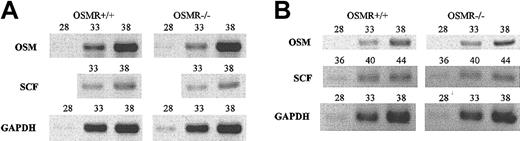
However, if OSM is regulated through the OSMR signaling itself, the interpretation of transplant experiments would be more complex. To test this possibility, we examined the expression of OSM in OSMR–/– BM and spleen by semiquantitative RT-PCR and found that OSM expression was unaltered in OSMR-deficient mice, precluding the possibility (Figure 8). Similarly, as stem cell factor (SCF) is an important cytokine for hematopoiesis including megakaryopoiesis,46 we compared the expression of SCF in OSMR+/+ and OSMR–/– BM and spleen and found no significant difference, suggesting that SCF expression is independent of OSMR signaling.
Semiquantitative RT-PCR analysis of OSM and SCF genes. One microgram of total RNA derived from BM (A) and spleen (B) of 3 mice was used to generate cDNA. Gene-specific primers for OSM, SCF, and GAPDH as control were used for PCR. The number represents PCR cycles of each sample.
Semiquantitative RT-PCR analysis of OSM and SCF genes. One microgram of total RNA derived from BM (A) and spleen (B) of 3 mice was used to generate cDNA. Gene-specific primers for OSM, SCF, and GAPDH as control were used for PCR. The number represents PCR cycles of each sample.
Discussion
Signal transduction of IL-6–family cytokines is elicited by homodimerization of gp130 by IL-6 and IL-11 and heterodimerization of gp130 and LIFR by LIF, CNTF, and cardiotrophin-1. While heterodimerization of gp130 and OSMR is induced by OSM,47 it remained possible that other cytokines also use the OSMR. Relatively mild defects were found in mice with a mutation of individual cytokines of this family due to functional overlap among the family members. In contrast, the disruption of the shared components of the receptors, gp130 or LIFR, leads to severe phenotype and mortality because this inactivates functions of multiple cytokines simultaneously.48,49 Since the box1 and box2 regions, which contribute to downstream signaling cascade, are present in the intracellular domain of OSMR,30 OSMR itself is also a signal transducing molecule. Therefore, it was possible that the disruption of OSMR might cause severe defects if OSMR is used by another novel cytokine. However, the hematologic characteristics in OSMR–/– mice were similar to those in OSM–/– mice (K. Minehata, manuscript in preparation), suggesting that OSMR is not shared by other cytokines.
The overlapping functions of the IL-6–family cytokines are explained by the shared signal transducer gp130.1 Especially OSM and LIF display many similar biologic activities.5,9,50 OSM and LIF are not only structurally related but their genes are also tightly linked in the same chromosomal location, suggesting that the 2 genes arose by duplication.4 Moreover, the OSMR gene was also found to be located in the vicinity of the LIFR locus in mouse.30 Actually, OSM and LIF share the same LIF receptor in human.12 Therefore, OSM had long been considered as just another LIF. It has been reported that LIF-deficient mice display female sterility due to defective blastocyst implantation, postnatal growth retardation, and defects in hematopoiesis and thymocyte proliferation.40 While the levels of circulating mature red blood cells and platelets, as well as the numbers of BFU-Es and GM-CFCs in the BM, were normal in LIF–/– mice, the numbers of BFU-Es and GM-CFCs in the spleen of LIF–/– mice were significantly decreased compared with wild-type mice. These results are quite different from what we found in OSMR–/– mice as shown in Figure 3 (ie, BFU-Es and CFU-Es in BM were reduced, whereas they were increased in spleen). Hematologic characteristics in IL-6–/– mice were also different from OSMR–/– mice38 (ie, the number of BFU-Es was increased in the BM of IL-6–/– mice compared with wild-type mice), whereas enhanced hematopoiesis in the IL-6–/– spleen was similar to OSMR–/– mice. Thus, mice lacking either LIF, IL-6, or OSMR display a distinct pattern of hematopoiesis, although all the IL-6–family cytokines use gp130 as the common signal transducer. The complex response is likely to reflect a complex interaction between the cytokines and their receptors in this family and compensatory mechanisms that affect hematopoietic systems directly or indirectly.
It is known that the IL-6–family cytokines are involved in megakaryopoiesis in vitro and in vivo. Although IL-6, IL-11, LIF, and OSM enhanced the megakaryocytic colony formation in synergy with other cytokines such as IL-3 or TPO, they had little or no such activity alone32-34,45 and they rather appear to stimulate the maturation of megakaryocytes in vitro by increasing size and ploidy. In vivo, the administration of these cytokines elevated numbers of megakaryocytes and platelets.35-37,45 However, mice lacking either IL-6, LIF, or the specific IL-11 receptor α-chain are not thrombocytopenic despite these activities,38-40 indicating functional redundancy among these cytokines in megakaryopoiesis. Unlike these mice, the number of peripheral platelets was reduced in OSMR–/– mice, suggesting that OSM plays an important role for megakaryopoiesis under the steady-state condition. Since transplantation of OSMR-deficient BM cells into wild-type mice resulted in the reduction of peripheral platelets, OSM might directly affect the maturation of megakaryocytes in vivo as described in the previous in vitro study.45 Consistently, immunohistochemical analysis revealed that CD41+ megakaryocytic cells expressed OSMR (K. Minehata, manuscript in preparation).
Recently, Broxmeyer et al51 reported that the number of hematopoietic progenitors in Stat4–/– mice was significantly reduced and recovered by T-cell–specific transgenic expression of Stat4. Furthermore, the injection of a Th1 cytokine OSM, but not other cytokines, into Stat4–/– mice recovered progenitor cell activity to wild-type levels. Based on these results they suggested that OSM is a potential mediator between T cells and hematopoietic progenitors. These data are also consistent with our findings that OSMR–/– mice exhibit impaired homeostasis.
Considering the results of BM transplantation, OSM is likely to be involved in hematopoietic homeostasis by acting on the BM stromal cells. OSM is a proinflammatory as well as an anti-inflammatory cytokine depending on the cellular context. An important characteristic of OSM is its potent activity to induce tissue inhibitor of metalloproteinases (TIMPs). Matrix metalloproteinases (MMPs) are involved in extracellular matrix breakdown during inflammatory reactions and TIMPs inhibit their function. Therefore, the balance between MMPs and TIMPs is important for the remodeling of the extracellular matrix. Among the IL-6–family cytokines, OSM selectively stimulates the expression of TIMP-1 in fibroblasts19,52 and synovial lining cells.53 The marked induction of TIMP-1 by OSM indicates that the OSM-specific receptor is required for the full spectrum of TIMP-1 expression. In fact, the recovery from liver injury was delayed in OSMR–/– mice that showed impaired TIMP-1 expression and prolonged activation of MMP-9 (K. Nakamura, submitted). Thus, the regulation of MMP-9 and TIMP-1 by OSM may play an important role for the maintenance of BM microenvironment. Recently, Heissig et al54 reported that MMP-9–/– mice failed to recover hematopoiesis after BM ablation because of the defective release of soluble SCF and HSC mobilization from BM. Therefore, we presumed alteration of the SCF level in mutant mice. However, OSMR signaling was not required for expression of SCF at transcriptional and serum protein levels (Figure 8; K. Minehata, manuscript in preparation). It is therefore likely that OSM regulates expression of the other cytokines or membrane-bound proteins that constitute the BM microenvironment.
In contrast to BM, hematopoietic progenitors in spleen of OSMR–/– were increased compared with OSMR+/+ mice. Hematopoietic progenitors in spleen were increased in OSMR-deficient mice that received either wild-type or OSMR-deficient BM cells, whereas they were not increased in the wild-type mice that received either wild-type or OSMR-deficient BM cells. These results suggest that OSM affects the splenic hematopoietic microenvironment rather negatively.
In conclusion, our results indicate that OSMR–/– mice exhibit unique hematologic characteristics that have not been recognized in mice defective in functions of either IL-6, IL-11, LIF, or CNTF and that OSM regulates hematopoiesis in BM and spleen by acting on both stromal cells and erythrocytic and megakaryocytic progenitors.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-02-0367.
Supported in part by Grants-in-Aid for Scientific Research and Special Coordination Funds from the Ministry of Education, Culture, Sports, Science and Technology of Japan; a research grant from the Ministry of Health, Labour and Welfare, and Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Corporation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Drs S. Takahashi and M. Okabe for providing us with vectors and GPF transgenic mice, respectively.

![Figure 6. Transplantation of BM cells in OSMR–/– mice. (A) To investigate whether wild-type hematopoietic cells are able to convert the hematologic profile of mutant mice to the wild type, BM cells were transplanted into busulfan-treated neonatal OSMR–/– mice. To distinguish donor cells from recipients, BM cells from GFP-transgenic mouse were used as input cells. –/– (–/–) indicates OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; –/– (+/+), OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; and +/+, GFP-transgenic OSMR+/+ mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Of fifteen recipient mice, 4 or 5 mice exhibiting relatively high proportion of donor type were used for the following hematologic analyses. (B) Representative FACS profiles of a mouse from each group. (i) –/– (–/–); (ii) –/– (+/+); (iii) +/+; and (iv) non-GFP control. (C-D) Hematologic analyses of the reconstituted OSMR–/– mice and wild-type GFP mice. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (C) and platelets (D) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (E-F) GFP-positive donor cells were sorted from BM (E) and spleen (F) of each engrafted recipient mouse. GFP-transgenic wild-type mouse was used as OSMR+/+ control. The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the sorted donor BM and spleen cells was assessed. The figure represents the mean ± SD [n = 4 for –/– (–/–), n = 4 for –/– (+/+) and n = 3 for +/+]. *P < .05 between wild-type and mutant mice. †P < .01 between wild-type and mutant mice.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2003-02-0367/6/m_h82135157006.jpeg?Expires=1769152049&Signature=P8FgFbUvW6QY466aDaRwi1InGPDaTy3AlhJNoGvbTZh4s03vAMx91N1wcdubKpDWPUO2DTP-euFZlwA3Gac1O~SsEY49NQFEoaK8tlRZUptPT2vnxjkYb5nmMhUd4bJEFWnQQAJcgnIepZAC84EbutHwKl1Nyxl9Ar3yFqoby76OFhObsJUqoYQUD5JubYH7m-Yj4xIM0RhaTiVziejMoTm6mY27S-DEkhQ4OaUYSA0fNPVbauhnNiI1upvdobqtMxNyH2sJCyvAWPqliAWhXuG0mK0dSfTHsu~zAymXrfFnP3z4MWr4710kQ5G3XCaj~w1C1JKEzSq7flMdTTfHEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Transplantation of BM cells in OSMR+/+ mice. OSMR+/+ or OSMR–/– GFP-positive BM cells were transplanted into busulfan-treated neonatal OSMR+/+ mice. +/+ (+/+) indicates OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; +/+ (–/–), OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; and –/–, GFP-transgenic OSMR–/– mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Four +/+ (+/+) mice exhibiting relatively high proportion of donor-type and 2 +/+ (–/–) mice (nos. 1 and 2) exhibiting high chimerism were used for the following hematologic analyses. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (A) and platelets (B) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (C-D) The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the BM (C) and spleen (D) cells were assessed. The figure represents the mean ± SD [n = 4 for +/+ (+/+) and –/–] and each value for +/+ (–/–).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2003-02-0367/6/m_h82135157007.jpeg?Expires=1769152049&Signature=ciz1d6HHgavPxIBPXtt9OHjZIiG2accSWtzQK7rrN831lzHYZv1A48Rh7HQze6kCK~a43Eq9nElvEEpGJkG0qWQrchRuFfvATKCELYAdnbRfq20DKY8mybBLtlZABdmELVNCTQCKKPaVcU8yb2IX-07CWjCXu8hKc8oWcGN3FjCxr-1J3p1dionwRqyC0yvEttufJ37UF5jTTWImDnjoW6vXilRtKAtAjHg69QetbTNnRe5pK04c3t~4YspHpukG~lsTj4Qy18PcqHgGncERlZFj90Lt7JASTqf-zQV7fQqOx3s0-K7ec~YFTLNPcCJTVluoDgr~VI7gO1oWCNU9YA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Transplantation of BM cells in OSMR–/– mice. (A) To investigate whether wild-type hematopoietic cells are able to convert the hematologic profile of mutant mice to the wild type, BM cells were transplanted into busulfan-treated neonatal OSMR–/– mice. To distinguish donor cells from recipients, BM cells from GFP-transgenic mouse were used as input cells. –/– (–/–) indicates OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; –/– (+/+), OSMR–/– mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; and +/+, GFP-transgenic OSMR+/+ mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Of fifteen recipient mice, 4 or 5 mice exhibiting relatively high proportion of donor type were used for the following hematologic analyses. (B) Representative FACS profiles of a mouse from each group. (i) –/– (–/–); (ii) –/– (+/+); (iii) +/+; and (iv) non-GFP control. (C-D) Hematologic analyses of the reconstituted OSMR–/– mice and wild-type GFP mice. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (C) and platelets (D) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (E-F) GFP-positive donor cells were sorted from BM (E) and spleen (F) of each engrafted recipient mouse. GFP-transgenic wild-type mouse was used as OSMR+/+ control. The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the sorted donor BM and spleen cells was assessed. The figure represents the mean ± SD [n = 4 for –/– (–/–), n = 4 for –/– (+/+) and n = 3 for +/+]. *P < .05 between wild-type and mutant mice. †P < .01 between wild-type and mutant mice.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2003-02-0367/6/m_h82135157006.jpeg?Expires=1769612828&Signature=ALj8~snngLZ9zBt5kl3kghjHwtgaqDsyA3gRLliVKdo3iSWcSLS0Y48Ln2lQTjsN0s0T0izpDXPtSv8GuUlLeK0uObJJdiySzLmcuVqNdscyf1C4zsMswQwKFk~21xXi8F4CombWkiPjRjB~M1IagR7Rl5CTcqoiO2q-xLmwUSus9hy9smX3Bgp9djyCy0QxU~ZdzXmHs1O2hWr4e4MKRu-YfecB3j-IJ5gy5EN4xS4Vy9Df-9cs02TqMOTnCp9tXwwGGfsBuC1ogIW8JNBtrOanbPQTwXll539Y8mozerovzeE58D8nrQZKfZAyIBD98UyW9uUTFSsL-Tm4OSNoQg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Transplantation of BM cells in OSMR+/+ mice. OSMR+/+ or OSMR–/– GFP-positive BM cells were transplanted into busulfan-treated neonatal OSMR+/+ mice. +/+ (+/+) indicates OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR+/+ mouse; +/+ (–/–), OSMR+/+ mice reconstituted with BM cells of GFP-transgenic OSMR–/– mouse; and –/–, GFP-transgenic OSMR–/– mice. Recipient mice were killed 4 months after transplantation and the levels of donor contribution in peripheral blood were determined by FACS. Four +/+ (+/+) mice exhibiting relatively high proportion of donor-type and 2 +/+ (–/–) mice (nos. 1 and 2) exhibiting high chimerism were used for the following hematologic analyses. Orbital plexus blood was collected from anesthetized mice. Peripheral RBCs (A) and platelets (B) were analyzed by using automated counter Sysmex K-4500. The horizontal bar represents the mean. (C-D) The hematopoietic progenitor content (GM-CFCs, left panel; and CFU-GEMMs/CFU-MEs/BFU-Es, right panel) of the BM (C) and spleen (D) cells were assessed. The figure represents the mean ± SD [n = 4 for +/+ (+/+) and –/–] and each value for +/+ (–/–).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2003-02-0367/6/m_h82135157007.jpeg?Expires=1769612828&Signature=GOmsB6IKHIM5wcMM8f-oMYhMa6jC6SVUyiJgijjOkpDECSxRZg55mF2xhBoUIJXnDoewcG0DdMrA3M71hSCdLVnGMTX~uaZse2yY3V~Gn9Vs4kXO4GmM0MBLWUFeecuMlQrOxnTXUqEdS5KSRa4y5svd4USW3phukDROS04gc~sS0T3Pv5qt9iJr5I65r6rCuF6mq3uKqOz1KAbYX5z0pXFfrRQo9DH3OT6NBy7b0WiKYiimm3e6tUtm-Mo1DyGED-2-VgYcQiVni5c2xQ~3VvDlYn2MzVTEQD5wJ~9I6ffsbm6j0Q2vClelV7DkMNIdTVMeOeARUQ374wH2D8ecAw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)