Abstract
Bcr-Abl proteins are effective inducers of the leukemic phenotype in chronic myeloid leukemia (CML) and distinct variants of acute lymphoblastic leukemia (ALL). Targeting bcr-abl by treatment with the selective tyrosine kinase inhibitor imatinib has proved to be highly efficient for controlling leukemic growth. However, it is unclear whether imatinib is sufficient to eradicate the disease because of primary or secondary resistance of leukemic cells. Therefore, targeting Bcr-Abl with an alternative approach is of great interest. We demonstrate that RNA interference (RNAi) with a breakpoint-specific short-interfering RNA (siRNA) is capable of decreasing Bcr-Abl protein expression and of antagonizing Bcr-Abl–induced biochemical activities. RNAi selectively inhibited Bcr-Abl–dependent cell growth. Furthermore, bcr-abl–homologous siRNA increased sensitivity to imatinib in Bcr-Abl–overexpressing cells and in a cell line expressing the imatinib-resistant Bcr-Abl kinase domain mutation His396Pro, thereby antagonizing 2 of the major mechanisms of resistance to imatinib.
Introduction
Chronic myeloid leukemia (CML) arises from the reciprocal translocation t(9;22) forming the highly stable, constitutively active tyrosine kinase Bcr-Abl. This kinase activity is assumed to be sufficient and necessary to initiate CML.1-3 Imatinib, a potent Abl-specific tyrosine kinase inhibitor recently approved by the United States Food and Drug Administration (FDA), represents a highly effective therapy for this disease.4,5 However, clinical resistance occurs frequently, in particular during the late stages of CML.6,7 As molecular mechanisms leading to resistance point mutations in the Abl kinase domain of Bcr-Abl, and Bcr-Abl overexpression have been described.8-10 Consequently, the investigation of alternative and supplementary therapies is still of great clinical importance.
Silencing bcr-abl using antisense oligonucleotides led to conflicting results concerning specificity and functionality.11,12 Because of the high stability of siRNAs and the effectiveness of the silencing process induced, RNA interference has been shown to be superior to conventional antisense strategies.13,14 RNAi is thought to be an evolutionarily conserved surveillance mechanism that responds to double-stranded RNA by sequence-specific silencing of homologous genes.15
Recently, Scherr et al16 demonstrated the possibility of interfering with the induction of Bcr-Abl expression in an inducible vector system using a breakpoint-specific siRNA. However, little is known about the biochemical and functional consequences of down-regulation of Bcr-Abl protein levels by means of RNAi.
We used an siRNA construct homologous to the b3a2 break-point to silence bcr-abl gene expression in a murine and a human Bcr-Abl–dependent cell line. Prolonged siRNA-treatment of Bcr-Abl–positive cells led to a significant down-regulation of constitutive Bcr-Abl protein levels. This caused a reversion of Bcr-Abl–dependent effects on cell cycle regulatory and antiapoptotic proteins, resulting in a selective inhibition of cell growth. Moreover, interference with bcr-abl expression enhanced the sensitivity of γ-irradiation and imatinib. Additionally, RNAi restored imatinib sensitivity in cells expressing the imatinib-resistant Bcr-Abl kinase domain mutant His396Pro.
Study design
Cell culture
The factor-independent 32Dp210 Bcr-Abl oligoclonal cell line was generated by transfection of the parental cells with the retroviral vector Migp210,17 Migp210-Thr315Ile, or Migp210-His396Pro, as previously described.18 The Bcr-Abl–positive cell line M07p210 was generated by electroporation of M07 cells with the pGD210 plasmid. When compared with primary CML cells, 32Dp210 and M07p210 cells are characterized by a 5- to 6-fold overexpression of the Bcr-Abl protein as revealed by Western blot analysis (not shown). Cells were cultivated in RPMI 1640/10% fetal calf serum (FCS) complemented with glutamine and 1 ng/mL recombinant interleukin-3 (IL-3) or 100 ng/mL granulocyte macrophage–colony-stimulating factor (GM-CSF), as indicated. CD34+/CML cells were purified as described.19
Reagents
Imatinib was kindly provided by Novartis (Basel, Switzerland).
Analysis of protein expression
Western blot analysis was performed 24 hours after the second siRNA treatment, as described.19-21
Survival assay
Cells were exposed to distinct concentrations of imatinib for 40 hours or were γ-irradiated and thereafter cultivated for 40 hours at 37°C. Cell survival was then measured by MTT (3,[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay as described.20
Detection of apoptosis
Annexin V staining was performed as described.21
siRNA variants
The siRNA constructs used (Table 1) were kindly provided by Ribopharma (Kulmbach, Germany). Asymmetric siRNAs were designed because they were found superior in our laboratory (not shown).
siRNA treatment
Cells were set to a density of 3.2 × 106/mL; 800 μL of this cell suspension were mixed with siRNA in a 4-mm electroporation cuvette. Cells were electroporated by means of an EasyJect-electroporator (Peqlab, Erlangen, Germany) using a single-pulse protocol (250 V, 1800 μF, ∞Ω) providing a transfection efficiency of 80% or more without significant reduction of viability (not shown). This treatment was repeated after 24 hours. Because of the long half-life of the Bcr-Abl protein, repeated treatment proved to be essential for optimal down-regulation of Bcr-Abl (not shown). This was not because of an increase in transduction rates but was most likely mediated by the sustained presence of siRNA inside the target cell. RNAi in mammalian cells is transient, and Bcr-Abl protein expression increased again after reaching maximal inhibition 24 hours after (repeated) electroporation (not shown). Both 32Dp210 and M07p210 exhibit strict dependence on the activity of Bcr-Abl in the absence of growth factors. Therefore, exogenous growth factor was added during siRNA treatment. After the second treatment, cells were washed and factor-deprived before the different examination procedures were begun.
Results and discussion
We analyzed the effects of prolonged siRNA treatment on Bcr-Abl protein levels in 32Dp210-wt, 32Dp210-Thr315Ile, 32Dp210-His396Pro, M07p210, and primary CD34+/CML cells. The cell lines used were characterized by Bcr-Abl protein overexpression when compared with primary CML cells (“Study design”). Cells were treated twice at intervals of 24 hours using either siRNA homologous to b3a2-fusion site (BAF7) or mismatch siRNA control (BAF8). BAF7 treatment resulted in a significant reduction of Bcr-Abl protein levels compared with cells treated with mismatch control siRNA (Figure 1A). The decrease of Bcr-Abl was similar in all BAF7-transduced cells, as revealed by analyses of the cotranscribed EGFP reporter gene (data not shown). Additionally, BAF7 antagonized regulatory effects of Bcr-Abl on expression of the antiapoptotic protein Bcl-XL and the cell cycle inhibitor p27. Bcl-XL expression is known to be induced by Bcr-Abl,19,22,23 whereas p27 gets down-regulated.24 As expected, BAF7-treated 32Dp210 showed a clear reduction of Bcl-XL and a concomitant increase of p27 when compared with BAF8 or with electroporation control (EPC; Figure 1B).
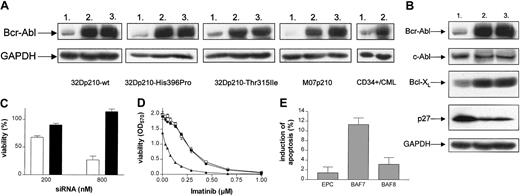
RNAi leads to specific down-regulation of Bcr-Abl protein levels, resulting in a significant reduction of viability and sensitization to imatinib mesylate. (A) Western blot analysis of Bcr-Abl in 32Dp210-wt, 32Dp210-Thr315Ile, 32Dp210-His396Pro, M07p210, and primary CD34+/CML cells after siRNA treatment (800 nM). Lane 1, BAF7, siRNA homologous to bcr-abl b3a2 fusion site; lane 2, BAF8, mismatch siRNA; lane 3, EPC. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level served as a loading control. The decrease in Bcr-Abl levels caused by BAF7 treatment was determined by densitometry—83% in 32Dp210-wt, 86.22% in 32Dp210-His396Pro, 71.83% in 32Dp210-Thr315Ile, 93.9% in M07p210, and 74.3% in primary CD34+/CML cells (because of the low expression of Bcr-Abl, a more sensitive technique was applied in primary CD34+ cells). (B) Western blot analysis of Bcr-Abl, c-Abl, Bcl-XL, and p27 in 32Dp210 cells after treatment with siRNA (800 nM), as indicated (lane1, BAF7; lane 2, BAF8; lane 3, EPC). GAPDH level served as a loading control. (C) BAF7 treatment led to a dose-dependent reduction of viability in 32Dp210 cells. Forty hours after the second treatment with 200 nM and 800 nM siRNA, respectively, viability of cells (relative to EPC) was determined by means of MTT (BAF7, white bars; BAF8, black bars). Values are means ± SD of 6 examinations. (D) Treatment with suboptimal amounts of breakpoint-specific siRNA (200 nM) results in sensitization to imatinib mesylate. After siRNA treatment, 32Dp210 cells were exposed to imatinib concentrations ranging from 0.03 μM to 1.0 μM. Cell viability was measured after 40 hours by means of MTT assay. BAF7-treated cells (▴) showed significantly higher sensitivity for imatinib than cells treated with BAF8 (•) or cells electroporated without any siRNA (EPC, □). The IC50 values for imatinib were 0.08 μM (BAF7), 0.27 μM (BAF8), and 0.27 μM (EPC), respectively. Values are means ± SD of triplicates. (E) After siRNA treatment, M07p210 cells were exposed to imatinib at a concentration of 0.05 μM for 21 hours. The percentage of apoptotic cells was then determined by Annexin V–FITC assay. Values are means ± SD of 3 independent experiments.
RNAi leads to specific down-regulation of Bcr-Abl protein levels, resulting in a significant reduction of viability and sensitization to imatinib mesylate. (A) Western blot analysis of Bcr-Abl in 32Dp210-wt, 32Dp210-Thr315Ile, 32Dp210-His396Pro, M07p210, and primary CD34+/CML cells after siRNA treatment (800 nM). Lane 1, BAF7, siRNA homologous to bcr-abl b3a2 fusion site; lane 2, BAF8, mismatch siRNA; lane 3, EPC. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level served as a loading control. The decrease in Bcr-Abl levels caused by BAF7 treatment was determined by densitometry—83% in 32Dp210-wt, 86.22% in 32Dp210-His396Pro, 71.83% in 32Dp210-Thr315Ile, 93.9% in M07p210, and 74.3% in primary CD34+/CML cells (because of the low expression of Bcr-Abl, a more sensitive technique was applied in primary CD34+ cells). (B) Western blot analysis of Bcr-Abl, c-Abl, Bcl-XL, and p27 in 32Dp210 cells after treatment with siRNA (800 nM), as indicated (lane1, BAF7; lane 2, BAF8; lane 3, EPC). GAPDH level served as a loading control. (C) BAF7 treatment led to a dose-dependent reduction of viability in 32Dp210 cells. Forty hours after the second treatment with 200 nM and 800 nM siRNA, respectively, viability of cells (relative to EPC) was determined by means of MTT (BAF7, white bars; BAF8, black bars). Values are means ± SD of 6 examinations. (D) Treatment with suboptimal amounts of breakpoint-specific siRNA (200 nM) results in sensitization to imatinib mesylate. After siRNA treatment, 32Dp210 cells were exposed to imatinib concentrations ranging from 0.03 μM to 1.0 μM. Cell viability was measured after 40 hours by means of MTT assay. BAF7-treated cells (▴) showed significantly higher sensitivity for imatinib than cells treated with BAF8 (•) or cells electroporated without any siRNA (EPC, □). The IC50 values for imatinib were 0.08 μM (BAF7), 0.27 μM (BAF8), and 0.27 μM (EPC), respectively. Values are means ± SD of triplicates. (E) After siRNA treatment, M07p210 cells were exposed to imatinib at a concentration of 0.05 μM for 21 hours. The percentage of apoptotic cells was then determined by Annexin V–FITC assay. Values are means ± SD of 3 independent experiments.
We then determined the biologic consequences of silencing Bcr-Abl. BAF7 treatment led to a significant dose-dependent reduction of viability in 32Dp210 cells (Figure 1C). To study synergisms between RNAi and imatinib, we used suboptimal amounts of siRNA. bcr-abl RNAi resulted in a reversion of the resistance to DNA damage in Bcr-Abl–positive 32D cells leading to an enhanced sensitivity to γ-irradiation. The γ-irradiation dose causing a 50% cell kill was 2.5 Gy in BAF7-treated cells, whereas cells treated with mismatch control tolerated approximately 2.5 times higher doses (6 Gy).
The quantity of Bcr-Abl protein also determined the sensitivity of these cells to imatinib. After reduction of the Bcr-Abl protein level with siRNA, a 3.4-fold drop of the IC50 value of imatinib was observed when compared with controls (Figure 1D). This phenomenon was also observed in human M07p210 cells—0.05 μM imatinib caused a significant induction of apoptosis in BAF7-treated cells, whereas the same concentration had no effect on controls (Figure 1E). These results are in contrast to previously published data that reported no observed additive effect of imatinib and Bcr-Abl RNAi treatment on the induction of apoptosis in K562 cells.25 These K562 cells, however, exhibited a high level of resistance to imatinib because only 8% underwent apoptosis after 48-hour treatment. Therefore, this relatively resistant cell system may be suboptimal for evaluating possible additive effects with other potential inhibitors.
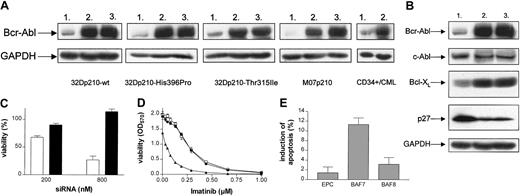
We further studied whether the inhibition of bcr-abl translation by siRNA is sufficient to overcome resistance to imatinib. Point mutations are one of the most frequent mechanisms of imatinib resistance in CML patients, in whom resistance develops after an initial response.26 Two imatinib-resistant Bcr-Abl kinase domain mutations described are Thr315Ile and His396Pro. Expression of p210-Thr315Ile in 32D cells confers complete resistance to imatinib, whereas p210-His396Pro renders the respective cells 5-fold less sensitive to imatinib when compared with 32Dp210-wt (Figure 2A). To assess whether RNAi treatment could be used to overcome this type of imatinib resistance, cells were treated as described in “Results and discussion.” The 32Dp210-His396Pro carrying a Bcr-Abl protein partially resistant to imatinib showed a 4-fold sensitization to imatinib when treated with BAF7, rendering these cells as sensitive as untreated 32Dp210-wt cells (Figure 2B). In contrast, imatinib sensitivity of 32Dp210-Thr315Ile cells highly resistant to imatinib was not affected by siRNA treatment (Figure 2B).
bcr-abl RNAi sensitizes cells expressing imatinib-resistant His396Pro-Bcr-Abl domain mutation to imatinib. (A) 32Dp210-wt (•), 32Dp210-Thr315Ile (▴), and 32Dp210-His396Pro (□) were analyzed using MTT assay to determine their sensitivity to imatinib. IC50 values were 0.3 μM in 32Dp210-wt, 1.4 μM in 32Dp210-His396Pro, and 5.2 μM in 32Dp210-Thr315Ile. Values are means ± SD of triplicates. (B) Breakpoint-specific siRNA treatment (200 nM) leads to reestablishment of sensitivity to imatinib in 32Dp210-His396Pro. After treatment, 32Dp210-His396Pro (i) and 32Dp210-Thr315Ile (ii) were exposed to imatinib mesylate. Sensitivity was analyzed using the MTT assay. In 32Dp210-His396Pro Bcr-Abl, RNAi resulted in a distinct down-regulation of imatinib IC50 values ranging from 1.2 μM in control cells treated with BAF8 (•) and 1.1 μM in EPC (□) reaching 0.3 μM in cells treated with BAF7 (▴). Despite this, the sensitivity of 32Dp210-Thr315Ile did not change after siRNA treatment (IC50: 4.9 [BAF7], 5.2 [BAF8], 4.6 [EPC]). Values are means ± SD of triplicates.
bcr-abl RNAi sensitizes cells expressing imatinib-resistant His396Pro-Bcr-Abl domain mutation to imatinib. (A) 32Dp210-wt (•), 32Dp210-Thr315Ile (▴), and 32Dp210-His396Pro (□) were analyzed using MTT assay to determine their sensitivity to imatinib. IC50 values were 0.3 μM in 32Dp210-wt, 1.4 μM in 32Dp210-His396Pro, and 5.2 μM in 32Dp210-Thr315Ile. Values are means ± SD of triplicates. (B) Breakpoint-specific siRNA treatment (200 nM) leads to reestablishment of sensitivity to imatinib in 32Dp210-His396Pro. After treatment, 32Dp210-His396Pro (i) and 32Dp210-Thr315Ile (ii) were exposed to imatinib mesylate. Sensitivity was analyzed using the MTT assay. In 32Dp210-His396Pro Bcr-Abl, RNAi resulted in a distinct down-regulation of imatinib IC50 values ranging from 1.2 μM in control cells treated with BAF8 (•) and 1.1 μM in EPC (□) reaching 0.3 μM in cells treated with BAF7 (▴). Despite this, the sensitivity of 32Dp210-Thr315Ile did not change after siRNA treatment (IC50: 4.9 [BAF7], 5.2 [BAF8], 4.6 [EPC]). Values are means ± SD of triplicates.
These experiments demonstrate that the biologic phenotype of Bcr-Abl–positive cells and their response to imatinib can be effectively modulated by breakpoint-specific siRNA. Two major mechanisms of imatinib resistance, overexpression of the protein and a proportion of point mutations in the adenosine triphosphate (ATP) binding site, are antagonized by decreasing the quantity of the Bcr-Abl protein using siRNA. We conclude that breakpoint-specific siRNA represents a promising principle for modulating transforming genes and should be further developed toward clinical application.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2002-12-3899.
Supported by the Robert Bosch Foundation (project nos. 010052 and 11.5.8002.0006.1) and by grants from the German Mildred-Scheel-Stiftung (J.D.) and the Bundesministerium für Forschung und Bildung, grant no. SFB456 (J.D.). C.M. is supported by a fellowship from the Deutsche José-Carreras Stiftung.
One of the authors (H.-P.V.) is employed by a company (Ribopharma AG) whose potential product (bcr-abl siRNA) was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr E. Buchdunger (Novartis, Basel, Switzerland) for the generous gift of imatinib and Ribopharma AG (Kulmbach, Germany) for providing the siRNAs.

![Figure 2. bcr-abl RNAi sensitizes cells expressing imatinib-resistant His396Pro-Bcr-Abl domain mutation to imatinib. (A) 32Dp210-wt (•), 32Dp210-Thr315Ile (▴), and 32Dp210-His396Pro (□) were analyzed using MTT assay to determine their sensitivity to imatinib. IC50 values were 0.3 μM in 32Dp210-wt, 1.4 μM in 32Dp210-His396Pro, and 5.2 μM in 32Dp210-Thr315Ile. Values are means ± SD of triplicates. (B) Breakpoint-specific siRNA treatment (200 nM) leads to reestablishment of sensitivity to imatinib in 32Dp210-His396Pro. After treatment, 32Dp210-His396Pro (i) and 32Dp210-Thr315Ile (ii) were exposed to imatinib mesylate. Sensitivity was analyzed using the MTT assay. In 32Dp210-His396Pro Bcr-Abl, RNAi resulted in a distinct down-regulation of imatinib IC50 values ranging from 1.2 μM in control cells treated with BAF8 (•) and 1.1 μM in EPC (□) reaching 0.3 μM in cells treated with BAF7 (▴). Despite this, the sensitivity of 32Dp210-Thr315Ile did not change after siRNA treatment (IC50: 4.9 [BAF7], 5.2 [BAF8], 4.6 [EPC]). Values are means ± SD of triplicates.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/6/10.1182_blood-2002-12-3899/6/m_h81834930002.jpeg?Expires=1768417141&Signature=RzeQc8f3C5jgMBdsgOz5cwOsj14nEHnP48z6eX5gb8IT0PYd1QP5H5pBJ2-IWQyVcIS4qDd9GgcViroddsXZlPqBMUp9kJJR6m6BEa3cWelRHT17XRfutQO9aBKA~NsOWfFWwQ99~m1tmny0O9mdBPwj-z8apFcKa1ktky4GutsK0g8Pf8fXS0isuV6WtKzUU78qWYaOy0qp5IChUiVmiXlZu0g5XW9-taUUB0QvIa5PNsd-QGmz9mn0rqLGj9Z4nV1f5XqN0C4cWQ0k5Ff0yN1Sjea97DiZLpoU0O6DQ-1NzVNm9aHBMNg4DSISI4y4tYu0JT~arNASdaJ0FJucpw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. bcr-abl RNAi sensitizes cells expressing imatinib-resistant His396Pro-Bcr-Abl domain mutation to imatinib. (A) 32Dp210-wt (•), 32Dp210-Thr315Ile (▴), and 32Dp210-His396Pro (□) were analyzed using MTT assay to determine their sensitivity to imatinib. IC50 values were 0.3 μM in 32Dp210-wt, 1.4 μM in 32Dp210-His396Pro, and 5.2 μM in 32Dp210-Thr315Ile. Values are means ± SD of triplicates. (B) Breakpoint-specific siRNA treatment (200 nM) leads to reestablishment of sensitivity to imatinib in 32Dp210-His396Pro. After treatment, 32Dp210-His396Pro (i) and 32Dp210-Thr315Ile (ii) were exposed to imatinib mesylate. Sensitivity was analyzed using the MTT assay. In 32Dp210-His396Pro Bcr-Abl, RNAi resulted in a distinct down-regulation of imatinib IC50 values ranging from 1.2 μM in control cells treated with BAF8 (•) and 1.1 μM in EPC (□) reaching 0.3 μM in cells treated with BAF7 (▴). Despite this, the sensitivity of 32Dp210-Thr315Ile did not change after siRNA treatment (IC50: 4.9 [BAF7], 5.2 [BAF8], 4.6 [EPC]). Values are means ± SD of triplicates.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/6/10.1182_blood-2002-12-3899/6/m_h81834930002.jpeg?Expires=1768417142&Signature=tlz~Gfw9HzO3XIVnk-TNTON9QsTzNW5xfQjeZqyhuSGXxJvSNIU7~FCy8KSQiTRd~WywSXyFjnmhw9VlhOiojoeirEHhOvj6zX3VvUGhWIl3fDYZvPFoktLqidQdZ6JhMuPM5G5lxWZyi8PK4DIAJrIpAtUQzEFtW-YxOAgv9M8pcLrDZOb4G~tzWD~qQ0c2nMWta1PzfLnJmj0gl5K2a5-IeOWYeQWc6aWw-i4oMrT6JS1JE9tI~5KHmxqsVsUGzDqF8~Z3GS~FXIsjYXh~hkFckbQ3AHQyl6oD~YdZWeuvx2OYbO2rF62PsCCdYFKtxUrWB-Mgn7QaC2y0uZVdpw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)