Nucleoside derivatives are currently used in the treatment of hematologic malignancies. Although intracellular events involved in the pharmacologic action of these compounds have been extensively studied, the role of plasma membrane transporters in nucleoside-derived drug bioavailability and action in leukemia cells has not been comprehensively addressed. We have monitored the amounts of mRNA for the 5 nucleoside transporter isoforms cloned so far (CNT1, CNT2, CNT3, ENT1, and ENT2) in several human cell types and in normal human leukocytes. We then examined the expression patterns of these plasma membrane proteins in patients with chronic lymphocytic leukemia (CLL) and correlated them with in vitro fludarabine cytotoxicity. Despite a huge individual variability in the mRNA amounts for every transporter gene expressed in CLL cells (CNT2, CNT3, ENT1, and ENT2), no relationship between mRNA levels and in vitro fludarabine cytotoxicity was observed. Fludarabine accumulation in CLL cells was mostly, if not exclusively, mediated by ENT-type transporters whose biologic activity was clearly correlated with fludarabine cytotoxicity, which reveals a role of ENT-mediated uptake in drug responsiveness in patients with CLL.
Introduction
Most nucleoside-derived drugs currently used in the treatment of hematologic malignancies are purine analogs. These compounds may directly repress DNA replication and interfere with the endogenous nucleotide metabolism, by inhibiting key enzymes like ribonucleotide reductase, thus promoting the incorporation of phosphorylated drugs into DNA. Nucleoside-derived drugs may also exert their cytotoxic action in a non–cycle-dependent manner because they are equally effective in the treatment of B-cell chronic lymphocytic leukemia (CLL).1 This disease is characterized by the accumulation of long-lived, functionally inactive, mature appearing, neoplastic B lymphocytes.2 The clonal excess of B cells is mainly caused by a decrease in cell death rather than by increased cell proliferation.3 Although no curative treatment is currently available for patients with CLL, several drugs, such as purine nucleoside analog fludarabine, have shown high activity against the disease4 and evidence indicates that in CLL cells these drugs exert their cytotoxic effect by promoting apoptosis.5-8
Although nucleoside metabolism appears to be a necessary step in the development of cytotoxicity, the major routes of nucleoside uptake in patients with CLL have not been studied at the molecular level. With the molecular cloning of concentrative, high-affinity, and equilibrative, low-affinity nucleoside transporters (CNTs and ENTs, respectively)9-12 a new panel of putative proteins determining nucleoside-derived drug bioavailability and pharmacologic action is now available for analysis in lymphoproliferative malignancies and other types of cancer.
Three isoforms of CNT transporters have been identified up to now9-12: CNT1, a pyrimidine nucleoside-preferring transporter; CNT2, a purine nucleoside-preferring carrier protein; and the recently cloned CNT3, a broad-specificity nucleoside transporter protein. Two equilibrative nucleoside transporters differing in their sensitivity to the nucleoside analog NBTI have been kinetically defined.10-13 NBTI-sensitive and -insensitive transport systems correspond to ENT1 and ENT2. Nucleoside transport processes across the plasma membrane are not constitutive.14 Neither is the pattern of isoform expression among different cell types and tissues.14,15 Selective loss of nucleoside isoform transporter expression has been reported in animal cancer models,16,17 and very recently in human tumors.18 (X. Farré, E. Guillén-Gómez, L. Sánchez, F.J.C., J. Palacios, M.P.-A., unpublished observations, October 2002).
Immune system cells and cells derived from selected lymphoproliferative malignancies are unique in their ability to relate nucleoside transporter gene expression with transporter biologic function in intact cells. The panel of nucleoside transporters expressed in immune system cells has been determined in our laboratory. Both concentrative and equilibrative transport systems were functionally detected in the human B-cell lines Raji and BLS.19,20 In these studies it was found that they were equally sensitive to cytokine treatment and cell activation. CNT1 and CNT2 transporter expression was also up-regulated after induction of bone marrow murine macrophage apoptosis by treatments with lipopolysaccharide (LPS) or tumor necrosis factor α (TNF-α), whereas the equilibrative transporter isoform ENT1 appeared to respond to cell proliferation induced by macrophage colony-stimulating factor (M-CSF).21,22 Recently, Dumontet and coworkers analyzed the potential mechanisms of resistance to cytarabine in patients with acute myeloid leukemia (AML) by measuring different relevant genes, including hENT1, by reverse transcription–polymerase chain reaction (RT-PCR).23Although these authors showed that reduced hENT1 mRNA in leukemic blasts at diagnosis may correlate with clinical outcome, at present there is no comprehensive study involving all the nucleoside transporter genes cloned so far (CNT1, CNT2,CNT3, ENT1, and ENT2) that aims to identify the role of nucleoside transport processes in nucleoside-derived drug cytotoxicity in CLL.
The aim of this study was to analyze the expression of these nucleoside transporters cloned so far in hematologic cell lines and CLL cells and to study whether transport of fludarabine into cells could explain the sensitivity of these cells to fludarabine-induced cytotoxicity.
Patients, materials, and methods
Patients
Twelve men and 10 women, with a median age of 69 years (range, 41-91 years), diagnosed with CLL and who had not been previously treated with fludarabine were included in the study. The diagnosis was established according to the World Health Organization classification.24 All patients were informed of the nature of this study and informed consent was obtained from each patient in accordance with Hospital Clinic Ethical Committee.
Cell lines
The following human cell lines were used: Raji and Daudi derived from a Burkitt lymphoma, Jurkat derived from an immature T-cell leukemia, Granta 519 and NCEB derived from mantle cell lymphoma, JVM-2 derived from a B-cell prolymphocytic leukemia, DOHH2 derived from a follicular lymphoma, NB4 derived from a promyelocytic leukemia, and SW480 derived from colon adenocarcinoma. Cell lines were purchased from the American Type Culture Collection (ATCC; Manassas, VA), except for Granta 519, NCEB, and JVM-2 that were generous gifts of Dr N. Andersen (Copenhagen, Denmark).
Reagents
Fludarabine monophosphate was obtained from Schering (Berlin, Germany). [5,6-3H]Uridine andd-[1-14C]mannitol were purchased form Amersham Pharmacia Biotech (Buckinghamshire, England) and [8-3H]guanosine, [5-3H(N)]cytidine and [8-3H]fludarabine from Moravek Biochemicals (Brea, CA). Natural nucleosides and fludarabine were from Sigma (St Louis, MO).
Isolation and culture of cells
Mononuclear cells were isolated from 5 mL anticoagulated venous blood by fractionation through a Ficoll/Hypaque (Seromed, Berlin, Germany) gradient. These cells typically were more than 95% positive for CD19 and CD5, as assessed by flow cytometry. CLL cells were used either immediately or after thawing samples that had been cryopreserved in liquid nitrogen in the presence of 10% dimethyl sulfoxide. Lymphocytes were cultured immediately after thawing at a concentration of 5 × 106 cells/mL in RPMI 1640 culture medium supplemented with 10% heat-inactivated fetal calf serum (Gibco BRL, Paisley, Scotland), 2 mM glutamine, and 0.04 mg/mL gentamicin, at 37°C in a humidified atmosphere containing 5% carbon dioxide. The human cell line derived from a prolymphocytic leukemia, JVM-2, was cultured in the same conditions.
RNA extraction and quantitative RT-PCR
Total RNA was isolated from each tumor sample and from cell lines JVM-2, Jurkat, Granta 519, NCEB, DOHH2, NB4, Raji, Daudi, and SW480, using the guanidinium thiocyanate method (Ultraspec; Biotecx Laboratories, Houston, TX).25 RNA was treated with DNAse (Ambion, Austin, TX) to eliminate contaminating DNA. For cDNA synthesis, 1 μg RNA and TaqMan reverse transcription reagents (including Multiscribe reverse transcriptase and random hexamers) were used, as described by the manufacturer (Applied Biosystems, Foster City, CA). The primers and probes used to amplify nucleoside transporters cDNA by real-time PCR were designed using Primer Express software and are listed in Table1. Real-time monitoring of PCR amplification of cDNAs was done using the TaqMan Universal master mix (Applied Biosystems) using 200 nM probe and 100 nM of each primer, in the ABI Prism 7700 sequence Detection System (Applied Biosystems). Relative quantification of gene expression was performed as described in the TaqMan user's manual using human β-glucuronidase (GUSB; Applied Biosystems) as an internal control. The threshold cycle (CT) is defined as the cycle number at which the fluorescence corresponding to the amplified PCR product is detected. The PCR arbitrary units of each transporter was defined as the mRNA levels of these genes normalized to the GUS expression level in each case. RNA from JVM-2 cell line was used as reference control to normalize each nucleoside transporter/GUS amplification ratios.
Nucleoside uptake measurements in JVM-2 cells and CLL cells from patients
Nucleoside uptake was measured using a rapid filtration method adapted from a technique previously characterized at our laboratory.26 Cells were washed and resuspended in either a sodium or a choline chloride buffer, as previously reported. Uptake assays were started by mixing cell suspensions with an equal volume of buffer, supplemented with a radionucleoside at a specific activity of 4000 dpm/pmol for JVM-2 cells and 17 500 dpm/pmol for CLL cells. Nucleosides were routinely used at a concentration of 1 μM, except for the determination of apparent Km values for fludarabine uptake into JVM-2 cells, when concentrations ranging from 0.5 to 500 μM were assayed. When the selected incubation times had elapsed, aliquots (containing 1.2 × 106 and 3-4 × 106 cells for JVM-2 and CLL cells, respectively) were taken and added to ice-cold 0.4-mL needle Eppendorf tubes, containing an upper buffer phase, an intermediate oil layer (dibutylphthalate/bis-3,5-trimethylhexyl)phthalate [3:2, vol/vol]), and a 10% HClO4/25% glycerol solution at the bottom. The tube was immediately centrifuged (15 000g for 60 seconds), thus enabling cells to be separated from the incubation medium and pelleted into the HClO4 layer.d-[1-14C]Mannitol (at a specific activity ranging from 900 to 3500 dpm/pmol) was included in the incubation medium to assess the amount of extracellular medium trapped in the bottom acid layer. To ensure that pelleted cells were fully recovered for radioactivity counting, tubes were blade cut at the oil layer level, releasing the bottom part into scintillation counting vials. Double counting allowed discrimination between the transported substrate (a tritiated nucleoside) and the extracellular marker (d-[1-14C]mannitol). Protein in the transport mixture was measured by the Bradford method (Bio-Rad, Hercules, CA).
Cell viability assay
Cell viability was determined by the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay as previously described.27 CLL lymphocytes (5 × 105 cells/well) were incubated in 96-well plates in the absence or in the presence of fludarabine (3, 7.5, and 15 μM) in a final volume of 100 μL. After 48 hours, 10 μL MTT (5 mg/mL in phosphate-buffered saline [PBS]) was added to each well for a further 6 hours. The blue MTT formazan precipitated was dissolved in 100 μL isopropanol/1 M HCl (24:1) and the absorbance values at 550 nm were determined on a multiwell plate reader.
Statistical analysis
Correlations between nucleoside transporter gene expression as well as correlations between nucleoside transport and fludarabine-induced cytotoxicity were analyzed using SPSS 10.0 software package (SPSS, Chicago, IL).
Results
Nucleoside transporter isoform expression pattern in normal human leukocytes and human cell lines
The patterns of nucleoside transporter expression were analyzed by quantitative RT-PCR in normal human leukocytes and human cell lines. Normalized CT values were considered for comparison purposes. This approach allowed a quantitative comparison among cells and cell lines for every studied nucleoside transporter gene:CNT1, CNT2, CNT3, ENT1, andENT2. In Table 2, normalized CT values have been substituted by symbols to reflect the extent of gene expression for every single nucleoside transporter gene. Positive symbols in a scale from 1 to 3 reflect increasing expression levels, as indicated in the footnote to Table 2.
CNT1, the high-affinity nucleoside transporter involved in fluoropyrimidine uptake, is not expressed in normal human leukocytes and human leukemic cell lines. A colon-derived cell line (SW480) and human kidney cDNA were used as positive controls, showing that the lack of expression of CNT1 is not the result of impaired amplification reaction. CNT2 was broadly expressed in all analyzed cells, whereas CNT3 had a low expression in 3 of 8 leukemic cell lines studied, and it was detected at low levels in normal leukocytes. In contrast, ENT transporters were expressed in all cases and no great variability among cells and cell lines in mRNA levels was observed.
CNT and ENT mRNA expression levels in patients with CLL
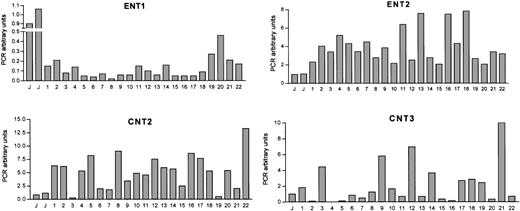
Figure 1 shows the amount of ENT1, ENT2, CNT2, and CNT3 mRNAs in CLL cells from 22 patients. The expression of each nucleoside transporter in JVM-2 cells was used as a relative calibrator and the expression levels of these cells were assigned the value of 1 as an arbitrary unit. No CNT1 expression was observed in any of CLL patients analyzed even when the number of amplification cycles was increased to 50, in agreement with the observations in most normal human leukocytes and leukemic cell lines (Table 2).
Nucleoside transporter mRNA expression levels in CLL cells.
CT values for nucleoside transporter RT-PCR amplification from CLL cDNA have been normalized to an endogenous reference gene (β-glucuronidase). mRNA expression levels are given in arbitrary units, using the JVM-2 cell line as reference control. PCR arbitrary units were defined as described in “Patients, materials, and methods.”
Nucleoside transporter mRNA expression levels in CLL cells.
CT values for nucleoside transporter RT-PCR amplification from CLL cDNA have been normalized to an endogenous reference gene (β-glucuronidase). mRNA expression levels are given in arbitrary units, using the JVM-2 cell line as reference control. PCR arbitrary units were defined as described in “Patients, materials, and methods.”
Significant heterogeneity among patients was observed in ENT1, ENT2, CNT2, and CNT3 mRNA levels. Indeed, the expression pattern of the 4 studied genes for every patient did not result in any significant partition into subgroups when a cluster analysis was performed (not shown). When every transporter gene was considered individually, the highest variability was found for the CNT3 carrier, whereas the isoform showing least heterogeneity in the population was ENT2. Extensive correlation analysis was performed to assess a possible relationship between these genes at the mRNA level but no correlation was found (data not shown).
Characterization of nucleoside transport into JVM-2 cells
A basic characterization of the transport processes involved in nucleoside uptake into the model cell line JVM-2 was performed. Uridine was initially chosen as a substrate for 2 reasons: first, it is broadly recognized by all cloned human nucleoside transporters, and, second, it is poorly metabolized. Uridine uptake into JVM-2 cells was clearly linear for up to 10 minutes, whereas a significant fraction of this transport activity appeared to be sodium dependent, according to the differences found in uridine accumulation measured either in a sodium chloride or a choline chloride medium (Figure2A). Some pyrimidine and purine nucleosides, such as cytidine and guanosine, appear to be transported independently by the pyrimidine-preferring and purine-preferring nucleoside carriers, CNT1 and CNT2, respectively. In principle, both may be substrates for the recently cloned isoform CNT3. To determine which specific nucleoside transporter isoforms would account for the uridine transport activity detected in JVM-2 cells, further experiments were performed using guanosine and cytidine as substrates. Uptake of both nucleosides was linear for several minutes (not shown) and was also partially sodium dependent, although a significant component of nucleoside transport into JVM-2 cells appeared to be equilibrative (Figure 2B,D). The NBTI-sensitive and -insensitive carriers ENT1 and ENT2 appear to be responsible for this uptake activity, in agreement with the pattern of mRNA expression detected in this cell line (Table2). Because the pyrimidine-preferring transporter CNT1 was not detected in JVM-2 cells in terms of mRNA, cytidine uptake may be the result of CNT3 expression. In that case, pyrimidine (cytidine) transport would be cross-inhibited by a purine nucleoside, such as guanosine. As shown in Figure 2E, 100 μM guanosine completely blocked sodium-dependent cytidine uptake, which strongly indicates that pyrimidine nucleoside in JVM-2 cells is CNT3 mediated. Cross-inhibition of guanosine transport by cytidine was equally detected (Figure 2C) although not fully achieved under similar experimental conditions, which would be in agreement with coexpression of CNT2, as anticipated from the gene expression analysis data (Table 2).
Characterization of nucleoside transport into JVM-2 cells.
(A) Time course of uridine uptake into JVM-2 cells. Cells were incubated with 1 μM uridine either in a NaCl (▪) or a choline chloride (▴) medium. Na+-dependent transport (○) was calculated by subtracting those rates measured in choline medium from those measured in Na+ medium. Transport of the purine-nucleoside guanosine (B-C) and the pyrimidine nucleoside cytidine (D-E) was similarly measured under initial velocity conditions (2 minutes). Cross-inhibitions of Na+-dependent uptake were also monitored to further elucidate which nucleoside transporters were responsible for concentrative uptake activity (CNT-related). Transport of 1 μM guanosine was inhibited with 100 μM cytidine (C) and transport of 1 μM cytidine with 100 μM guanosine (E). To determine the contribution of equilibrative transport activity (ENT), 1 μM NBTI was used to discriminate between NBTI-sensitive (ENT1) and NBTI-insensitive (ENT2) transport rates. Results are the mean ± SE of 3 to 6 experiments measured in triplicate.
Characterization of nucleoside transport into JVM-2 cells.
(A) Time course of uridine uptake into JVM-2 cells. Cells were incubated with 1 μM uridine either in a NaCl (▪) or a choline chloride (▴) medium. Na+-dependent transport (○) was calculated by subtracting those rates measured in choline medium from those measured in Na+ medium. Transport of the purine-nucleoside guanosine (B-C) and the pyrimidine nucleoside cytidine (D-E) was similarly measured under initial velocity conditions (2 minutes). Cross-inhibitions of Na+-dependent uptake were also monitored to further elucidate which nucleoside transporters were responsible for concentrative uptake activity (CNT-related). Transport of 1 μM guanosine was inhibited with 100 μM cytidine (C) and transport of 1 μM cytidine with 100 μM guanosine (E). To determine the contribution of equilibrative transport activity (ENT), 1 μM NBTI was used to discriminate between NBTI-sensitive (ENT1) and NBTI-insensitive (ENT2) transport rates. Results are the mean ± SE of 3 to 6 experiments measured in triplicate.
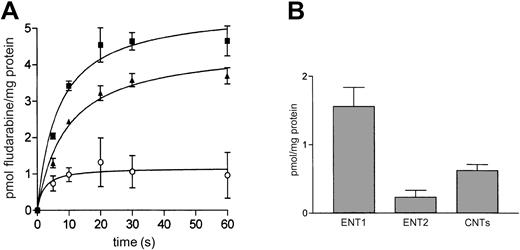
Fludarabine uptake into JVM-2 cells
The same method (see “Characterization of nucleoside transport into JVM-2 cells”) was used to monitor fludarabine transport into JVM-2 cells. Interestingly, this cell line showed a much higher capacity to accumulate fludarabine than natural nucleosides. In fact, transport processes were extremely rapid, and thus linear velocity conditions were lost before the first minute of incubation (Figure 3A). Although this feature would compromise conventional determinations of substrate uptake kinetics, fludarabine accumulation into JVM-2 cells could be measured reliably at short incubation time points (ie, 5 seconds) and reflects, as for the natural substrates, the pattern of gene expression found for JVM-2 cells shown in Table 2. As for all other nucleosides analyzed in this study, fludarabine uptake was also partially dependent on the sodium transmembrane gradient. A significant sodium-dependent component of transport was found along with an equilibrative uptake, which was mostly NBTI sensitive and, thus, associated with ENT1 expression (Figure 3B). Because the major component of fludarabine uptake was equilibrative, apparent Km values for fludarabine transport into JVM-2 cells were derived from uptake measurements (data not shown) performed in choline chloride medium, either in the presence or in the absence of 1 μM NBTI. Apparent Km for ENT1-mediated fludarabine transport was 82 μM. Thus, drug concentrations used are below the apparent Km. Vmax values ranged from 22 to 37 pmol/s/mg protein.
Fludarabine uptake into JVM-2 cells.
(A) Time course of fludarabine uptake. Cells were incubated with 1 μM fludarabine either in an NaCl (▪) or a choline chloride (▴) medium. Na+-dependent transport (○) was calculated by subtracting those rates measured in choline medium from those measured in Na+ medium. (B) Discrimination among the different nucleoside transporter systems was performed as described in Figure 2. Transport rates were measured at 5 seconds. Results are the mean ± SE of 3 different determinations measured in triplicate.
Fludarabine uptake into JVM-2 cells.
(A) Time course of fludarabine uptake. Cells were incubated with 1 μM fludarabine either in an NaCl (▪) or a choline chloride (▴) medium. Na+-dependent transport (○) was calculated by subtracting those rates measured in choline medium from those measured in Na+ medium. (B) Discrimination among the different nucleoside transporter systems was performed as described in Figure 2. Transport rates were measured at 5 seconds. Results are the mean ± SE of 3 different determinations measured in triplicate.
Nucleoside uptake into cells from patients with CLL
Once the basal uptake measurement conditions had been established for natural nucleosides and for fludarabine, nucleoside transport activity was analyzed in CLL cells. Because both cryopreserved and fresh samples have been used in this study, the putative effect of freezing and storing on nucleoside transporter performance was evaluated in randomly selected patients. Uptake rates were identical in fresh and cryopreserved CLL cells (not shown). Although mRNA expression of CNT2 or CNT3 (or both) was detected in all patients, sodium-dependent guanosine transport activity was only detected in 12 of 22 patients, and only 5 of these 12 patients showed relatively high transport activity (not shown). Intracellular accumulation of this nucleoside did not reveal any significant correlation with the CNT and ENT gene expression pattern. Interestingly, when short-term accumulation of fludarabine was analyzed (at 5-10 seconds after exposing cells to the drug) the sodium transmembrane gradient did not concentrate fludarabine even in those patients who retained sodium-dependent guanosine transport activity. This means that fludarabine transport into CLL cells is mostly, if not uniquely, mediated by ENT-type transporters, which is indeed consistent with the results obtained in JVM-2 cells (Figure 3) in which most of the uptake of fludarabine was mediated by ENT1, despite a fast sodium-dependent component of drug accumulation. Nevertheless, the possibility that sodium-dependent accumulation of fludarabine occurs at longer incubation times in CLL cells showing sodium-dependent guanosine uptake cannot be ruled out.
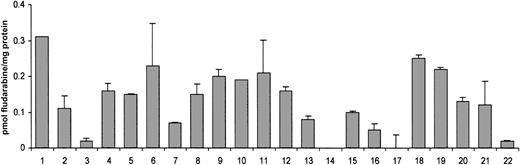
Figure 4 shows the variability in equilibrative fludarabine uptake into CLL cells from the 22 patients studied. Two patients did not show any significant fludarabine transport activity at short-time incubation points, whereas the other 20 retained this ability in a range of individual variation that was close to 20-fold.
Fludarabine uptake into cells from patients with CLL.
CLL cells were incubated as described in “Patients, materials, and methods” in the presence of 1 μM fludarabine for 10 seconds, either in an NaCl or a choline chloride medium. No Na+-dependent uptake was detected. Results are expressed as equilibrative uptake of fludarabine, measured in duplicate for each patient (mean ± SEM).
Fludarabine uptake into cells from patients with CLL.
CLL cells were incubated as described in “Patients, materials, and methods” in the presence of 1 μM fludarabine for 10 seconds, either in an NaCl or a choline chloride medium. No Na+-dependent uptake was detected. Results are expressed as equilibrative uptake of fludarabine, measured in duplicate for each patient (mean ± SEM).
Ex vivo sensitivity to fludarabine and its relationship with fludarabine uptake
In 20 patients in vitro sensitivity of CLL cells to fludarabine was analyzed. As expected, incubation of cells with pharmacologic doses of fludarabine (3 μM) and higher (7.5 and 15 μM) induced a variable response among cells from different patients (data not shown) with some cases showing a high fludarabine-induced cytotoxic effect and others being resistant. In most cases in which mild and strong resistance were observed, no reliable median effective concentration (EC50) values could be calculated because a significant fraction of cells did not undergo apoptosis. For this reason cell viability values were calculated in the range of drug concentrations considered to be pharmacologically relevant. There was no correlation between CNT or ENT gene expression levels and fludarabine-induced cytotoxicity. However, there was a negative correlation between the equilibrative transport activity for the drug (always measured at 1 μM) and the in vitro fludarabine-induced cytotoxicity (Figure5A-B). A significant correlation was observed at 3 different concentrations of fludarabine:P = .008, r = 0.576 for fludarabine 15 μM (not shown);P = .006, r = 0.591 for fludarabine 7.5 μM (Figure5B); and P = .029, r = 0.488 for fludarabine 3 μM (Figure 5A).
Correlation between fludarabine uptake and ex vivo sensitivity to fludarabine.
CLL lymphocytes were incubated for 48 hours either in the absence or in the presence of fludarabine at different concentrations (3 μM [A] and 7.5 μM [B]). Cell viability was determined by an MTT assay. Data are shown as the mean value of triplicate cultures. Correlation coefficient and P values are given in “Results.”
Correlation between fludarabine uptake and ex vivo sensitivity to fludarabine.
CLL lymphocytes were incubated for 48 hours either in the absence or in the presence of fludarabine at different concentrations (3 μM [A] and 7.5 μM [B]). Cell viability was determined by an MTT assay. Data are shown as the mean value of triplicate cultures. Correlation coefficient and P values are given in “Results.”
Discussion
This study demonstrates a relationship between fludarabine uptake via equilibrative transport systems into cells from patients with CLL and their in vitro sensitivity to this drug. This observation is relevant because several studies have found a correlation between in vitro and in vivo treatment results.7,28-31 Furthermore, although CNT-type transporter genes appear to be expressed in CLL cells, most, if not all, of the fludarabine uptake, as well as the natural nucleoside transport of CLL cells, rely on ENT-type carriers. Indeed, ENT1-mediated activity has been shown to be required for gemcitabine cytotoxicity in a variety of leukemia cell lines.32 This results strongly suggest that transporter-associated biologic activity might be a more reliable predictor of response to fludarabine in patients with CLL than the quantitative analysis of transporter mRNAs. This may not be a general rule because other types of cancer and other nucleoside-derived drugs may show other types of correlations between CNT and ENT biologic function and mRNA expression levels. Thus, as recently reported, low ENT1 mRNA levels at diagnosis, as determined by quantitative RT-PCR analysis, appear to play a significant role in resistance to cytarabine in patients with AML.23 As discussed, this is not the case for patients with CLL treated with fludarabine.
The extent to which mRNA levels of nucleoside transporter genes relate with their corresponding protein amounts and the latter with biologic activity is unknown due to the lack of appropriate antibodies against the whole set of cloned human transporters. Polyclonal monospecific antibodies against the rat isoforms of CNT1 and CNT2 have been generated in our laboratory.15,33 A lack of correlation between CNT1 mRNA levels and its corresponding protein amounts was initially found in rat liver.33 Comprehensive CNT1 and CNT2 distribution analysis of rat tissues using Western blot also revealed this lack of correlation with CNT mRNA pattern distribution.15 In rat cell and in vivo models of hepatocarcinogenesis it has been shown that nucleoside transporter expression can be lost selectively.16,17 More recently, an antibody against human ENT1 has been characterized by others and used to monitor nucleoside transporter expression in breast cancer patients.18 Although several ENT1−individuals were detected, this was not correlated with response to treatment or outcome. A polyclonal monospecific antibody against human CNT1 suitable for immunohistochemistry analysis in paraffin-embedded tissues has recently been raised and characterized in our laboratory.34 Tissue array technology applied to gynecologic tumors has revealed similar heterogeneous patterns in CNT1 expression (X. Farré, E. Guillén-Gómez, L. Sánchez, F.J.C., J. Palacios, M.P.-A., unpublished observations, October 2002). To what extent protein levels will correlate with biologic activity is also unclear because nucleoside carrier proteins appear to be located also intracellularly35 and posttranslational modification of transporters has also been reported.36 Intracellular trafficking defects may help to explain the lack of correlation between CNT2 and CNT3 mRNA levels with nucleoside uptake in CLL cells, but the lack of antibodies against these 2 human isoforms (not available commercially and not even reported in the literature) makes it difficult to address this point. But then, why do some CLL cells show sodium-dependent transport activity but lack sodium-dependent fludarabine accumulation? Guanosine is a good substrate for both CNT237,38 and CNT339 transporters, whereas fludarabine does not appear to be transported by the human isoform of CNT2,40 although it is a substrate for CNT3.39 Thus, those cells showing concentrative guanosine uptake would express CNT2- but not CNT3-related transport activity. The high variability in CNT3 mRNA levels among patients, along with negligible concentrative uptake of fludarabine, would be consistent with the idea that CNT3 is a highly inducible transporter. Moreover, its regulation from the gene to the active protein inserted at the plasma membrane would require a precise set of still unknown events in lymphoid cells, which are probably differentially altered in CLL cells. With the recent cloning of this isoform, it has been shown that CNT3 mRNA is present in HL60 cells at very low levels, but it is highly inducible (up to 17-fold) after cells are induced to differentiate.39
Altogether, ENT-mediated fludarabine accumulation appears to be the only pathway for carrier-mediated drug uptake into cells from CLL patients. Equilibrative uptake of substrates into cells can be driven unidirectionally in an efficient manner if strong coupling with metabolism is achieved. This is the most likely explanation for the fast accumulation of fludarabine in JVM-2 cells. The role of carrier-mediated processes in this event is clearly demonstrated by its sodium dependence (at least in JVM-2 cells) and its high NBTI sensitivity. Nevertheless, even with incubation time periods as short as 5 to 10 seconds, metabolism cannot be ruled out. Fludarabine uptake data should then not be interpreted as initial velocity measurements to be used for kinetic analysis, but rather as estimations of carrier-mediated accumulation of drug inside cells. The interesting point of this contribution is that measurements of drug accumulation performed a few seconds after fludarabine addition clearly correlate with in vitro cytotoxicity tests in which cells were incubated in the presence of the drug for 48 hours. Assuming that passive nonmediated diffusion can occur, the relevance here demonstrated of carrier-mediated processes in determining drug responsiveness would be probably related to metabolic channeling of substrates into specific intracellular drug targets, a situation that would not be achieved by passive diffusion of substrates across the plasma membrane.
In summary, this study shows that cells from patients with CLL express 4 nucleoside transporter genes, ENT1, ENT2,CNT2, and CNT3, with very different mRNA levels lacking any significant correlation with in vitro sensitivity to fludarabine treatment. This is probably due to the fact that nucleoside transport activity, directly measured in CLL cells, is mostly equilibrative (ENT related), although in some cases a CNT2 concentrative sodium-dependent transporter activity was found. Because fludarabine is not a CNT2 substrate, drug uptake is mediated by ENT transporters (mostly ENT1). In addition, ENT-mediated fludarabine transport significantly correlates with in vitro sensitivity to the drug. Therefore, we conclude that transporter-associated biologic activity is a much more reliable predictive marker of therapeutic response to fludarabine in patients with CLL than the quantitative analysis of transporter mRNAs.
The authors would like to thank Mari Carmen Vela for her excellent technical assistance, and Robin Rycroft for his editorial help. The current affiliation for B.B. is Servei de Patologia, Hospital del Mar, Barcelona, Spain.
Prepublished online as Blood First Edition Paper, October 31, 2002; DOI 10.1182/blood-2002-07-2236.
Supported in part by grants SAF99-0115, SAF2002-00717 (Ministerio de Ciencia y Tecnologı́a, Spain) and a Grant-in-Aid from Fundación Ramón Areces (Spain), and Generalitat de Catalunya (M.P.-A.), and grant 00-946 (Fondo de Investigación Sanitaria, Ministerio de Sanidad y Consumo, Spain) and a grant-in-aid from AECC (Asociación Española Contra el Cáncer; D.C.) and a grant from Jose Carreras International Foundation Against Leukemia (EM-P-01; E.M.). M.M.-A. was an AECC fellow and now holds a research fellowship from the Ministerio de Educación, Cultura y Deportes, Spain. B.B. was recipient of a research fellowship from the Instituto de Salud Carlos III.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dolors Colomer, Unitat Hematopatologia, Hospital Clinic, Villarroel 170, 08036 Barcelona, Spain; e-mail:dcolomer@medicina.ub.es; or Marçal Pastor-Anglada, Departament de Bioquı́mica i Biologia Molecular, Universitat de Barcelona, Diagonal 645, 08028 Barcelona, Spain; e-mail:mpastor@bio.ub.es.


![Fig. 5. Correlation between fludarabine uptake and ex vivo sensitivity to fludarabine. / CLL lymphocytes were incubated for 48 hours either in the absence or in the presence of fludarabine at different concentrations (3 μM [A] and 7.5 μM [B]). Cell viability was determined by an MTT assay. Data are shown as the mean value of triplicate cultures. Correlation coefficient and P values are given in “Results.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/6/10.1182_blood-2002-07-2236/4/m_h80633978005.jpeg?Expires=1764998410&Signature=Q-i3F9cQlgjZrflBpsuHHe6XwKt2xtCmNhFUETQ4uZTgbouUjpJdcxYnvzAMiZzeTfduqVKAlDkks2ES~js9VoD0peCKCDe~C~n3YV5nXd7OwDLw0zYD5rjb5nDWS345tnMK2jaMTkmmgWpZEchGkYooYoWrAUJY0uS7CvJFALgLtkFbxH-PYdZZ3STlyY6S8KZJjPE~~1UUfAjWGIVDJPICFOfGPTQNYrLpB1i4kYGT9WI2iT9SAXAVDGSLau~zkKj-Uya0zWAvx2owFGYLuM~YQ543jZZc6Kb-zLYABl4IhMNiuIse6-3hFjToBr-dy7fVnbgNgXYh9Vl1he~jfQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
