Hematopoietic stem and progenitor cells have distinct differentiation stage-specific immortalization potentials that involve Bahcc1.
Results suggest a novel MLL-fusion-Bahcc1 axis in MLL-fusion–mediated leukemogenesis.
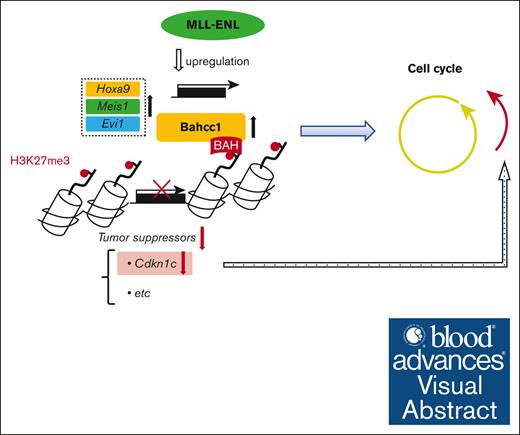
Visual Abstract
In leukemogenesis, genotoxic stress in hematopoietic stem and progenitor cells (HSPCs) drives individual context-dependent programs of malignant transformation. In light of the various differentiation stages of HSPCs based on a recently revised definition using CD150/CD48, our analyses showed that a subpopulation of long-term repopulating HSCs was most susceptible to MLL-ENL–mediated transformation. An analysis of the molecular mechanism identified Bromo-adjacent homology domain and coiled-coil containing 1 (Bahcc1), which encodes a reader molecule of trimethylated histone H3 lysine 27 (H3K27me3), as a candidate gene involved in distinct susceptibility to leukemic transformation. Interestingly, Bahcc1 was previously reported to be highly expressed in acute myeloid leukemia (AML) with an unfavorable prognosis, including some cases of MLL-rearranged AML. We found that MLL-ENL upregulated Bahcc1 through binding to its promoter, and that Bahcc1 was involved in MLL-ENL–mediated immortalization at least partly through repression of H3K27me3-marked Cdkn1c. Analyses using bone marrow transplantation in mice showed that depletion of Bahcc1 suppressed the leukemogenic activity of MLL-ENL. In a public database, high BAHCC1 expression was found to be associated with a poor prognosis in pediatric AML, in which BAHCC1 expression was significantly lower in MLL-AF9-AML than in other MLL-fusion-AML. These findings shed light on the distinct immortalization potential of HSPCs and suggest a novel MLL-fusion-Bahcc1 axis, which may lead to development of molecular targeted therapy against MLL-fusion–mediated leukemia.
Introduction
Acute myeloid leukemia (AML) is a hematological malignancy arising from a subpopulation of leukemic stem cells.1-4 Previous studies5-8 have shown that oncogenic events may transform normal hematopoietic stem cells (HSCs) and progenitor cells into leukemic stem cells. Interestingly, we have shown9 that conditional expression of MLL-ENL leads to leukemic transformation of CD34−c-KithighSca-1highLineage− (KSL) cells (previously defined as murine long-term repopulating HSCs [LT-HSCs]), but not that of CD34+KSL cells (previously defined as murine short-term repopulating HSCs [ST-HSCs]) or myeloid progenitor cells. Recent advances10-13 have also led to revision of the definition of HSCs and immature progenitor cells (HSPCs), using markers including CD150/CD48. However, little is known about the molecular mechanism of leukemic transformation in the “revised” HSPCs.
A class of epigenetic regulators14-18 has been found to play a key role in leukemogenesis. One epigenetic regulatory mechanism is suppression of expression of genes marked with trimethylated histone H3 lysine 27 (H3K27me3),19-22 which is recognized by reader molecules, leading to chromatin compaction. In addition to the canonical readers, Bromo-adjacent homology (BAH) domain and coiled-coil containing 1 (BAHCC1), a large protein of 2608 amino acids, recognizes H3K27me3 via the BAH domain,23,24 which is shared with BAH domain containing 1 (BAHD1). Both molecules mediate gene silencing by recruiting transcriptional repressor complexes. Interestingly, BAHCC1 is highly expressed in AML with an unfavorable prognosis, including some cases of MLL-rearranged leukemia.25 In addition, BAHCC1 depletion suppresses growth of human leukemia cell lines expressing MLL-AF4 in vitro, while BAHCC1/Bahcc1 is critical for several types of leukemogenesis in vitro and in vivo but not for BCR-ABL–mediated leukemogenesis.23 Thus, the precise role of BAHCC1/Bahcc1 in MLL-fusion–mediated leukemogenesis remains unclear.
To investigate the molecular mechanism of MLL-fusion–mediated leukemogenesis in HSPCs more closely, we first examined the immortalization potential of MLL-ENL in “revised” HSPCs. The analyses showed that conditionally expressed MLL-ENL definitively immortalized “revised” LT-HSCs but did not have this effect on more differentiated multiple potent progenitor (MPP) cells and did not always immortalize immature progenitor cells. Comprehensive gene expression analyses of the HSPCs revealed increased expression of Bahcc1 in the immortalized cells. In vitro analyses of the role of Bahcc1 in leukemogenesis showed that MLL-ENL upregulated Bahcc1 through promoter binding, and that Bahcc1 was involved in MLL-ENL–mediated immortalization, at least partly through repression of H3K27me3-marked cyclin-dependent kinase inhibitor 1c (Cdkn1c). Furthermore, in vivo analyses showed that depletion of Bahcc1 suppressed the leukemogenic activity of MLL-ENL, suggesting that Bahcc1 may be a target for treatment of MLL-rearranged AML. Collectively, this study reveals distinct differentiation stage-specific immortalization potentials in HSPCs and describes a novel MLL-fusion-Bahcc1 axis in MLL-fusion–mediated leukemogenesis.
Methods
Construct
pMYs retroviral vectors harboring a FLAG (DYKDDDDK peptide)-tagged MLL-ENL internal ribosomal entry site (IRES)-neor or -EGFP have been described elsewhere.9 A 5'-fragment of Bahcc1 generated with polymerase chain reaction (PCR) products using complementary DNA (cDNA) derived from immortalized LT-HSCs (described below) was combined with a fragment derived from a mouse cDNA clone M5C1031K16 (Dnaform, Tokyo, Japan) to produce full-length Bahcc1 with a 3XFLAG tag at the C-terminus in pcDNA3.1 (Thermo Fisher Scientific Inc, Waltham, MA).
Mice
Compound C57BL/6 (B6) mice conditionally expressing cytomegalovirus early enhancer/chicken β actin (CAG) promoter–driven MLL-ENL by inducible Cre-estrogen receptor α chain (CreER) fusion protein expressed from the Rosa26 locus (C/Tg mice) have been previously described.9,26 All animal studies were approved by the Animal Care Committee of Mie University (no. 24-34-re5).
Purification of HSPCs
LT-HSCs (CD150+/CD48−KSL), ST-HSCs (CD150−/CD48−KSL), MPP2 (CD150+/CD48+KSL) cells, and MPP3/4 (CD150−/CD48+KSL) cells of C/Tg mice, and KSL and c-KithighSca-1−Lin− (hereafter designated MP) cells of wild-type B6 mice were purified from the respective bone marrow (BM) cells using a fluorescence-activated cell sorting (FACS) Aria II (BD Biosciences, Franklin Lakes, NJ).9,26 In brief, lineage-depleted cells were isolated from BM mononuclear cells labeled with Lineage Cell Detection Cocktail-Biotin using a MACS cell separation system (Miltenyi Biotec, Auburn, CA). For purification of LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells derived from C/Tg mice, lineage-depleted cells were stained with peridinin chlorophyll protein (PerCP)-Cy5.5-conjugated streptavidin, and phycoerythrin (PE)–conjugated anti-c-Kit (2B8), PE-Cy7–conjugated anti-Sca-1 (D7), allophycocyanin (APC)–conjugated anti-CD150 (TC15-12FL12.2), and brilliant violet 421 (BV421)–conjugated anti-CD48 (HM48-1) monoclonal antibodies. For purification of KSL and MP cells derived from wild-type B6 mice, lineage-depleted cells were stained with PerCP-Cy5.5–conjugated streptavidin, and PE-conjugated anti-Sca-1 (D7) and APC-conjugated anti-c-Kit (2B8) monoclonal antibodies. All monoclonal antibodies were purchased from BioLegend (San Diego, CA).
Retroviral transduction
Myeloid immortalization assay
Myeloid immortalization assays via serial replating were performed as described elsewhere.26 In brief, purified LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells (150 cells) were directly sorted into 1.5 mL of methylcellulose medium prepared from M3234 (StemCell Technologies, Vancouver, Canada) according to the manufacturer’s protocol, supplemented with 25 ng/mL mouse stem cell factor, 10 ng/mL each of human interleukin 6, mouse interleukin 3, and mouse granulocyte-macrophage colony–stimulating factor (Miltenyi Biotec), and 0.05 μM of 4-hydroxytamoxifen (Sigma-Aldrich, St Louis, MO) dissolved in ethanol or an equal amount of ethanol as a vehicle control. Sorted cells in 1 mL of the mixture were immediately plated in 35-mm dishes. KSL and MP cells retrovirally transduced with MLL-ENL-IRES-neor were also seeded in the same M3234-based methylcellulose medium supplemented with the same kinds of cytokines and 1 mg/mL G418 (Thermo Fisher Scientific Inc).
After culturing for 5 to 7 days, colonies were enumerated and single-cell suspensions (1 × 104 cells) of colonies were subsequently replated in α-MEM (Thermo Fisher Scientific Inc)–based medium containing 1% methylcellulose (Shin-Etsu Chemical Co, Ltd, Tokyo, Japan) supplemented with the same cytokines without 4-hydroxytamoxifen, ethanol, or G418. After the third round of plating, 5 × 103 of the cells forming >5 colonies were additionally replated. The colony-forming cells were harvested at the end of the fourth round of plating and were cultured in α-MEM supplemented with 20% fetal bovine serum and the same cytokines to obtain immortalized cells that were capable of unlimited proliferation under liquid culture.
Gene silencing
The target sequences for Bahcc1 and luciferase were 5′-GAAGAACCTGCTGAAATAC-3′ (shB-2) and 5′-AAAGTGGCCAACAAAGAGG-3′ (shB-8), and the shLuc sequence described elsewhere.29 Short hairpin RNAs (shRNAs) sequences were inserted into pMXsU6-Kusabira Orange (KO)9 or pMXsU6-puro (a kind gift from Toshio Kitamura, The Institute of Medical Science, University of Tokyo). Cells retrovirally transduced with pMXsU6-KO harboring an shRNA sequence were subjected to sorting according to expression of KO on the FACS Aria II 2-3 days after transduction. Cells transfected with pMXsU6-puro harboring an shRNA sequence were selected with 30 μg/mL puromycin (Sigma-Aldrich) for 48 hours at 2 days after transfection.
BMT
Assessment of leukemogenic activity in vivo was performed using BM transplantation (BMT).9,30 Briefly, splenocytes were harvested from leukemic B6 mice in primary BMT using retrovirally MLL-ENL-IRES-EGFP–transduced KSL cells and were retrovirally transduced with shRNA/KO expressors in pMXsU6-KO. At 48 hours after transduction, 900 green fluorescent protein (GFP)+ cells with high KO expression were sorted using the FACS Aria II and immediately transplanted into sublethally irradiated (5.25 Gy) recipient B6 mice. BM and spleen tissues from morbid mice were analyzed as described elsewhere.30
RNA seq and data analysis
Candidate genes involved in the differential modes of myeloid immortalization among HSPCs were found on the basis of their increased expression levels (log2 fold change [FC] >1 in induced ST-HSCs and log2 FC >0 in induced MPP2 cells, or log2 FC >1 in induced MPP2 cells and log2 FC >0 in induced ST-HSCs, with adjusted P values < .05) in RNA sequencing (RNA seq). Genes that were abundantly detected (count per million >10 in induced ST-HSCs and MPP2 cells and their respective controls) were focused on, followed by selection of genes with exclusive enhancement of expression in CD34− KSL cells, compared with that in CD34+ KSL cells, in a previous cDNA microarray analysis.9
Statistical analysis
An unpaired Welch t test and 1-way analysis of variance followed by a Tukey-Kramer or Dunnett test as a post hoc test were used to compare 2 groups and >2 groups, respectively. Quantitative PCR (qPCR) data were log2-transformed before statistical analysis. The probability of overall survival of the mice, and that of patients with AML from public databases, was estimated using the Kaplan-Meier method and compared using log-rank tests, with Bonferroni correction in comparison among 3 groups. All statistical tests were performed using GraphPad Prism 9 (GraphPad Software, San Diego, CA).
FACS, immunoprecipitation (IP), western blotting, RNA seq, reverse transcription/chromatin immunoprecipitation (ChIP)–qPCR, gene silencing in human leukemic cell lines, and bioinformatics analysis are described in the supplemental Methods.
All animal studies were approved by the Animal Care Committees of Mie University (No. 24-34-re5)
Results
LT-HSCs are immortalized by conditional expression of MLL-ENL, but MPP3/4 cells are not, whereas ST-HSCs and MPP2 cells are not always immortalized
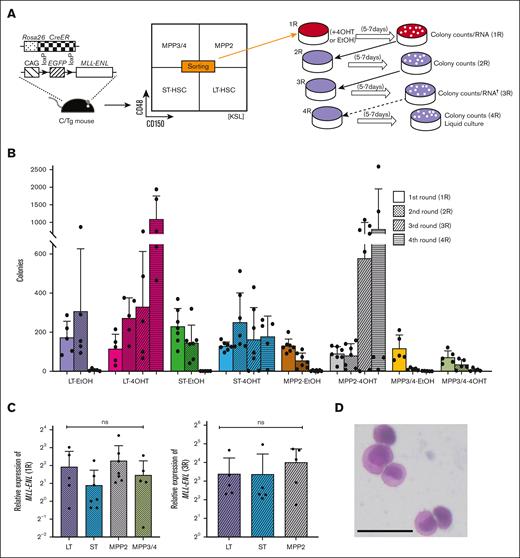
To examine the immortalization potential of MLL-ENL in the “revised” HSPCs, LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells from C/Tg mice were subjected to myeloid immortalization assays by conditional expression of MLL-ENL (Figure 1A-B; supplemental Figure 1). Although nearly equal expression levels of MLL-ENL were induced at the end of the initial plating (Figure 1C), only LT-HSCs definitely formed compact colonies composed of morphologically myelomonocytic blasts (Figure 1D) after serial replating, whereas MPP3/4 cells did not do so. ST-HSCs and MPP2 cells formed colonies in 5 of 7 trials after serial replating (Figure 1B). In addition, in transferring cells harvested from the fourth plating into liquid culture for generating immortalized cells, the cells harvested from LT-HSC-derived replatable colonies always proliferated under liquid culture, whereas those harvested from ST-HSC- or MPP2-derived replatable colonies did so in only 2 of 7 trials (Figure 1B-C).
Myeloid immortalization potential of LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells with conditional expression of MLL-ENL. (A) Experimental strategy for myeloid immortalization assays using serial replating. LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells were purified from BM cells derived from C/Tg mice conditionally expressing CAG promoter-driven MLL-ENL by inducible Cre-estrogen receptor α chain (CreER) fusion protein expressed from the Rosa26 locus by sorting with CD150/CD48 expression in KSL-gated cells. Initially, 100 cells from sorted cells were seeded with 4-hydroxytamoxifen (4OHT) or EtOH, followed by serial replating of 104 cells without drug. In cases with >5 colonies at the end of the third round (3R) of plating, 5 × 103 cells were replated for 4R plating. †RNA was extracted only from initially 4OHT-treated cells. (B) Myeloid immortalization assays. (C) Reverse transcription (RT)-qPCR of MLL-ENL in colony-forming cells at the end of initial plating (1R) and third plating (3R) in myeloid immortalization assays. (D) Typical morphology of immortalized LT-HSCs constituting the colonies. Cells with Wright-Giemsa staining were viewed with an Olympus CKX41 microscope using a ×4/0.13 objective lens, and an Olympus BX41 microscope using a ×20/0.5 objective lens. Images were acquired with Olympus DP21 software. Original magnification, ×200; bar, 50 μm. (E) RT-qPCR of Hoxa9, Meis1, and Evi1 in colony-forming cells at the end of initial plating (with treatment of EtOH or 4OHT) and third plating (only cells derived from initially 4OHT-treated LT-HSCs, ST-HSCs, and MPP2 cells) in myeloid immortalization assays. In several samples from MPP3/4 cells, expression levels of Evi1 were below the limit of detection. Colors and patterns of bars are the same as shown in panel B. (F) Representative FACS plots of immortalized LT-HSCs, ST-HSCs, and MPP2 cells. Bar graphs show the mean with standard deviation (SD) of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (analysis of variance [ANOVA] followed by Tukey-Kramer multiple comparisons for panels C and E and unpaired Welch t tests for panel E).
Myeloid immortalization potential of LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells with conditional expression of MLL-ENL. (A) Experimental strategy for myeloid immortalization assays using serial replating. LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells were purified from BM cells derived from C/Tg mice conditionally expressing CAG promoter-driven MLL-ENL by inducible Cre-estrogen receptor α chain (CreER) fusion protein expressed from the Rosa26 locus by sorting with CD150/CD48 expression in KSL-gated cells. Initially, 100 cells from sorted cells were seeded with 4-hydroxytamoxifen (4OHT) or EtOH, followed by serial replating of 104 cells without drug. In cases with >5 colonies at the end of the third round (3R) of plating, 5 × 103 cells were replated for 4R plating. †RNA was extracted only from initially 4OHT-treated cells. (B) Myeloid immortalization assays. (C) Reverse transcription (RT)-qPCR of MLL-ENL in colony-forming cells at the end of initial plating (1R) and third plating (3R) in myeloid immortalization assays. (D) Typical morphology of immortalized LT-HSCs constituting the colonies. Cells with Wright-Giemsa staining were viewed with an Olympus CKX41 microscope using a ×4/0.13 objective lens, and an Olympus BX41 microscope using a ×20/0.5 objective lens. Images were acquired with Olympus DP21 software. Original magnification, ×200; bar, 50 μm. (E) RT-qPCR of Hoxa9, Meis1, and Evi1 in colony-forming cells at the end of initial plating (with treatment of EtOH or 4OHT) and third plating (only cells derived from initially 4OHT-treated LT-HSCs, ST-HSCs, and MPP2 cells) in myeloid immortalization assays. In several samples from MPP3/4 cells, expression levels of Evi1 were below the limit of detection. Colors and patterns of bars are the same as shown in panel B. (F) Representative FACS plots of immortalized LT-HSCs, ST-HSCs, and MPP2 cells. Bar graphs show the mean with standard deviation (SD) of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (analysis of variance [ANOVA] followed by Tukey-Kramer multiple comparisons for panels C and E and unpaired Welch t tests for panel E).
We next examined the expression levels of critical genes regulated by MLL-fusion genes in myeloid immortalization (Figure 1E). Initially, Hoxa9 and Evi1, and Hoxa9 alone, were significantly upregulated in LT-HSCs, and MPP2 cells, respectively. Meis1 was also upregulated but did not reach statistical difference. After serial replating, expression levels were significantly elevated in the respective colony-forming cells derived from LT-HSCs, ST-HSCs, and MPP2 cells, except for that of Meis1 in ST-HSC-derived cells. Interestingly, in contrast to immortalized ST-HSCs, immortalized LT-HSCs and MPP2 cells exhibited similar immunophenotypes of surface antigens, such as Gr-1/CD11b, but no differences in apoptotic subpopulations were found among these 3 types of immortalized cells (Figure 1F). These findings suggest that LT-HSCs were most susceptible to MLL-ENL–mediated transformation.
Conditional expression of MLL-ENL enhances expression of Bahcc1 in LT-HSCs, but not in ST-HSCs or MPP2 cells, during myeloid immortalization
To examine the molecular mechanism involved in the differences in immortalization potential among HSPCs, we searched for differentially expressed genes among the induced LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells at the end of initial plating in myeloid immortalization assays. At first, RNA extracted from induced ST-HSCs and MPP2 cells (at the end of initial plating) that would later be found to acquire colony-forming ability after the fourth round of plating were compared with the corresponding EtOH–treated control cells. RNA seq analysis of ST-HSCs and MPP2 cells revealed several differentially expressed genes (Figure 2A-B). Among these genes, expression of Bahcc1 (more enhanced) and Pik3r6 was exclusively enhanced in CD34− KSL cells in comparison to CD34+ KSL cells, in a reanalysis of our previous cDNA microarray results9 using conditional expression of MLL-ENL by retroviral transduction of CreER (Figure 2C; supplemental Table 1). Since LT-HSCs and MPP3/4 cells were enriched for CD34− and CD34+ KSL cells, respectively, in C/Tg mice (Figure 2D), we examined expression of Bahcc1 in induced HSPCs. Unexpectedly, Bahcc1 expression at the initial plating was unchanged in the induced cells compared with corresponding controls and did not differ among induced HSPCs. However, Bahcc1 expression at the third plating was significantly increased in induced LT-HSCs, ST-HSCs, and MPP2 cells. Significant enhancement (compared with that at initial plating) was only found in induced LT-HSCs (Figure 2E). These results suggest that MLL-ENL immortalized LT-HSCs with enhanced expression of Bahcc1 but did not always immortalize ST-HSCs or MPP2 cells, despite moderate (but not significant) enhancement of Bahcc1 expression.
Possible involvement of Bahcc1 in myeloid immortalization by conditional expression of MLL-ENL. (A) Initial screening strategy of candidate genes involved in differential modes of myeloid immortalization using induced ST-HSCs and MPP2 cells by RNA seq. (B) Numbers of differential expression genes (DEGs) screened by RNA seq in comparisons 1 (log2 FC>1 in induced ST-HSCs and log2 FC >0 in induced MPP2 cells, with adjusted P values <.05) and 2 (log2 FC >1 in induced MPP2 cells and log2 FC >0 in induced ST-HSCs, with adjusted P values <.05). Genes abundantly detected (count per million [CPM] >10 in induced ST-HSCs and MPP2 cells, and their respective controls) were focused on. (C) Refinement of focused genes based on exclusive enhancement in CD34 (−) KSL cells conditionally expressing MLL-ENL in the previous study.9Bahcc1 and Pik3r6 were selected from the focused genes among DEGs. Overlap of the focused genes in comparisons 1 and 2 is also shown. (D) Representative FACS plots and quantification of HSPCs based on CD150/CD48 expression in CD34(−) and CD34(+) KSL cells. (E) RT-qPCR of Bahcc1 in myeloid immortalization assays using the same samples as shown in Figure 1E. Colors and patterns of bars are the same as shown in Figure 1E. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ns, not significant (ANOVA followed by Tukey-Kramer multiple comparisons and unpaired Welch t test for panel E).
Possible involvement of Bahcc1 in myeloid immortalization by conditional expression of MLL-ENL. (A) Initial screening strategy of candidate genes involved in differential modes of myeloid immortalization using induced ST-HSCs and MPP2 cells by RNA seq. (B) Numbers of differential expression genes (DEGs) screened by RNA seq in comparisons 1 (log2 FC>1 in induced ST-HSCs and log2 FC >0 in induced MPP2 cells, with adjusted P values <.05) and 2 (log2 FC >1 in induced MPP2 cells and log2 FC >0 in induced ST-HSCs, with adjusted P values <.05). Genes abundantly detected (count per million [CPM] >10 in induced ST-HSCs and MPP2 cells, and their respective controls) were focused on. (C) Refinement of focused genes based on exclusive enhancement in CD34 (−) KSL cells conditionally expressing MLL-ENL in the previous study.9Bahcc1 and Pik3r6 were selected from the focused genes among DEGs. Overlap of the focused genes in comparisons 1 and 2 is also shown. (D) Representative FACS plots and quantification of HSPCs based on CD150/CD48 expression in CD34(−) and CD34(+) KSL cells. (E) RT-qPCR of Bahcc1 in myeloid immortalization assays using the same samples as shown in Figure 1E. Colors and patterns of bars are the same as shown in Figure 1E. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ns, not significant (ANOVA followed by Tukey-Kramer multiple comparisons and unpaired Welch t test for panel E).
Bahcc1 is critical for MLL-ENL–mediated immortalization of LT-HSCs
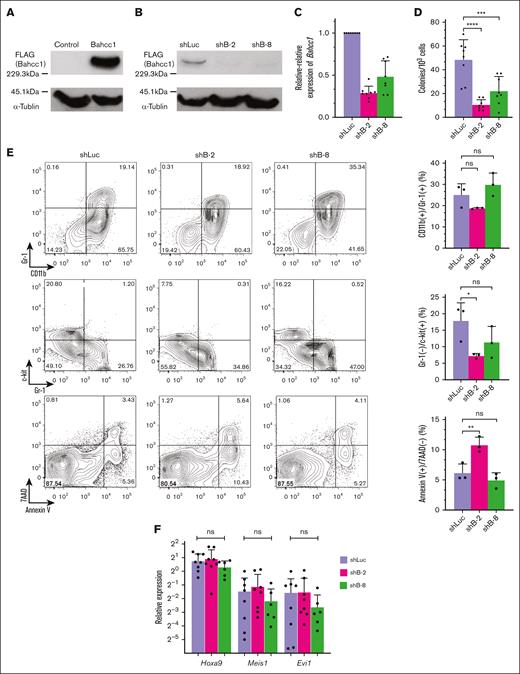
Next, to examine the involvement of Bahcc1 in myeloid immortalization of LT-HSCs by MLL-ENL, Bahcc1 was retrovirally depleted in immortalized LT-HSCs using shRNAs (shB-2 and shB-8) that could deplete Bahcc1 in 293 cells stably expressing Bahcc1 (Figure 3A-C). The Bahcc1-depleted cells exhibited a significant reduction in clonogenicity, and the reduction was more marked in shB-2–transduced cells than in shB-8 transduced cells (Figure 3D). In addition, we found a significant increase in the apoptotic subpopulation as well as a significant reduction in the Gr-1−/c-kit+ subpopulation with high potential for colony replating31 in colony-forming cells derived from shB-2-transduced cells (Figure 3E). In colony-forming cells derived from shB-8-transduced cells, we found a tendency toward a reduction in the Gr-1−/c-kit+ subpopulation, but no increase in the apoptotic subpopulation. In contrast, there were no significant changes in the Gr-1+/CD11b+ subpopulation with myeloid differentiation, or in Hoxa9, Meis1, and Evi1 expression (Figure 3F). These results suggest that MLL-ENL conferred aberrant self-renewal and anti-apoptotic activities on LT-HSCs, at least in part through Bahcc1, without affecting myeloid differentiation.
Bahcc1 depletion suppressed myeloid immortalization of LT-HSCs by conditional expression of MLL-ENL. (A-B) Western blot analyses of Bahcc1 in 293 cells (293Bahcc1) that stably expressed 3XFLAG-tagged Bahcc1 (A), and in shRNA-transduced 293Bahcc1 cells (B). 293 cells transduced with pcDNA3.1 vector alone were used as a negative control in panel A. shRNAs against luciferase (shLuc, a negative control) and Bahcc1 (shB-2 and shB-8) were transduced into 293Bahcc1 cells in panel B. Immunoprecipitants with anti-FLAG antibody were blotted with anti-DDDDK-tag antibody to detect expression of Bahcc1 (upper panels). α-Tubulin was analyzed in total lysates as an internal control (bottom panels). (C-D) Relative-relative expression levels of Bahcc1 (C) in immortalized LT-HSCs with shRNA transduction and clonogenicity (D) of the cells. (E-F) Representative FACS plots and quantification of CD11b+/Gr-1+, Gr-1−/c-kit+, and apoptotic (annexin V+/7AAD−) subpopulations by FACS (E), and RT-qPCR of Hoxa9, Meis1, and Evi1 (F) in colony-forming cells derived from immortalized LT-HSCs with depletion of Bahcc1. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (ANOVA followed by Dunnett multiple comparisons for panels D-F).
Bahcc1 depletion suppressed myeloid immortalization of LT-HSCs by conditional expression of MLL-ENL. (A-B) Western blot analyses of Bahcc1 in 293 cells (293Bahcc1) that stably expressed 3XFLAG-tagged Bahcc1 (A), and in shRNA-transduced 293Bahcc1 cells (B). 293 cells transduced with pcDNA3.1 vector alone were used as a negative control in panel A. shRNAs against luciferase (shLuc, a negative control) and Bahcc1 (shB-2 and shB-8) were transduced into 293Bahcc1 cells in panel B. Immunoprecipitants with anti-FLAG antibody were blotted with anti-DDDDK-tag antibody to detect expression of Bahcc1 (upper panels). α-Tubulin was analyzed in total lysates as an internal control (bottom panels). (C-D) Relative-relative expression levels of Bahcc1 (C) in immortalized LT-HSCs with shRNA transduction and clonogenicity (D) of the cells. (E-F) Representative FACS plots and quantification of CD11b+/Gr-1+, Gr-1−/c-kit+, and apoptotic (annexin V+/7AAD−) subpopulations by FACS (E), and RT-qPCR of Hoxa9, Meis1, and Evi1 (F) in colony-forming cells derived from immortalized LT-HSCs with depletion of Bahcc1. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (ANOVA followed by Dunnett multiple comparisons for panels D-F).
MLL-ENL upregulates Bahcc1 via promoter binding
High expression of BAHCC1 has been found in some AML cases with an unfavorable prognosis,25 but little is understood about the molecular mechanism of upregulation of BAHCC1. Because a recent study32 showed that Mll-deficiency reduces Bahcc1 expression and that Mll binds just upstream of Bahcc1 exon 2 in intestinal stem cells, we examined whether MLL-ENL upregulates Bahcc1 via promoter binding. In myeloid immortalization assays using wild-type KSL and MP cells retrovirally transduced with FLAG-tagged-MLL-ENL, expression of Bahcc1 was significantly increased at the initial and third plating, and was also significantly more enhanced at the third plating in MLL-ENL-transduced-KSL cells (Figure 4A), similar to the enhancement in induced LT-HSCs. ChIP-qPCR assays of immortalized KSL cells showed a significant increase in MLL-ENL binding to regions around the putative transcription start sites of Bahcc1, as found in public ChIP-seq data showing enrichment of trimethylated histone H3 lysine 4 and RNA polymerase II in thymus, BM, and small intestine cells (Figure 4B-C). In addition, public ChIP-seq data (DRA00487133 and GSE7989934) in several human MLL-rearranged leukemic cell lines, including HB1119 cells expressing MLL-ENL, also exhibited enrichment of MLL fusion proteins around the corresponding regions of human BAHCC1, accompanied with transcriptionally active histone marks and RNA polymerase II (supplemental Figure 2). Therefore, these results suggest that MLL-ENL upregulated Bahcc1 via promoter binding during immortalization of HSPCs.
MLL-ENL upregulates Bahcc1 via promoter binding. (A) RT-qPCR of Bahcc1 in myeloid immortalization assays using wild-type MLL-ENL- (ME) or empty vector- (IN) transduced KSL and MP cells. (B) Overview of the genomic region of the Bahcc1 locus in an adapted UCSC Genome Browser view. ChIP-seq data from mouse thymus, BM, and small intestine (SmInt) are shown in the LICR tracks (H3K4m3 and input control, Pol II, and input control). (C) ChIP-qPCR of MLL-ENL around exons 1 and 2 of Bahcc1 using retrovirally FLAG-tagged-MLL-ENL-immortalized KSL cells. Primer sets (1-5) are shown (top). Hoxa9 and Hbb-b1 were used as controls. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (ANOVA followed by Tukey-Kramer multiple comparisons for panel A and unpaired Welch t tests for panel C). IgG1, IgG subclass 1.
MLL-ENL upregulates Bahcc1 via promoter binding. (A) RT-qPCR of Bahcc1 in myeloid immortalization assays using wild-type MLL-ENL- (ME) or empty vector- (IN) transduced KSL and MP cells. (B) Overview of the genomic region of the Bahcc1 locus in an adapted UCSC Genome Browser view. ChIP-seq data from mouse thymus, BM, and small intestine (SmInt) are shown in the LICR tracks (H3K4m3 and input control, Pol II, and input control). (C) ChIP-qPCR of MLL-ENL around exons 1 and 2 of Bahcc1 using retrovirally FLAG-tagged-MLL-ENL-immortalized KSL cells. Primer sets (1-5) are shown (top). Hoxa9 and Hbb-b1 were used as controls. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (ANOVA followed by Tukey-Kramer multiple comparisons for panel A and unpaired Welch t tests for panel C). IgG1, IgG subclass 1.
Bahcc1 is involved in MLL-ENL–mediated immortalization, at least partly through repression of H3K27me3-marked Cdkn1c
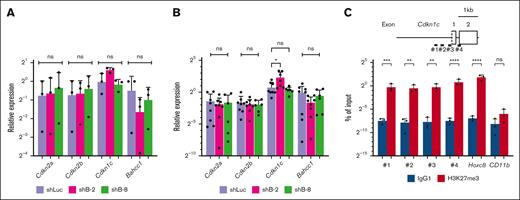
To examine the role of Bahcc1 in myeloid immortalization of HSPCs by MLL-ENL, we focused on several genes, including Cdkn1c, Cdkn2a, and Cdkn2b, which were found to be epigenetically regulated by Bahcc1.23 Considering the results of our Bahcc1 knockdown experiments, the expression levels of Cdkn1c, Cdkn2a, and Cdkn2b, which are involved in regulation of the cell cycle and apoptosis, were analyzed in immortalized LT-HSCs with Bahcc1 depletion. For analysis in the early phase after Bahcc1 depletion, shRNA-transduced cells were sorted and directly analyzed at 72 hours after transduction because it was difficult to analyze cells that were cultured for 24 hours after sorting at the regular time (48 hours after transduction). Although the condition of less efficient depletion of Bahcc1 should be noted, expression of Cdkn1c tended to be increased in shB-2–transduced cells, but not in shB-8-transduced cells, whereas expression of Cdkn2a and Cdkn2b was not changed in either group of cells (Figure 5A).
Derepression of Cdkn1c in immortalized LT-HSCs with Bahcc1 depletion. (A-B) RT-qPCR of Cdkn2a, Cdkn2b, Cdkn1c, and Bahcc1 in immortalized LT-HSCs transduced with shRNA/KO expressors. Cells sorted on the basis of high KO expression 72 hours after transduction (A), and colony-forming cells (B) derived from the shRNA-transduced cells shown in Figure 3C were analyzed. (C) ChIP-qPCR of H3K27me3 around exon 1 of Cdkn1c using FLAG-tagged-MLL-ENL-immortalized KSL cells. Primer sets (1-4) are shown (top). Hoxc8 and CD11b were used as controls. shLuc, shRNA against luciferase; shB-2 and shB-8, different shRNAs against Bahcc1. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant. ANOVA followed by Dunnett multiple comparisons for panels A-B and unpaired Welch t tests for panel C.
Derepression of Cdkn1c in immortalized LT-HSCs with Bahcc1 depletion. (A-B) RT-qPCR of Cdkn2a, Cdkn2b, Cdkn1c, and Bahcc1 in immortalized LT-HSCs transduced with shRNA/KO expressors. Cells sorted on the basis of high KO expression 72 hours after transduction (A), and colony-forming cells (B) derived from the shRNA-transduced cells shown in Figure 3C were analyzed. (C) ChIP-qPCR of H3K27me3 around exon 1 of Cdkn1c using FLAG-tagged-MLL-ENL-immortalized KSL cells. Primer sets (1-4) are shown (top). Hoxc8 and CD11b were used as controls. shLuc, shRNA against luciferase; shB-2 and shB-8, different shRNAs against Bahcc1. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant. ANOVA followed by Dunnett multiple comparisons for panels A-B and unpaired Welch t tests for panel C.
In line with the results obtained in the early phase, expression of Cdkn1c was significantly increased in colony-forming cells derived from shB-2–transduced cells but not in cells derived from shB-8 transduced cells, whereas expression of Cdkn2a and Cdkn2b was not changed in either group of cells (Figure 5B). Interestingly, Bahcc1 depletion was abrogated in the colony-forming cells, indicating that cells escaping from Bahcc1 knockdown were dominant during culture after sorting. In addition, to examine whether Cdkn1c was epigenetically regulated by Bahcc1 by binding to H3K27me3 in MLL-ENL-immortalized HSPCs, ChIP-qPCR assays of immortalized KSL cells were performed around previously described regions.35 These assays showed a significant increase in H3K27me3 around the tested regions of Cdkn1c (Figure 5C). Thus, these results suggest that depletion of Bahcc1 led to derepression of Cdkn1c through reduction of its binding to H3K27me3 marks in the regulatory genomic regions of Cdkn1c in MLL-ENL-immortalized cells.
Bahcc1 is also critical for MLL-fusion–mediated leukemogenesis of HSPCs in vivo
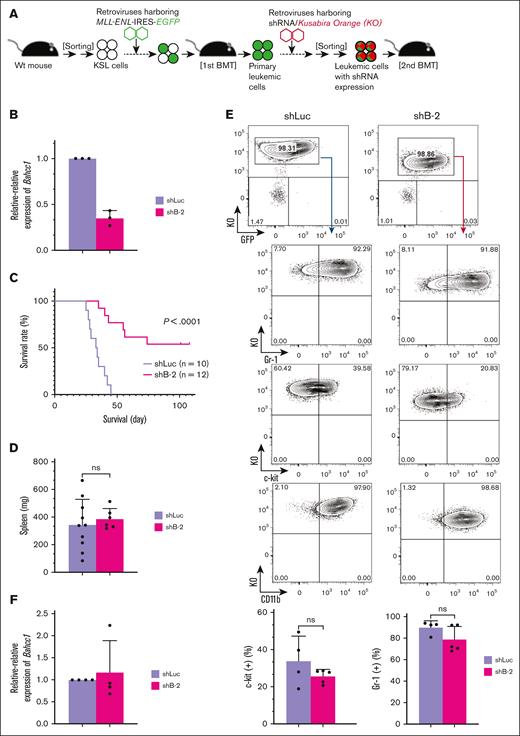
Finally, to further examine the role of Bahcc1 in the molecular mechanism of MLL-ENL-mediated leukemogenesis in vivo, we analyzed the leukemogenic activity of Bahcc1-depleted leukemic cells using a secondary BMT model (Figure 6A). Primary MLL-ENL-transduced leukemic cells were efficiently depleted with Bahcc1 by shB-2 and were transplanted into recipient mice (Figure 6B). In secondary BMT within an observation period of ∼100 days, 6 of 12 recipient mice who underwent transplantation with Bahcc1-depleted leukemic cells died, whereas 10 of 10 control mice died (Figure 6C). Both groups of moribund mice similarly exhibited splenomegaly (Figure 6D) and most of their BM cells were GFP/KO-positive myelomonocytic cells (Figure 6E), indicating development of secondary AML. However, in spite of expression of KO in most BM cells, expression of Bahcc1 in the BM cells of the 2 groups did not differ (Figure 6F), indicating that leukemic cells escaping from depletion of Bahcc1 were dominant during development and/or progression of secondary AML. Interestingly, in some of the recipient mice who underwent transplantation with Bahcc1-depleted cells, the fluorescence intensity of KO was lower than that in control mice (Figure 6E). Expression of KO in peripheral white blood cells and BM cells was not detected at 53 and 100 days after BMT, respectively, in surviving mice transplanted with Bahcc1-depleted cells.
Bahcc1 depletion prolongs development of MLL-ENL-induced leukemia. (A) Experimental strategy using secondary BMT. Primary leukemic cells expressing MLL-ENL together with EGFP were harvested, followed by retroviral transduction with shRNA/Kusabira-Orange (KO) expressors. Leukemic cells highly expressing KO were sorted and immediately transplanted into secondary recipient mice. (B) Relative-relative expression levels of Bahcc1 in shRNA-transduced leukemic cells. shLuc, shRNA against luciferase; shB-2, shRNA against Bahcc1. (C) Survival curves of mice transplanted with Bahcc1-depleted (shB-2; n = 12) or control (shLuc; n = 10) leukemic cells. Data from 3 independent experiments were combined. (D) Spleen weights of moribund mice. (E) Representative FACS plots of BM cells from moribund mice. GFP/KO doubly positive cells were gated as shown in the top panels, followed by subsequent analyses for c-kit, Gr-1, and CD11b expression. (F) Relative-relative expression levels of Bahcc1 by RT-qPCR in BM cells from moribund mice. Bar graphs show the mean with SD of at least 3 independent experiments. ns, not significant (log-rank test for panel C and unpaired Welch t tests for panels D-E). Wt, wild-type.
Bahcc1 depletion prolongs development of MLL-ENL-induced leukemia. (A) Experimental strategy using secondary BMT. Primary leukemic cells expressing MLL-ENL together with EGFP were harvested, followed by retroviral transduction with shRNA/Kusabira-Orange (KO) expressors. Leukemic cells highly expressing KO were sorted and immediately transplanted into secondary recipient mice. (B) Relative-relative expression levels of Bahcc1 in shRNA-transduced leukemic cells. shLuc, shRNA against luciferase; shB-2, shRNA against Bahcc1. (C) Survival curves of mice transplanted with Bahcc1-depleted (shB-2; n = 12) or control (shLuc; n = 10) leukemic cells. Data from 3 independent experiments were combined. (D) Spleen weights of moribund mice. (E) Representative FACS plots of BM cells from moribund mice. GFP/KO doubly positive cells were gated as shown in the top panels, followed by subsequent analyses for c-kit, Gr-1, and CD11b expression. (F) Relative-relative expression levels of Bahcc1 by RT-qPCR in BM cells from moribund mice. Bar graphs show the mean with SD of at least 3 independent experiments. ns, not significant (log-rank test for panel C and unpaired Welch t tests for panels D-E). Wt, wild-type.
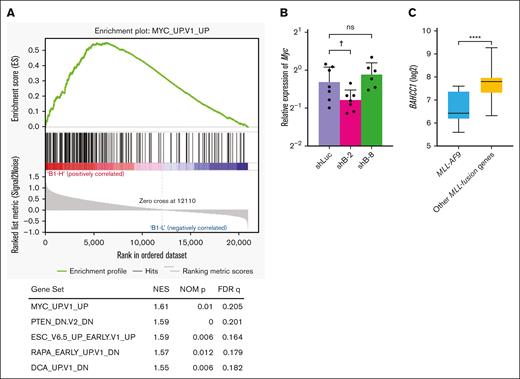
Analyses of the link between BAHCC1 and human AML in public databases revealed an association of high expression of BAHCC1 with a poor prognosis in pediatric Therapeutically Applicable Research to Generate Effective Treatments (TARGET) AML cohorts, but not in the adult Cancer Genome Atlas AML project (TCGA LAML) cohort (supplemental Figure 3A-D). A similar association was found in MLL-rearranged AML (supplemental Figure 3E) and in nonrearranged AML (supplemental Figure 3F) in the recent TARGET AML cohort. Gene set enrichment analyses of MLL-rearranged AML cohorts (GSE1785536 and GSE6180437) revealed that high expression of BAHCC1 was most closely associated with a gene set upregulated by MYC38 in C6 oncogenic signature gene sets (Figure 7A; supplemental Figure 4A). The enrichment in the gene set upregulated by MYC was also found, although not significantly, in another AML data set (GSE1957739; supplemental Figure 4B). In line with this finding, expression of Myc tended to be reduced in colony-forming cells derived from immortalized LT-HSCs with Bahcc1 depletion by shB-2 (Figure 7B). Interestingly, BAHCC1 expression was significantly lower in AML with MLL-AF9 than in AML with other MLL-fusion genes, in MLL-rearranged AML in childhood (GSE1957739; Figure 7B) and the recent TARGET AML cohort (supplemental Figure 4C). BAHCC1 depletion suppressed cell proliferation in human leukemic cell lines, HB1119 cells expressing MLL-ENL and THP-1 cells expressing MLL-AF9, although derepression of CDKN1C was limited (supplemental Figure 5).
BAHCC1 is critical for MLL-rearranged AML. (A) Gene set enrichment analysis (GSEA) of MLL-rearranged AML samples (GSE1785536) with high (B1-H, n = 9) and low (B1-L, n = 9) expression of BAHCC1 using C6 oncogenic signature gene sets. GSEA plot using the most enriched gene set, MYC_UP.V1_UP,38 and a list of the top 5 enriched gene sets are shown (filtered by NOM (nominal) P < .05 and FDR q < 0.25). NES, normalized enrichment score. (B) RT-qPCR of Myc in colony-forming cells derived from the Bahcc1-depleted cells shown in Figure 3C. Bar graphs show the mean with SD of at least 3 independent experiments. (C) Expression levels of BAHCC1 in AML samples (GSE1957739) with MLL-AF9 (n = 11) and other MLL-fusion genes (n = 31). Data are shown in box and whisker plots. †P = .054; ∗∗∗∗P < .0001; ns, not significant ANOVA followed by Dunnett multiple comparison for panel B and Mann-Whitney U test for panel C.
BAHCC1 is critical for MLL-rearranged AML. (A) Gene set enrichment analysis (GSEA) of MLL-rearranged AML samples (GSE1785536) with high (B1-H, n = 9) and low (B1-L, n = 9) expression of BAHCC1 using C6 oncogenic signature gene sets. GSEA plot using the most enriched gene set, MYC_UP.V1_UP,38 and a list of the top 5 enriched gene sets are shown (filtered by NOM (nominal) P < .05 and FDR q < 0.25). NES, normalized enrichment score. (B) RT-qPCR of Myc in colony-forming cells derived from the Bahcc1-depleted cells shown in Figure 3C. Bar graphs show the mean with SD of at least 3 independent experiments. (C) Expression levels of BAHCC1 in AML samples (GSE1957739) with MLL-AF9 (n = 11) and other MLL-fusion genes (n = 31). Data are shown in box and whisker plots. †P = .054; ∗∗∗∗P < .0001; ns, not significant ANOVA followed by Dunnett multiple comparison for panel B and Mann-Whitney U test for panel C.
Taken together with the results for MLL-ENL-immortalized cells with Bahcc-1 depletion in vitro, these in vivo results suggest that Bahcc-1 depletion suppressed development and/or progression of MLL-ENL–mediated AML and that BAHCC1 had an important role in MLL-fusion–mediated myeloid leukemogenesis, although a decrease in engraftment of depleted cells should be noted. Therefore, our results suggest that upregulation of Bahcc1 by MLL-ENL is critical for MLL-ENL–mediated leukemogenesis, at least partly through epigenetic repression of expression of the negative cell-cycle regulator Cdkn1c.
Discussion
Here, we showed that CD150+/CD48−KSL cells (LT-HSCs) were the most susceptible targets for leukemic immortalization by MLL-ENL among HSPCs in our conditional transgenic mouse model. This finding is compatible with our previous results9 showing that CD34−KSL cells enriched for LT-HSCs are exclusively transformed by conditionally expressed MLL-ENL. However, this study also showed that the induced MLL-ENL could confer leukemic immortalization on ST-HSCs and MPP2 cells, albeit not in every experiment. These results imply that MPP cells might harbor fewer intrinsic advantages to MLL-fusion–mediated leukemogenesis than LT-HSCs. This is partly consistent with previous studies7,40 showing more aggressive leukemogenesis by MLL-AF9 or MLL-ENL in LT-HSCs, but not with another study41 that showed intrinsic protection of LT-HSCs against MLL-ENL–mediated leukemogenesis. These differences might be explained by the differences in mouse models, including the promoters used and the mode of conditional or inducible expression42,43; however, further analyses are needed.
This study also revealed the role of the MLL-ENL-Bahcc1 axis in leukemogenesis by MLL-ENL, at least partly through repression of H3K27me3-marked Cdkn1c.44 This finding is reminiscent of a recent study45 suggesting that the DNA methylation reader MBD2 developed MLL-AF9–driven leukemogenesis through repression of Cdkn1c transcription by binding to its methylated genomic promoter region. BAHCC1 depletion suppressed the growth of human leukemic cell lines expressing MLL-ENL or MLL-AF9, but with limited derepression of CDKN1C. These findings suggest the possible involvement of BAHCC1-mediated repression of other tumor suppressor genes as well in MLL-fusion–mediated leukemogenesis, which needs further investigation. Interestingly, Bahcc1 depletion did not change the expression of Hoxa9, Meis1, or Evi1 in immortalized LT-HSCs, implying that Bahcc1 is an independent key player in MLL-ENL–mediated leukemogenesis. In addition, we found a close association of high expression of BAHCC1 with a gene set upregulated by MYC by gene set enrichment analysis, and a tendency for reduction in expression of Myc in immortalized LT-HSCs with Bahcc1 depletion. This finding does not agree with results showing no alterations in MYC expression in T-cell leukemic/lymphoma cell lines transduced with missense mutations abrogating BAH function by genome editing of BAHCC1.23 These differences might imply possible functions of the BAHCC1/Bahcc1 region other than the BAH domain, which occupies only ∼4.6% of the protein.
Our results in vivo showed prolonged latencies in BMT assays of Bahcc1-depleted leukemic cells, which is consistent with the association of high BAHCC1 expression with aggressive MLL-rearranged AML in childhood, but careful interpretation is still required. Xenograft BMT assays with BAHCC1 depletion also had prolonged latencies,23 but all mice tested developed lethal disease, in contrast to our results. However, bioluminescent signals of BAHCC1-depleted cells in vivo were much weaker at the early time point after BMT, suggesting reduced homing and/or engraftment in BMT assays. The significantly lower expression of BAHCC1 in MLL-AF9-AML might be involved, at least partly, in the relatively better prognosis in MLL-AF9-AML,46 but further analysis, including elucidation of the mechanism of the MLL-AF9–associated lower expression, is required. Furthermore, the precise role of Bahcc1 in normal hematopoiesis should be addressed for development of novel therapies targeting the MLL-fusion-BAHCC1 axis, although knock-in mice harboring the mutation abrogating BAH function did not exhibit alterations in hematopoiesis.23
In conclusion, LT-HSCs among HSPCs showed greatest susceptibility to MLL-ENL–mediated immortalization. In an analysis of immortalization potential, Bahcc1 was found to be a key molecule. MLL-ENL upregulated Bahcc1, and Bahcc1 depletion suppressed MLL-ENL–mediated leukemogenesis in vivo and in vitro, at least partly through derepression of H3K27me3-marked Cdkn1c. Taken together, these results demonstrate an important role of the MLL-fusion-Bahcc1 axis that may lead to applications in molecular targeted therapy.
Acknowledgments
The authors thank Toshio Kitamura (The Institute of Medical Science, University of Tokyo) for providing pMXsU6-puro and THP-1 cells, Akihiko Yokoyama (Tsuruoka Metabolomics Laboratory, National Cancer Center) for providing HB1119 cells, Japan Medical Communication (Fukuoka, Japan) and PALABRA Inc. (Kyoto, Japan) for language assistance, and Rukia Iwayama for technical support.
This work was supported in part by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology in Japan (TN; Basic-B 17H04227, RO; Basic-C 21K08411).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Authorship
Contribution: A.N., T.N., and R.O. designed the study, analyzed the data, interpreted the results, and wrote the manuscript; A.N., M.M., M.S., and R.O. performed in vitro experiments; A.N., I.T., and R.O. performed in vivo experiments; and I.T., T.N., and R.O. contributed to study supervision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ryoichi Ono, Department of Microbiology and Molecular Genetics, Mie University Graduate School of Medicine, 2-174 Edobashi, Tsu, Mie 514-8507, Japan; email: ronomie@doc.medic.mie-u.ac.jp.
References
Author notes
Next-generation sequencing data were deposited to the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE229484.
The full-text version of this article contains a data supplement.

![Myeloid immortalization potential of LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells with conditional expression of MLL-ENL. (A) Experimental strategy for myeloid immortalization assays using serial replating. LT-HSCs, ST-HSCs, MPP2 cells, and MPP3/4 cells were purified from BM cells derived from C/Tg mice conditionally expressing CAG promoter-driven MLL-ENL by inducible Cre-estrogen receptor α chain (CreER) fusion protein expressed from the Rosa26 locus by sorting with CD150/CD48 expression in KSL-gated cells. Initially, 100 cells from sorted cells were seeded with 4-hydroxytamoxifen (4OHT) or EtOH, followed by serial replating of 104 cells without drug. In cases with >5 colonies at the end of the third round (3R) of plating, 5 × 103 cells were replated for 4R plating. †RNA was extracted only from initially 4OHT-treated cells. (B) Myeloid immortalization assays. (C) Reverse transcription (RT)-qPCR of MLL-ENL in colony-forming cells at the end of initial plating (1R) and third plating (3R) in myeloid immortalization assays. (D) Typical morphology of immortalized LT-HSCs constituting the colonies. Cells with Wright-Giemsa staining were viewed with an Olympus CKX41 microscope using a ×4/0.13 objective lens, and an Olympus BX41 microscope using a ×20/0.5 objective lens. Images were acquired with Olympus DP21 software. Original magnification, ×200; bar, 50 μm. (E) RT-qPCR of Hoxa9, Meis1, and Evi1 in colony-forming cells at the end of initial plating (with treatment of EtOH or 4OHT) and third plating (only cells derived from initially 4OHT-treated LT-HSCs, ST-HSCs, and MPP2 cells) in myeloid immortalization assays. In several samples from MPP3/4 cells, expression levels of Evi1 were below the limit of detection. Colors and patterns of bars are the same as shown in panel B. (F) Representative FACS plots of immortalized LT-HSCs, ST-HSCs, and MPP2 cells. Bar graphs show the mean with standard deviation (SD) of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001; ns, not significant (analysis of variance [ANOVA] followed by Tukey-Kramer multiple comparisons for panels C and E and unpaired Welch t tests for panel E).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/9/10.1182_bloodadvances.2023011320/2/m_blooda_adv-2023-011320-gr1b.jpeg?Expires=1769300188&Signature=4TPUFpOuFX-dHqMNitttuwxlxlzc94~60PFpvs4gn8jf5qn0ejrGk5hDFIpNDHHKfU4LvovWJN1JowDrWSLN89nm-nV3buRbPN7xGTq1~amqFru64JtuZoxd2sAwEipWrAY45CivaiTu~iR6aFzrJusHA2Tq6PhuW6mOPl4C9KJuUYF82a64Eq7LsvdQM5KuSrwwNw3ouekOvi~RG2-wGAqSttxr4GNGJM62eDqRMBaJH9NnAOYIj5zUBrVNB0iAJmdCRZ-xrTf2SSb2M1Te0WVhxuiLPLpeE~sTtppcfKFn4JSiRPiGemaP2zJFZpmz03O3pQvgwLJOgYrxylCZMg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Possible involvement of Bahcc1 in myeloid immortalization by conditional expression of MLL-ENL. (A) Initial screening strategy of candidate genes involved in differential modes of myeloid immortalization using induced ST-HSCs and MPP2 cells by RNA seq. (B) Numbers of differential expression genes (DEGs) screened by RNA seq in comparisons 1 (log2 FC>1 in induced ST-HSCs and log2 FC >0 in induced MPP2 cells, with adjusted P values <.05) and 2 (log2 FC >1 in induced MPP2 cells and log2 FC >0 in induced ST-HSCs, with adjusted P values <.05). Genes abundantly detected (count per million [CPM] >10 in induced ST-HSCs and MPP2 cells, and their respective controls) were focused on. (C) Refinement of focused genes based on exclusive enhancement in CD34 (−) KSL cells conditionally expressing MLL-ENL in the previous study.9Bahcc1 and Pik3r6 were selected from the focused genes among DEGs. Overlap of the focused genes in comparisons 1 and 2 is also shown. (D) Representative FACS plots and quantification of HSPCs based on CD150/CD48 expression in CD34(−) and CD34(+) KSL cells. (E) RT-qPCR of Bahcc1 in myeloid immortalization assays using the same samples as shown in Figure 1E. Colors and patterns of bars are the same as shown in Figure 1E. Bar graphs show the mean with SD of at least 3 independent experiments. ∗P < .05; ∗∗P < .01; ns, not significant (ANOVA followed by Tukey-Kramer multiple comparisons and unpaired Welch t test for panel E).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/9/10.1182_bloodadvances.2023011320/2/m_blooda_adv-2023-011320-gr2.jpeg?Expires=1769300188&Signature=eB8dEyIS-xUHOUXez3CPQ~lrUIontla~sbnGLAtzA-deW8h5vbti31bMRnXvDW4z8oT4OWmOCUP06YzOM~mmGMUg2oCRu5Vf-ynD0aufApfCiQHhFWbtBpkqZj7XMmK55Pz7ES--J6t2lHaLiM1CxyCa3CG1IYidmpziEy2j4pVmt44GDPNhaLs2z7mr8pLjEFHEzQ-NBPXjXSIh9CI7ZHglQo-EAlnj3byWNZ5K~41Oh5Z3xtsuEPJxRShUpssXAvQAdTbCoXiCaBe0ghFdRH41wGyTzn~H9FCdn4aLyB3DbSZ1u4KT7nSZ~j11O7v88mZO7sD8ex8j9Sz9uQBM-A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)