Extensive genetic analysis of EBV+ nPTCL revealed frequent TET2 and DNMT3A mutations with a possible association with clonal hematopoiesis.
EBV+ nPTCL carried recurrent intragenic deletions in the viral genome that may promote lymphomagenesis.
Visual Abstract
Epstein-Barr virus (EBV)-positive (EBV+) nodal T- and natural killer (NK)-cell lymphoma is a peripheral T-cell lymphoma (EBV+ nPTCL) that presents as a primary nodal disease with T-cell phenotype and EBV-harboring tumor cells. To date, the genetic aspect of EBV+ nPTCL has not been fully investigated. In this study, whole-exome and/or whole-genome sequencing was performed on 22 cases of EBV+ nPTCL. TET2 (68%) and DNMT3A (32%) were observed to be the most frequently mutated genes whose presence was associated with poor overall survival (P = .004). The RHOA p.Gly17Val mutation was identified in 2 patients who had TET2 and/or DNMT3A mutations. In 4 patients with TET2/DNMT3A alterations, blood cell–rich tissues (the bone marrow [BM] or spleen) were available as paired normal samples. Of 4 cases, 3 had at least 1 identical TET2/DNMT3A mutation in the BM or spleen. Additionally, the whole part of the EBV genome was sequenced and structural variations (SVs) were found frequent among the EBV genomes (63%). The most frequently identified type of SV was deletion. In 1 patient, 4 pieces of human chromosome 9, including programmed death-ligand 1 gene (PD-L1) were identified to be tandemly incorporated into the EBV genome. The 3′ untranslated region of PD-L1 was truncated, causing a high-level of PD-L1 protein expression. Overall, the frequent TET2 and DNMT3A mutations in EBV+ nPTCL seem to be closely associated with clonal hematopoiesis and, together with the EBV genome deletions, may contribute to the pathogenesis of this intractable lymphoma.
Introduction
Epstein-Barr virus (EBV)-positive (EBV+) nodal T- and natural killer (NK)-cell lymphoma is a peripheral T-cell lymphoma (EBV+ nPTCL) that presents primary nodal disease, T-cell phenotype, and EBV-harboring tumor cells.1,2 This lymphoma is associated with a highly aggressive clinical course with median overall survival (OS) of 2.5 to 8.0 months and with no standard of treatment. In the Fourth World Health Organization (WHO) classification released in 2008, it had been unclear whether EBV+ nPTCL should be classified as PTCL or extranodal NK/T-cell lymphoma (NKTL) because both EBV+ nPTCL and NKTL are characterized by EBV positivity and cytotoxic phenotype. However, we previously demonstrated that EBV+ nPTCL presents as a lymph node–based disease without nasal involvement, usually with a CD8+/CD56− phenotype, distinct from NKTL.1,3 The clinicopathological uniqueness of EBV+ nPTCL was further supported by its T-cell origin in most reported cases.1 Therefore, this nodal lymphoma was considered to be PTCL, not otherwise specified (PTCL-NOS) in the revised Fourth WHO classification4 and is listed as a new entity in the Fifth WHO classification and as a provisional entity in the international consensus classification.5,6 The genetic aspect of EBV+ nPTCL has not been fully investigated to date. Wai et al recently reported that 9 of 14 EBV+ nPTCL cases (64%) carried TET2 mutations based on targeted sequencing.7 However, the association of clonal hematopoiesis of indeterminate potential (CHIP), disease-specific driver mutations, or mutations in the viral genome remains to be elucidated because the previous study focused on known driver mutations and did not use paired germ line samples. This study enrolled 22 patients with EBV+ nPTCL and performed whole-exome sequencing (WES) and/or whole-genome sequencing (WGS), with paired normal samples in 13 of these cases, to disclose the genomic landscape of this peculiar disease.
Methods
Patients
This study enrolled 22 patients with EBV+ nPTCL with available DNA samples, of whom 17 were examined in our previous studies.7-9 Of these 17 cases, 5 (UPN114, UPN232, UPN242, UPN992, and UPN994) were subjected to targeted mutation analysis with next-generation sequencing in a previous report.7 Furthermore, targeted-capture sequencing for programmed death-ligand 1 gene (PD-L1) was performed in 2 of 17 patients (UPN312 and UPN857).9 However, this series includes none of the patients previously analyzed by WES and/or genome sequencing.
All enrolled patients were diagnosed with PTCL-NOS from 1996 to 2018 following the 2017 WHO classification.4 All patients were clinically evaluated with a primary nodal disease. Immunohistochemistry revealed that all EBV+ nPTCL cases were positive for at least 1 T-cell antigen (CD3, CD4, CD5, or CD8) and negative for CD20. The presence of EBV small ribonucleic acids was determined by in situ hybridization (ISH) using EBV-encoded small nuclear early region (EBER-ISH). EBER-ISH was considered positive when ≥50% of the neoplastic cells stained positive. Patients with upper respiratory tract involvement were diagnosed with NKTL and excluded from our series. This study was conducted following the principles of all relevant ethical regulations, including the Declaration of Helsinki. The institutional review boards of Aichi Cancer Center Hospital and Fujita Health University approved this study.
DNA extraction
We extracted DNA from 5 to 10 formalin-fixed, paraffin-embedded (FFPE) slides of the bone marrow (BM), spleen, skin, stomach, intestine, and salivary gland using a QIAamp DNA FFPE Tissue Kit (catalogue no. 56404, Qiagen, Hilden, Germany) for germ line analyses. We extracted DNA from 2 to 5 FFPE slides per patient using a GeneRead DNA FFPE Kit (catalogue no. 180134, Qiagen) for tumor samples, following the manufacturer’s instructions. We analyzed all except 1 tumor tissues using lymph nodes. A Qubit fluorometer and a Qubit HS dsDNA kit (catalogue nos. Q33238 and Q32850, Thermo Fisher Scientific, Waltham, MA) were used to quantify DNA concentration.
WES
We sonicated DNA to an average of 200–base pair (bp) fragments using a Covaris M220 (Covaris, Woburn, MA). Additionally, we constructed prep libraries using the KAPA HyperPrep Kit (catalogue no. KK8504, Nippon Genetics, Tokyo, Japan). Briefly, fragmented DNA was end-repaired and ligated with the Adapter Oligo Mix in SureSelect XT Reagents (G9611B, Agilent, Santa Clara, CA). The ligated product was purified using Ampure XP beads (Beckman Coulter, Indianapolis, IN) according to the manufacturer’s protocol, and amplified with KAPA HiFi HotStart Ready Mix, SureSelect Primer, and SureSelect ILM Indexing Pre-Capture PCR Reverse Primer. We captured the exome sequences from the prep library using Human All Exon version 7 bait. The library was sequenced using a HiSeqX platform (Illumina, San Diego, CA) with the “2 × 150 bp reads” option to obtain an average of 10 gigabases per sample.
Detection of somatic point mutations from WES data
Mutation detection was performed essentially as previously described.10 Briefly, the sequence reads were aligned to the hg19 reference genome using the Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net/) with default parameters and a “–mem” option. Polymerase chain reaction (PCR) duplicates were removed using the Picard tools (https://broadinstitute.github.io/picard/).
To identify somatic point mutations, paired tumor-normal data were analyzed using VarScan2.11 Subsequently, we called candidate variants in the coding region that had a detection P value < .01, ≥9 reads with the variant, and minor allele frequencies of <0.0025 in single-nucleotide polymorphism databases (ESP6500, 1000 genomes, ExAC, and Kaviar). A candidate variant was filtered out if the identical variant was present in 12 irrelevant germ line samples with an average variant allele frequency (VAF) of >0.01. The variants were then annotated using ANNOVAR (https://annovar.openbioinformatics.org/).
For the tumor samples for which the corresponding germ line samples were not available (tumor-only samples), we also used VarScan2 to call candidate variants from tumor sample data with a detection threshold of P < .01. We picked up driver mutations in known driver genes, by referring to the Catalogue of Somatic Mutations in Cancer (https://cancer.sanger.ac.uk/cosmic) database. As a result, we identified driver mutations in DDX3X, DNMT3A, FYN, and TET2 from tumor-only samples.
Using VarScan2, we picked up CHIP-related mutations from tumor sample data (not paired with normal sample data) with a detection threshold of P < .01. A DNMT3A or TET2 variant was considered related to CHIP when the VAF was >0.05 for the identical variant in the corresponding normal sample.
WGS
WGS libraries were prepared starting from 50 to 100 ng of DNA using an NEBNext Ultra II DNA Prep Kit for Illumina (New England Biolabs, Ipswich, MA) according to the manufacturer’s instructions. Somatic variants were detected using the same pipeline used in WES. A 10-kilobase bin copy number estimate was obtained from the number of reads within the bin divided by the mean coverage of the whole sample. Structural variations (SVs) were detected using GRIDSS (https://github.com/PapenfussLab/gridss) and default parameters.
RHOA p.Gly17Val detection by droplet digital PCR
A QX200 Droplet Digital PCR system (Bio-Rad, Hercules, CA) with a droplet digital PCR assay was used to analyze the samples. Bio-Rad provided the predesigned PCR primer/probe mix for the RHOA p.Gly17Val mutation. PCR amplification was performed as follows: initial enzyme activation at 95°C for 10 minutes, 40 cycles of denaturation and annealing/extension at 94°C for 30 seconds, hold at 55°C for 2 minutes, and then enzyme deactivation at 98°C for 10 minutes. QuantaSoft software (Bio-Rad) was used for results analysis.
Target capture–based WGS of EBV
WGS of EBV was performed as described previously.12 Briefly, a custom SureSelect bait targeting the whole genome of EBV was used to construct sequencing libraries. We identified single-nucleotide variants, copy number alterations, and SVs using our in-house pipeline. We confirmed the performance of the target capture–based sequencing using data obtained by WGS in 6 patients.
We also determined the EBV type (1 or 2) using reference sequences for EBV nuclear antigen 2 and EBV nuclear antigen 3. Additionally, we performed average linkage hierarchical clustering of the EBV genomes based on the number of nucleotide alterations using R (https://www.r-project.org/). The likelihood of the accumulation of deletions was calculated as described by a Monte-Carlo–based simulation.
Histopathology
Tissue samples were fixed in 10% formalin and embedded in paraffin. Three pathologists (S.K., D.Y., and S.N.) reviewed the cases and divided them into 4 morphological groups based on the cell nucleus shape, including centroblastoid, pleomorphic, mixed, or unspecified.1
Immunophenotypic and ISH analysis
FFPE sections were subjected to immunoperoxidase analysis with the following monoclonal antibodies: CD4, CD5, and CD56 (Novocastra Laboratories, Newcastle, United Kingdom); CD3, CD8, CD10, L26/CD20, Ber-H2/CD30, B-cell lymphoma 6 (BCL6), and 22C3/PD-L1 (Dako, Santa Clara, CA); CXCL13 (R&D Systems, Minneapolis, MN); granzyme B (Monosan, Uden, The Netherlands); SP98/inducible T-cell costimulator (ICOS) and γ 3.20/ T-cell receptor γ (TCR-γ; Thermo Fisher Scientific); programmed cell death protein 1 (PD-1; Abcam, Cambridge, United Kingdom); βF1 (TCR β chain; T Cell Science, Cambridge, MA); H-41/TCRδ (Santa Cruz Biotechnology, Dallas, TX); and T-cell intracellular antigen 1 (TIA-; Coulter Immunology, Hialeah, FL). The reactions were considered positive with a cutoff of 30%. Neoplastic PD-L1 expression (nPD-L1) was considered positive if ≥10% of the tumor cells had membranous and/or cytoplasmic PD-L1 staining. A case was considered positive for PD-L1 in the microenvironment when ≥20% comprised nonmalignant cells with moderate or strong membrane or cytoplasmic PD-L1–specific staining, among the total tissue cellularity.13
We subjected FFPE sections to ISH using EBER oligonucleotides as previously reported to evaluate the presence of EBV small ribonucleic acids.3
TCRγ PCR analysis
DNA was extracted from formalin-fixed tissues, and TCRγ PCR analysis was conducted using the BIOMED2 protocol.14
Statistical analysis
The Fisher exact test was used to determine the correlations between the 2 groups. The Kaplan-Meier method and the log-rank test were used to analyze and compare patient survival data, respectively. The Cox proportional hazard regression model was used for univariate and multivariate analysis. The STATA software package version 16 (Stata Corporation, College Station, TX) was used for all statistical analyses.
Results
Study cohort and design
WES was performed on 22 tumor samples from 22 patients with EBV+ nPTCL. Of these cases, 13 were compared with DNA from normal tissues, and 6 were subjected to WGS. We identified a total of 587 (15-77 per patient) somatic point mutations in the exome of 13 patients (supplemental Table 1). We checked the presence of driver mutation in the mixture of germ line and somatic point mutations in the remaining 9 patients for whom no germ line samples were available.
Genomic landscape
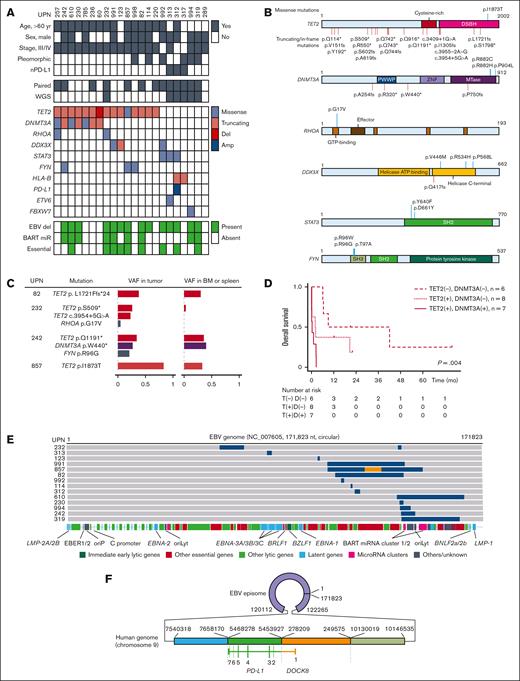
Mutations in known driver genes were identified in 20 of 22 patients (Figure 1A). The most frequently mutated genes were TET2 (15 cases, 68%) and DNMT3A (7 cases, 32%), both of which are associated with CHIP (Figure 1B).15-17 All DNMT3A mutations were co-occurrent with TET2 mutations (P = .051). Two patients demonstrated the RHOA p.Gly17Val hot spot mutation. Both of these patients were associated with TET2 and/or DNMT3A mutations. Each of the 3 additional genes (DDX3X, STAT3, and FYN) was mutated in 3 patients. The 3 identified STAT3 mutations were known hot spot mutations with implied gain-of-function. The 3 FYN alterations were found in the 2 amino acid residues of the SH3 domain (p.Arg96Gly, p.Arg96Trp, and p.Thr97Ala), which appear to be a mutation hot spot. Less frequently mutated genes included HLA-B (2 cases), ETV6 (1 case), FBXW7 (1 case), and PD-L1 (1 case). We did not identify TP53 mutations.
Genomic findings of EBV+ nPTCL. (A) The mutational landscape of EBV+ nPTCL. The rows contain clinical information, somatic mutation data, and EBV genome deletions. Del, deletions; Amp, amplifications. (B) The distribution of somatic mutations. (C) Presence of TET2 and DNMT3A mutations in the BM or spleen. The identical TET2 mutation was identified in the BM that histopathologically carried no tumor cells in UPN82. TET2 mutations were not obvious in the BM in UPN232. Both TET2 and DNMT3A mutations were identified in the BM in UPN242. The TET2 mutation was present both in the tumor and the spleen in UPN857, whereas the mutation in the tumor was a subject of loss of heterozygosity. (D) Kaplan-Meier plot of the OS of patients with EBV+ nPTCL stratified by TET2 (T) and DNMT3A (D) mutations at diagnosis (n = 21). P = .004 by log-rank test. The median OS of patients with DNMT3A mutations, patients with TET2 but without DNMT3A mutations, and patients without TET2 mutations were 0.2, 2.0, and 9 months, respectively. (E) Summary of intragenic deletions identified in EBV genomes of patients with EBV+ nPTCL. Each gray bar indicates an EBV genome from a patient with EBV+ nPTCL. Blue regions indicate deletions. An orange region indicates an inverted region of an EBV genome. The locations of EBV genome components are also indicated. oriP, replication origin used in latent infection; oriLyt, replication origin used in lytic infection. (F) A complex SV involving both EBV and human genomes identified in UPN312. The patient’s EBV genome incorporated 4 different parts of chromosome 9. Two adjacent pieces of the chromosome encoded DOCK8 and PD-L1, thereby forming DOCK8/PD-L1 fusion. All coding PD-L1 sequences were retained with an initial exon of DOCK8 on the 5′ side, indicating that messenger RNA transcription is driven by the DOCK8 promoter. A large part of the 3′ untranslated region (UTR) of PD-L1 was deleted, which upregulates PD-L1 expression.
Genomic findings of EBV+ nPTCL. (A) The mutational landscape of EBV+ nPTCL. The rows contain clinical information, somatic mutation data, and EBV genome deletions. Del, deletions; Amp, amplifications. (B) The distribution of somatic mutations. (C) Presence of TET2 and DNMT3A mutations in the BM or spleen. The identical TET2 mutation was identified in the BM that histopathologically carried no tumor cells in UPN82. TET2 mutations were not obvious in the BM in UPN232. Both TET2 and DNMT3A mutations were identified in the BM in UPN242. The TET2 mutation was present both in the tumor and the spleen in UPN857, whereas the mutation in the tumor was a subject of loss of heterozygosity. (D) Kaplan-Meier plot of the OS of patients with EBV+ nPTCL stratified by TET2 (T) and DNMT3A (D) mutations at diagnosis (n = 21). P = .004 by log-rank test. The median OS of patients with DNMT3A mutations, patients with TET2 but without DNMT3A mutations, and patients without TET2 mutations were 0.2, 2.0, and 9 months, respectively. (E) Summary of intragenic deletions identified in EBV genomes of patients with EBV+ nPTCL. Each gray bar indicates an EBV genome from a patient with EBV+ nPTCL. Blue regions indicate deletions. An orange region indicates an inverted region of an EBV genome. The locations of EBV genome components are also indicated. oriP, replication origin used in latent infection; oriLyt, replication origin used in lytic infection. (F) A complex SV involving both EBV and human genomes identified in UPN312. The patient’s EBV genome incorporated 4 different parts of chromosome 9. Two adjacent pieces of the chromosome encoded DOCK8 and PD-L1, thereby forming DOCK8/PD-L1 fusion. All coding PD-L1 sequences were retained with an initial exon of DOCK8 on the 5′ side, indicating that messenger RNA transcription is driven by the DOCK8 promoter. A large part of the 3′ untranslated region (UTR) of PD-L1 was deleted, which upregulates PD-L1 expression.
Blood cell–rich tissues (the BM or spleen) were available for assessing VAFs of identical mutations in 4 patients with TET2/DNMT3A alterations, that is UPN82, UPN232, UPN242, and UPN857 (Figure 1C). The ratio of EBER-positive cells to total cells in the BM or spleen was 0% for UPN82, UPN232, and UPN857, and <1% for UPN242 (supplemental Figure 1). Notably, 3 of 4 cases had at least 1 identical TET2 and/or DNMT3A mutation in the BM or spleen tissue with VAF of >0.05, which strongly indicates the presence of CHIP behind EBV+ nPTCL. In UPN242, although lymphoma cell infiltration was minimal, the VAF of TET2 and DNMT3A mutations in the BM was >36 times higher than the rate in EBER-positive cells. Based on these findings, the presence of CHIP is also suggested in UPN242. The presence of TET2 or DNMT3A mutations was associated with poorer OS (P = .004; Figure 1D). Furthermore, multivariate analysis indicated that TET2 and/or DNMT3A mutations (P = .034) were adverse prognostic factors, whereas age of >70 years at diagnosis was not (P = .58; supplemental Table 4). As all cases, except 1 with TET2 and/or DNMT3A mutations, were aged >60 years, an age cutoff of 70 years was used for the multivariate analysis.
EBV genome alterations
The whole of the EBV genome was sequenced using a target capture–based method or WGS. All identified viral genomes were that of type 1 EBV (the prevalent type in Japan), and the EBV genomes from patients with EBV+ nPTCL coclustered well in the other EBV genomes of other diseases in Japan, indicating no specific EBV strain that causes EBV+ nPTCL (supplemental Figure 2). SV was frequent among the EBV+ nPTCL EBV genomes (14 of 22 [63%]; Figure 1E). The most often found SV was deletions (727-32 074 nucleotides, 0.4%-18% of the entire EBV genome). The deletions accumulated in the genome are of BamHI A rightward transcripts microRNA clusters. Several essential genes for viral particle production were affected. Complex SVs were identified in 2 patients: a viral genome inversion sandwiched by 2 adjacent deletions in UPN857, and 4 pieces of the part of human chromosome 9 (2.6 megabases in total) tandemly incorporated into the EBV genome in UPN312. The combination of the 2 pieces of chromosome 9 formed the fusion of the promoter plus exon 1 of DOCK8 and exons 2-7 of PD-L1 (DOCK8/PD-L1 fusion).
The truncation of the 3′ untranslated region of PD-L1, which possibly upregulate the protein’s expression, was also observed. Combined with the high copy number (∼10-100 per cell) of EBV genomes compared with the human genome, the SV strongly suggests very high expression of the PD-L1 protein, which was actually detected in UPN312 (supplemental Figure 3).
Clinicopathologic features and TET2 and/or DNMT3A mutations
We compared the clinicopathological features between EBV+ nPTCL cases with and without TET2 mutations at presentation (Table 1; supplemental Table 5). The patients with TET2 mutations were more likely to have older age (median: 68 vs 59 years, P = .037), clinical stages III/IV (100% vs 57%, P = .023), high-intermediate–risk or high-risk class of International Prognostic Index (93% vs 33%, P = .014) disease, and less frequently demonstrated neoplastic PD-L1 expression (nPD-L1) positivity (0% vs 43%, P = .023). Pleomorphic appearance tended to be more frequently observed in patients without TET2 mutations compared with in those with mutations (57% vs 13%, P = .054; Figure 2). Patients with TET2 mutations, especially with concurrent DNMT3A mutations, were associated with a shorter OS (Figure 1D; hazard ratio, 3.95; P = .035; and hazard ratio, 5.31, P = .005, respectively). T follicular helper (TFH) cell markers were positive in 3 patients with EBV+ nPTCL. The positivity for TFH cell markers between TET2 mutation–positive and –negative cases demonstrated no significant difference.
Clinicopathological characteristics of EBV+ nodal T- and NK-cell lymphoma
| . | EBV+ nodal T- and NK-cell lymphoma . | P∗ . | ||
|---|---|---|---|---|
| All cases N = 22 (n [%]) . | TET2 mutations . | |||
| Present n = 15 (n [%]) . | Absent n = 7 (n [%]) . | |||
| Age at diagnosis (median [range]), y | 68 (21-83) | 68 (40-83) | 59 (21-75) | .037 |
| Age at diagnosis, >60 y | 16/22 (73) | 14/15 (93) | 2/7 (29) | .004 |
| Sex (male/female) | 14/8 | 8/7 | 6/1 | .19 |
| Performance status score of >1 | 10/19 (53) | 8/13 (62) | 2/6 (33) | .35 |
| Clinical stage Ⅲ/Ⅳ | 19/22 (86) | 15/15 (100) | 4/7 (57) | .023 |
| B symptoms present | 11/18 (61) | 8/11 (73) | 3/7 (43) | .33 |
| Extranodal involvement at >1 site | 1/22 (5) | 1/15 (7) | 0/7 (0) | 1.0 |
| IPI high-intermediate/high | 15/20 (75) | 13/14 (93) | 2/6 (33) | .014 |
| Platelets <130 × 10⁹/L | 15/20 (75) | 11/13 (85) | 4/7 (57) | .29 |
| Serum LDH level greater than normal | 19/21 (90) | 14/14 (100) | 5/7 (71) | .10 |
| Morphology | ||||
| Centroblastoid | 14/22 (64) | 11/15 (73) | 3/7 (43) | .34 |
| Pleomorphic | 6/22 (27) | 2/15 (13) | 4/7 (57) | .054 |
| Mixed | 2/22 (9) | 2/15 (13) | 0/7 (0) | 1.0 |
| Immunophenotype | ||||
| Cytotoxic molecules | 21/22 (95) | 14/15 (93) | 7/7 (100) | 1.0 |
| CD4 | 2/21 (10) | 2/15 (13) | 0/6 (0) | 1.0 |
| CD5 | 4/20 (20) | 2/13 (15) | 2/7 (29) | .59 |
| CD8 | 15/21 (71) | 12/15 (80) | 3/6 (50) | .29 |
| CD56 | 2/22 (9) | 0/15 (0) | 2/7 (29) | .091 |
| nPD-L1 | 3/22 (14) | 0/15 (0) | 3/7 (43) | .023 |
| miPD-L1 | 10/22 (45) | 7/15 (47) | 3/7 (43) | 1.0 |
| TFH cell marker† | 3/20 (15) | 2/14 (14) | 1/6 (17) | 1.0 |
| ICOS | 1/22 (5) | 1/15 (7) | 0/7 (0) | 1.0 |
| PD-1 | 3/22 (14) | 2/15 (13) | 1/7 (14) | 1.0 |
| T-cell type‡ | 20/20 (100) | 14/14 (100) | 6/6 (100) | - |
| . | EBV+ nodal T- and NK-cell lymphoma . | P∗ . | ||
|---|---|---|---|---|
| All cases N = 22 (n [%]) . | TET2 mutations . | |||
| Present n = 15 (n [%]) . | Absent n = 7 (n [%]) . | |||
| Age at diagnosis (median [range]), y | 68 (21-83) | 68 (40-83) | 59 (21-75) | .037 |
| Age at diagnosis, >60 y | 16/22 (73) | 14/15 (93) | 2/7 (29) | .004 |
| Sex (male/female) | 14/8 | 8/7 | 6/1 | .19 |
| Performance status score of >1 | 10/19 (53) | 8/13 (62) | 2/6 (33) | .35 |
| Clinical stage Ⅲ/Ⅳ | 19/22 (86) | 15/15 (100) | 4/7 (57) | .023 |
| B symptoms present | 11/18 (61) | 8/11 (73) | 3/7 (43) | .33 |
| Extranodal involvement at >1 site | 1/22 (5) | 1/15 (7) | 0/7 (0) | 1.0 |
| IPI high-intermediate/high | 15/20 (75) | 13/14 (93) | 2/6 (33) | .014 |
| Platelets <130 × 10⁹/L | 15/20 (75) | 11/13 (85) | 4/7 (57) | .29 |
| Serum LDH level greater than normal | 19/21 (90) | 14/14 (100) | 5/7 (71) | .10 |
| Morphology | ||||
| Centroblastoid | 14/22 (64) | 11/15 (73) | 3/7 (43) | .34 |
| Pleomorphic | 6/22 (27) | 2/15 (13) | 4/7 (57) | .054 |
| Mixed | 2/22 (9) | 2/15 (13) | 0/7 (0) | 1.0 |
| Immunophenotype | ||||
| Cytotoxic molecules | 21/22 (95) | 14/15 (93) | 7/7 (100) | 1.0 |
| CD4 | 2/21 (10) | 2/15 (13) | 0/6 (0) | 1.0 |
| CD5 | 4/20 (20) | 2/13 (15) | 2/7 (29) | .59 |
| CD8 | 15/21 (71) | 12/15 (80) | 3/6 (50) | .29 |
| CD56 | 2/22 (9) | 0/15 (0) | 2/7 (29) | .091 |
| nPD-L1 | 3/22 (14) | 0/15 (0) | 3/7 (43) | .023 |
| miPD-L1 | 10/22 (45) | 7/15 (47) | 3/7 (43) | 1.0 |
| TFH cell marker† | 3/20 (15) | 2/14 (14) | 1/6 (17) | 1.0 |
| ICOS | 1/22 (5) | 1/15 (7) | 0/7 (0) | 1.0 |
| PD-1 | 3/22 (14) | 2/15 (13) | 1/7 (14) | 1.0 |
| T-cell type‡ | 20/20 (100) | 14/14 (100) | 6/6 (100) | - |
IPI, International Prognostic Index; LDH, lactate dehydrogenase; miPD-L1, microenvironmental PD-L1; nPD-L1, neoplastic programmed cell-death ligand 1.
TET2 mutations present vs absent.
All of the evaluable 20 cases were negative for CD10 and BCL6, and all 22 evaluable cases were negative for CXCL13.
Patients with T-cell type showed positivity for TCR protein expression and/or TCRγ rearrangement.
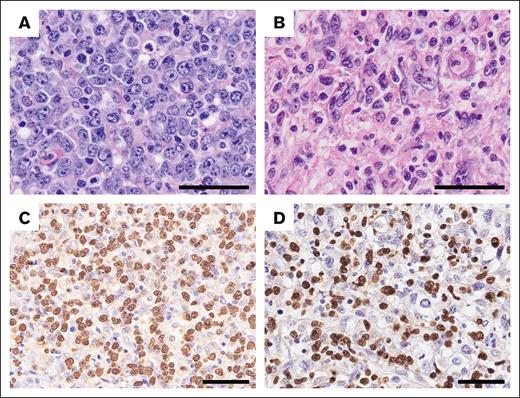
Light microscopy images of EBV+ nPTCL. Hematoxylin and eosin staining and EBV-encoded small RNA (EBER) ISH were performed to examine nuclear morphology, revealing a (A) centroblastoid (UPN857) or (B) pleomorphic appearance (UPN317). (C-D) Tumor cells are positive for EBER (C, UPN857; D, UPN317). Scale bar, 50 μm.
Light microscopy images of EBV+ nPTCL. Hematoxylin and eosin staining and EBV-encoded small RNA (EBER) ISH were performed to examine nuclear morphology, revealing a (A) centroblastoid (UPN857) or (B) pleomorphic appearance (UPN317). (C-D) Tumor cells are positive for EBER (C, UPN857; D, UPN317). Scale bar, 50 μm.
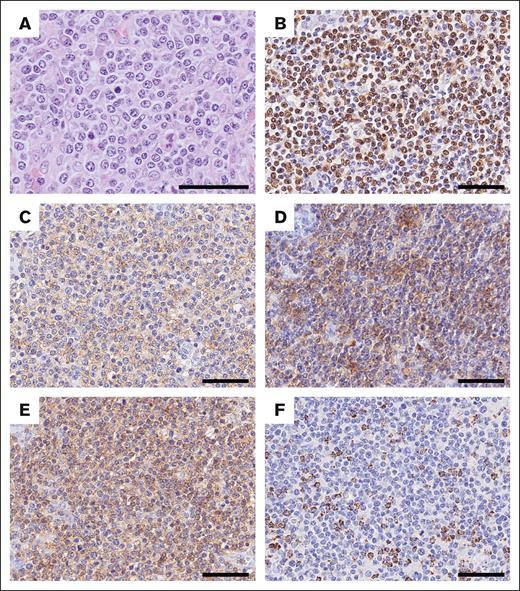
UPN257 was unique because neoplastic T cells were positive for CD4, ICOS, PD-1, and EBV-encoded small RNA, and negative for CD8, granzyme B, perforin, and TIA-1, showing overlapping features of both EBV+ nPTCL and nodal TFH cell lymphoma (nTFHL; Figure 3). This case demonstrated a diffuse infiltrate of medium to large T lymphoid cell with mild proliferation of high endothelial venules. Furthermore, WES revealed RHOA p.Gly17Val as well as DNMT3A and TET2 mutations in this tumor.
Morphological and phenotypic features of UPN257. (A) Tumor cells are medium to large in size and have a centroblastoid appearance. They are positive for (B) EBV-encoded small RNA (EBER), (C) PD-1, (D) ICOS, and (E) CD4, but negative for (F) TIA-1. Scale bar, 50 μm.
Morphological and phenotypic features of UPN257. (A) Tumor cells are medium to large in size and have a centroblastoid appearance. They are positive for (B) EBV-encoded small RNA (EBER), (C) PD-1, (D) ICOS, and (E) CD4, but negative for (F) TIA-1. Scale bar, 50 μm.
Discussion
TET2 and DNMT3A are genes frequently mutated in healthy individuals with CHIP.18 These 2 mutations were detected in 15 (68%) and 7 (32%) cases of EBV+ nPTCL, respectively, in this study. These frequencies were comparable with those found in angioimmunoblastic T-cell lymphoma (AITL; ∼80% and 20%-40%, respectively).19-21 Recently, Wai et al reported that 64% of EBV+ nPTCL cases harbored TET2 mutations.7 In addition, among 6 patients with EBV+ nPTCL analyzed in the EA4HP/SH lymphoma workshop, TET2 mutations were detected in 4 patients, and DNMT3A mutations in 3 patients.22 However, because their series demonstrated no paired normal blood samples, these previous studies were unable to determine whether these mutations were caused by CHIP.7,22 Our study revealed that 3 of 4 patients with available normal blood or spleen tissue had at least 1 identical TET2 and/or DNMT3A mutation in their normal samples as found in tumor tissues. Our findings indicate that most of the TET2 and DNMT3A mutations found in the current series were CHIP related, and a subset of EBV+ nPTCL originates from a TET2- and/or DNMT3A-mutated hematopoietic stem/progenitor cells. CHIP is an age-associated phenomenon,18 and the older age distribution of patients with TET2 mutations supports that these alterations were CHIP related in our series. Patients with EBV+ nPTCL with TET2 mutations, especially with DNMT3A alterations, were associated with a fulminant clinical course in this study. DNMT3A mutation was an adverse prognostic factor among patients with acute myeloid leukemia,23 which may be consistent with our results.
EBV genome in EBV-associated lymphoma/lymphoproliferative disorder is reported to harbor frequent SV.12 Our series identified intragenic deletion in 63% of patients with EBV+ nPTCL, which was more frequent than other EBV-associated lymphoid neoplasms examined, for example, 43% in NKTL and 35% in chronic active EBV infection.12 In our cohort, the 30-bp LMP1 deletion reported in Montes-Mojarro et al’s study of NKTL was not identified.24 The most frequently deleted region in this study was BamHI A rightward transcripts microRNA cluster, as previously reported in other EBV+ lymphomas.12 The previous study detected no intragenic EBV deletions in patients with infectious mononucleosis or posttransplant lymphoproliferative disorder.12 For EBV-infected lymphocytes detected in the background of AITL, Bahri et al failed to find the EBV genome deletions.25 Additionally, the deletion of the essential gene in the EBV genome was reported to promote lymphomagenesis in a xenograft model.12 These findings indicate that EBV infection is not ancillary in EBV+ nPTCL, and that EBV genome mutations play an oncogenic role in our series. In this context, it is plausible to assume that in most cases, EBV+ nPTCL developed as a result of EBV infection of cytotoxic T cells associated with CHIP and further deletion of the EBV genome.
RHOA p.Gly17Val mutation was observed in 50% to 70% of AITL and other nodal TFH cell lymphoma (nTFHL), which was hardly detected in other lymphomas in the past.26,27 In the current series, this mutation was detected in 2 cases (9%) of EBV+ nPTCL with TET2 mutations. These 2 cases had no history of hematolymphoid malignancy, including nTFHL (data not shown). Of these 2 patients, 1 patient (UPN257) was unique because neoplastic T cells were positive for CD4 and TFH cell markers as well as EBER; however, this case lacked cytotoxic molecule expression. Recently, Zhang et al reported 2 cases of nPTCL that were positive for CD4, TFH cell markers, and EBER-ISH.28 Cytotoxic molecules were expressed in 1 of these 2 cases, whereas TIA1 and granzyme B were negative in the other. These 2 cases also showed mutations in TET2 and RHOA p.Gly17Val. UPN257 in our series and the cases in Zhang et al showed intermediate features between EBV+ nPTCL and nTFHL. Langer et al reported a case of EBV+ nPTCL during the course of an EBV-negative PTCL; furthermore, both tumors were clonally related, suggesting that EBV infected the neoplastic cells of the EBV-negative PTCL.29 Regarding the role of RHOA p.Gly17Val mutation, Cortes et al demonstrated that mice transplanted with Tet2−/− BM progenitor cells infected with RHOA G17V-expressing retroviruses developed lymphomas with TFH cell–like features, including CXCR5, PD-1, BCL6, and ICOS expressions.30 Alternatively, mice transplanted with Tet2−/− hematopoietic progenitors infected with retroviruses expressing wild-type RHOA primarily developed myeloid malignancies but not AITL. These findings suggest that an additional RHOA p.Gly17Val mutation on a TET2−/− null background is crucial to induce TFH cell lineage specification on T cells. It is possible that UPN257 in our series and cases reported by Zhang et al either originated as nTFHL and were infected with EBV during the disease or were originally EBV+ nPTCL and developed nTFHL features with the addition of the RHOA p.Gly17Val mutation during the disease course. To clarify the relationship between EBV+ nPTCL and nTFHL, further investigations are warranted.
Frequently mutated genes in NKTL include BCOR (10%-32%), STAT3 (8%-27%), DDX3X (7%-20%), and TP53 (8%-16%).24,31-35 Furthermore, TP53 mutations were found in >40% of PTCL-NOS without TFH cell phenotype.36 In our EBV+ nPTCL cases, BCOR and TP53 mutations were absent, with TET2 being the most commonly mutated gene. Conversely, TET2 mutations were identified in only 10% to 12.2%, and 20% of NKTL and PTCL-NOS cases, respectively.35-37 These findings highlight the distinct mutation profile of EBV+ nPTCL compared with NKTL or PTCL-NOS.
Previously, Ng et al compared the clinicopathologic characteristics between the cases of T-cell origin in EBV+ nPTCL and those in NKTL. They demonstrated that older age, CD8 expression, and poor outcome remained substantially associated with T cell–type cases of EBV+ nPTCL compared with NKTL of T-cell origin.2 In addition, Xiong et al reported that in NKTL cases showing TCR rearrangements, EPHA1 (26%), TP53 (18%), ARID1A (17%), and BCOR (14%) were the main mutated genes.38 The mutation profile of NKTL with TCR rearrangement appears to differ from that of EBV+ nPTCL in our series. These findings provide further support that EBV+ nPTCL is clinicopathologically and pathogenetically distinct from NKTL beyond the cell of origin.
Recently, Nicolae et al demonstrated that TET2 or DNMT3A mutations were detected in 61% and 39%, respectively, among 33 patients with nPTCL-NOS with cytotoxic phenotype (nodal cytotoxic T-cell lymphoma [nodal CTL]) based on targeted deep sequencing.39 Of 33 patients with nodal CTL, 7 demonstrated EBV positivity in almost all tumor cells. Among these 7 cases of nodal EBV+ CTL, 3 presented with upper respiratory tract involvement. Interestingly, all 3 patients had CD8+ and CD56− phenotypes.39 These 3 cases demonstrated monoclonal TCR rearrangements and TET2 and DNMT3A mutations. Despite the limited number of cases, the clinicopathologic characteristics of nodal EBV+ CTL with upper aerodigestive tract involvement resemble those of the current series of EBV+ nPTCL, which had no nasal involvement.
Three EBV+ nPTCL cases demonstrated nPD-L1 expression, which positively correlated with STAT3 mutations (P = .038; data not shown). Of 3 nPD-L1–positive cases, 2 cases demonstrated HLA-B truncating mutations. Complex SV of the EBV genome with tandem incorporation of the human genome was identified in 1 (UPN312) of 2 patients, causing DOCK8/PD-L1 fusion. This case was the same as case 7 in Yamashita et al’s report.9 The 3′ untranslated region of PD-L1 was truncated in UPN312, as previously reported.9 These alterations in our series seem to contribute to immune escape in some of EBV+ nPTCL cases without TET2 mutation. Interestingly, none of the nPD-L1–positive cases in our series had TET2 mutations. Pan et al reported that the deletion of Tet2 in myeloid cells reduces melanoma tumor burden in mouse models,40 suggesting that wild-type Tet2 functions to sustain an immunosuppressive program in myeloid cells. To our knowledge, there are no reports indicating that TET2 mutations contribute to immune escape.
In conclusion, we identified frequent TET2 and DNMT3A mutations in EBV+ nPTCL, which seemed to be closely associated with CHIP. Recurrent EBV genome deletions are suggested to play an oncogenic role in the pathogenesis of EBV+ nPTCL. Further studies are warranted to use these findings to engineer novel therapeutic strategies for this intractable lymphoma.
Acknowledgments
The authors thank N. Ueda, K. Nakashima, and Y. Katayama for technical assistance. Computations were partially performed on the National Institute of Genetics (NIG) supercomputer at ROIS National Institute of Genetics.
This work was supported partly by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) (grant numbers JP21K06920 and JP19K16597), and the Daiko Foundation (to S.K.); and the Takeda Science Foundation (to Y.O.).
Authorship
Contribution: S.K. and Y.O. conceived the study; Y.O., M.H., and H.A. performed mutational analyses; S.K., A.O., D.Y., H.M., T.M.-T., K.O., A.T., W.H., and S.N. performed the research and analyzed the data; A.S., Y.G., Y.S., Y.T., K.T., N.A., and E.T. recruited patients and collected and provided clinical data; S.K. and Y.O. wrote the manuscript; and all of the authors critically read and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seiichi Kato, Center for Clinical Pathology, Fujita Health University Hospital, 1-98 Dengakugakubo, Kutsukake-cho, Toyoake 470-1192, Japan; email: seiichi.kato@fujita-hu.ac.jp; and Yusuke Okuno, Department of Virology, Nagoya City University Graduate School of Medical Sciences, Nagoya 467-8601, Japan; email: yusukeo@med.nagoya-cu.ac.jp.
References
Author notes
The variant reported in here is available in the Clinvar repository, with accession numbers: SCV004217846-SCV004218416.
The full-text version of this article contains a data supplement.