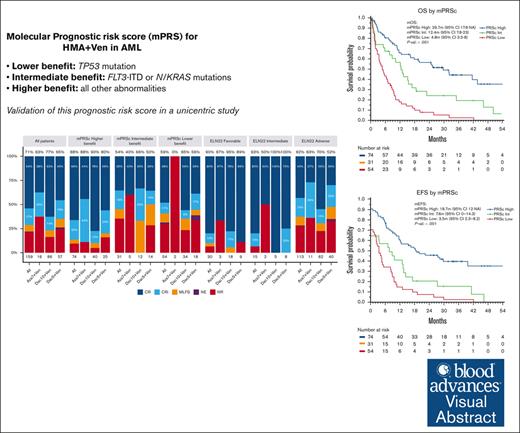
The mPRS can accurately segregate 3 groups of patients with AML with different outcomes with HMA and Ven.
Patients with FLT3-ITD and NRAS/KRAS mutations have a relatively unfavorable outcome with HMA and Ven, for which new strategies are needed.
Visual Abstract
Hypomethylating agents (HMAs) and venetoclax (Ven) represent the standard of care for patients with acute myeloid leukemia (AML) who are ineligible for intensive chemotherapy. However, the European LeukemiaNet (ELN) risk classifications have been validated for patients treated with intensive therapy. In this study, we validate a recently proposed new molecular prognostic risk signature (mPRS) for patients with AML treated with HMAs and Ven. This classification allocated patients to favorable, intermediate (N/KRAS or FLT3–internal tandem duplication mutations), and lower (TP53 mutations) benefit groups. We retrospectively analyzed 159 patients treated with HMA and Ven. The mPRS classification allocated 74 (47%), 31 (19%), and 54 (34%) patients to the higher, intermediate, and lower-benefit groups, respectively. The overall response rate was 71% (86%, 54%, and 59% in the higher, intermediate, and lower-benefit groups, respectively). The median overall survival (OS) and event-free survival (EFS) times were 30 and 19 months, respectively, in the higher-benefit group; 12 and 8 months in the intermediate-benefit group; and 5 and 4 months in the lower-benefit group (P < .001). The C-index for OS and EFS was higher when stratifying patients according to mPRS classification than with the ELN 2022 classification. The 2-year cumulative incidence of relapse was 35%, 70%, and 60% in the higher, intermediate, and lower-benefit groups, respectively (P = .005). The mPRS classification accurately segregated groups of patients with AML treated with HMA plus Ven. In these patients, N/KRAS and TP53 mutations appear to negatively affect outcomes; therefore, new treatment approaches are warranted.
Introduction
Acute myeloid leukemia (AML) is an aggressive hematopoietic stem cell neoplasm that predominantly affects older adults.1,2 The standard treatment consists of intensive chemotherapy, and in most cases, allogeneic hematopoietic stem cell transplantation (allo-HSCT) is needed to maintain disease remission.3 However, many patients are not eligible for intensive treatment owing to advanced age or comorbidities.4 Such patients are candidates for low-intensity treatments, which include low-dose cytarabine and hypomethylating agents (HMAs).5 In recent years, the addition of venetoclax (Ven) to low-intensity treatments has improved outcomes in these patients and, thus, has become an important part of the treatment of AML.6-8 This led to the approval by the US Food and Drug Administration of these combinations for patients aged >75 years or unfit for intensive chemotherapy.9
AML usually presents with recurrent genetic abnormalities, some of which have an evident impact on outcome. Therefore, several groups have proposed distinct prognostic classifications based on genetic findings to predict outcomes of AML.3,10,11 The most recent risk classification developed by a panel of worldwide experts is the European LeukemiaNet 2022 (ELN22).3 Briefly, this classification system defines 3 risk groups (favorable, intermediate, and adverse) based on cytogenetic and molecular findings at AML diagnosis. Researchers have validated this version as well as previous versions of the ELN risk classification system in cohorts of patients given intensive therapy.12-14
The ELN risk classification system was originally designed to help predict outcomes of patients with AML who were treated with intensive chemotherapy. However, the performance of these classifications in predicting survival in patients administered with HMAs and Ven has been suboptimal.15 This may be explained by the different mechanism of action of the combination of HMA and Ven compared with the cytotoxic effect of intensive chemotherapy. Furthermore, the patient population receiving lower-intensity treatment represent a unique cohort, characterized by older age and different disease biology, including higher rates of secondary AML and distinct genetic abnormalities.16,17 Recently, Döhner et al proposed the use of a 4-gene molecular prognostic risk signature (mPRS) to assess outcomes in patients with AML treated with HMAs and Ven.15 They developed this mPRS using clinical and biological data on patients given azacitidine and Ven in the phase 3 VIALE-A clinical trial.6 This mPRS classification uses N/KRAS, FLT3–internal tandem duplication (ITD), and TP53 mutations to stratify patients into 3 groups (higher, intermediate, and lower-benefit). With this simple 4-gene approach, the authors demonstrated clear segregation of 3 prognostic cohorts of patients with AML with different response rates and outcomes that appeared to perform better than ELN22. As described herein, we validated the mPRS in a large cohort of patients with AML given an HMA (decitabine or azacitidine) and Ven and performed a comprehensive survival analysis based on the results of using this classification.
Methods
Study design, risk group allocation, and response assessment
This was a single-center retrospective study performed at The University of Texas MD Anderson Cancer Center that included patients aged >60 years who had AML diagnosed from November 2014 to December 2021. Patients received HMAs and Ven in the setting of a clinical trial or off protocol. Cytogenetic analysis was performed using conventional karyotyping and fluorescence in situ hybridization. Mutational analysis was performed using a 28-gene next-generation sequencing (NGS) panel (for patients with AML diagnosed from 2014 to 2017) or an 81-gene NGS panel (for patients with AML diagnosed from 2017 to 2021). The NGS coverage for both panels is described in supplemental Tables 1 and 2.
FLT3-ITD mutations were detected using polymerase chain reaction–based DNA analysis. Patients were allocated to the intermediate-benefit group according to the mPRS if they had an FLT3-ITD, NRAS, or KRAS mutation. Patients with TP53 mutations were allocated to the lower-benefit group. The remaining patients were allocated to the higher-benefit group. Response to therapy was assessed using the ELN22 criteria for patients with AML.3 The overall response rate (ORR) was defined as the percentage of patients experiencing complete remission (CR) or CR with incomplete hematological recovery (CRi).
This study was approved by the MD Anderson Institutional Review Board and was conducted in accordance with the Declaration of Helsinki.
Statistical methods
Baseline characteristics were analyzed using descriptive statistics. The Student t test and Mann-Whitney U test were used for comparison of continuous variables with the normal and nonnormal distribution, respectively. For categorical variables, a χ2 test and the Fisher exact test were used. Median follow-up times were evaluated using the Kaplan-Meier estimate of potential follow-up.18 Overall survival (OS) time was measured from diagnosis to death. Event-free survival (EFS) time was measured from diagnosis to treatment failure, relapse, or death. Treatment failure was defined as not achieving CR or CRi. Patients who were evaluable for response and presented with treatment failure and those who died before any response assessment were considered to have had an event at day 1 of treatment. Patients who were alive and had nonevaluable response to treatment were censored on day 1 of treatment. The OS and EFS distributions were estimated using the Kaplan-Meier method and were compared using a log-rank test. For patients achieving CR or CRi, the cumulative incidence was calculated from the date of best response using death as the primary event and relapse as a competing event. Gray test was used to compare the cumulative incidence distributions. Cox proportional hazards regression was used for univariate and multivariate analysis. All statistical analyses were performed using the R computing language (version 4.2.2).
Results
Patient and treatment characteristics
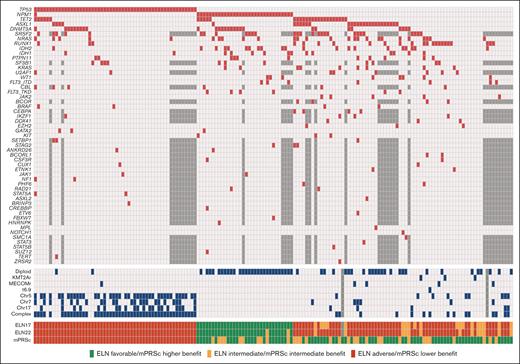
We identified a total of 179 patients with newly diagnosed AML who were treated with HMAs and Ven. Among them, 159 had available molecular data. Their baseline characteristics are summarized in Table 1. The median age was 75 years (range, 61-89 years), and 92 patients (58%) were male. Twenty-nine (18%) patients had secondary AML, and 33 patients (21%) had therapy-related AML. One patient with secondary AML received HMA because of a previous myelodysplastic syndrome diagnosis. According to cytogenetic analysis, 56 patients (31%) had normal karyotype, and 63 (40%) had complex karyotype. NGS results were available using an 81-gene panel (n = 121 [76%]) or a 28-gene panel (n = 38 [24%]). The most common mutations were those of TP53 (n = 54 [34%]), NPM1 (n = 33 [21%]), and TET2 (n = 29 [18%]). Mutational analysis results are detailed in Figure 1.
Baseline characteristics of the study patients
Characteristic . | All patients (N = 159) . | mPRS group . | P . | ||
|---|---|---|---|---|---|
| Higher benefit (n = 74) . | Intermediate benefit (n = 31) . | Lower benefit (n = 54) . | |||
| Age, median (range), y | 75 (61-89) | 74 (61-89) | 75 (62-86) | 75 (61-86) | .934 |
| Male sex, n (%) | 92 (57.9) | 45 (61) | 20 (65) | 27 (50) | .334 |
| Ethnicity, n (%) | .002 | ||||
| White | 126 (79.2) | 68 (91.9) | 24 (77.4) | 34 (63) | |
| Black | 7 (4.4) | 1 (1.4) | 1 (3.2) | 5 (9.3) | |
| Hispanic | 11 (7) | 3 (4.1) | 2 (6.5) | 6 (11.1) | |
| Asian | 6 (3.8) | 2 (2.7) | 0 | 4 (7.4) | |
| Median hemoglobin level, g/L (range) | 9.0 (7.1-12.7) | 9.2 (7.2-12.7) | 8.9 (7.1-10.9) | 8.9 (7.1-10.9) | .042 |
| Median WBC count, ×109 cells per L (range) | 2.9 (0.3-81.1) | 2.6 (0.6-81.1) | 3.9 (0.7-43.1) | 3.05 (0.30-72.60) | .875 |
| Median platelet count, ×109 cells per L (range) | 41 (4-368) | 54 (4-310) | 35 (5-368) | 36 (5-174) | .035 |
| Median bone marrow blast percentage (range) | 39 (11-95) | 39 (16-95) | 47 (22-86) | 36 (11-88) | .151 |
| Cytogenetics, n (%) | |||||
| Normal karyotype | 56 (35.2) | 37 (50) | 17 (54.8) | 2 (3.7) | <.001 |
| Chromosome 5/5q abnormality | 48 (30.2) | 9 (12.2) | 0 | 39 (72.2) | <.001 |
| Chromosome 7 abnormality | 29 (18.2) | 4 (5.4) | 2 (6.5) | 23 (42.6) | <.001 |
| Chromosome 17 abnormality | 26 (16.4) | 3 (4.1) | 2 (6.5) | 21 (38.9) | <.001 |
| t(6;9) | 1 (0.6) | 0 | 1 (3.2) | 0 | .129 |
| MECOMr | 5 (3.1) | 2 (2.7) | 2 (6.5) | 1 (1.9) | .491 |
| KMT2Ar | 3 (1.9) | 3 (4.1) | 0 | 0 | .164 |
| Complex karyotype | 63 (39.6) | 10 (13.5) | 5 (16.1) | 48 (89) | <.001 |
| Mutation, n (%) | |||||
| TP53 | 54 (34) | 0 | 0 | 54 (100) | <.001 |
| NPM1 | 33 (20.6) | 22 (29.7) | 10 (32.3) | 1 (1.9) | <.001 |
| TET2 | 29 (18.2) | 17 (23) | 6 (19.4) | 6 (11.1) | .227 |
| ASXL1 | 28 (17.6) | 13 (17.6) | 11 (35.5) | 4 (7.4) | .006 |
| DNMT3A | 28 (17.6) | 11 (14.9) | 8 (25.8) | 9 (16.7) | .381 |
| SRSF2 | 28 (17.6) | 18 (24.3) | 5 (16.1) | 5 (9.3) | .054 |
| NRAS/KRAS | 31 (19.5) | 0 | 26 (83.9) | 5 (9.3) | <.001 |
| RUNX1 | 23 (14.5) | 15 (20.3) | 5 (16.1) | 3 (5.6) | .048 |
| IDH1/2 | 34 (21.4) | 22 (29.7) | 9 (29) | 3 (5.6) | .001 |
| FLT3-ITD | 7 (3.9) | 0 | 7 (22.6) | 0 | <.001 |
| Therapy-related AML, n (%) | 33 (20.8) | 13 (17.6) | 3 (9.7) | 17 (31.5) | .038 |
| Secondary AML, n (%) | 29 (18.2) | 15 (20.3) | 6 (19.4) | 8 (14.8) | .721 |
| Treatment protocol, n (%) | .130 | ||||
| Aza7Ven | 16 (10.1) | 9 (12.2) | 5 (16.1) | 2 (3.7) | |
| Dec10Ven | 86 (54.1) | 40 (54.1) | 12 (38.7) | 34 (63) | |
| Dec5Ven | 57 (35.8) | 25 (33.8) | 14 (45.2) | 18 (33.3) | |
Characteristic . | All patients (N = 159) . | mPRS group . | P . | ||
|---|---|---|---|---|---|
| Higher benefit (n = 74) . | Intermediate benefit (n = 31) . | Lower benefit (n = 54) . | |||
| Age, median (range), y | 75 (61-89) | 74 (61-89) | 75 (62-86) | 75 (61-86) | .934 |
| Male sex, n (%) | 92 (57.9) | 45 (61) | 20 (65) | 27 (50) | .334 |
| Ethnicity, n (%) | .002 | ||||
| White | 126 (79.2) | 68 (91.9) | 24 (77.4) | 34 (63) | |
| Black | 7 (4.4) | 1 (1.4) | 1 (3.2) | 5 (9.3) | |
| Hispanic | 11 (7) | 3 (4.1) | 2 (6.5) | 6 (11.1) | |
| Asian | 6 (3.8) | 2 (2.7) | 0 | 4 (7.4) | |
| Median hemoglobin level, g/L (range) | 9.0 (7.1-12.7) | 9.2 (7.2-12.7) | 8.9 (7.1-10.9) | 8.9 (7.1-10.9) | .042 |
| Median WBC count, ×109 cells per L (range) | 2.9 (0.3-81.1) | 2.6 (0.6-81.1) | 3.9 (0.7-43.1) | 3.05 (0.30-72.60) | .875 |
| Median platelet count, ×109 cells per L (range) | 41 (4-368) | 54 (4-310) | 35 (5-368) | 36 (5-174) | .035 |
| Median bone marrow blast percentage (range) | 39 (11-95) | 39 (16-95) | 47 (22-86) | 36 (11-88) | .151 |
| Cytogenetics, n (%) | |||||
| Normal karyotype | 56 (35.2) | 37 (50) | 17 (54.8) | 2 (3.7) | <.001 |
| Chromosome 5/5q abnormality | 48 (30.2) | 9 (12.2) | 0 | 39 (72.2) | <.001 |
| Chromosome 7 abnormality | 29 (18.2) | 4 (5.4) | 2 (6.5) | 23 (42.6) | <.001 |
| Chromosome 17 abnormality | 26 (16.4) | 3 (4.1) | 2 (6.5) | 21 (38.9) | <.001 |
| t(6;9) | 1 (0.6) | 0 | 1 (3.2) | 0 | .129 |
| MECOMr | 5 (3.1) | 2 (2.7) | 2 (6.5) | 1 (1.9) | .491 |
| KMT2Ar | 3 (1.9) | 3 (4.1) | 0 | 0 | .164 |
| Complex karyotype | 63 (39.6) | 10 (13.5) | 5 (16.1) | 48 (89) | <.001 |
| Mutation, n (%) | |||||
| TP53 | 54 (34) | 0 | 0 | 54 (100) | <.001 |
| NPM1 | 33 (20.6) | 22 (29.7) | 10 (32.3) | 1 (1.9) | <.001 |
| TET2 | 29 (18.2) | 17 (23) | 6 (19.4) | 6 (11.1) | .227 |
| ASXL1 | 28 (17.6) | 13 (17.6) | 11 (35.5) | 4 (7.4) | .006 |
| DNMT3A | 28 (17.6) | 11 (14.9) | 8 (25.8) | 9 (16.7) | .381 |
| SRSF2 | 28 (17.6) | 18 (24.3) | 5 (16.1) | 5 (9.3) | .054 |
| NRAS/KRAS | 31 (19.5) | 0 | 26 (83.9) | 5 (9.3) | <.001 |
| RUNX1 | 23 (14.5) | 15 (20.3) | 5 (16.1) | 3 (5.6) | .048 |
| IDH1/2 | 34 (21.4) | 22 (29.7) | 9 (29) | 3 (5.6) | .001 |
| FLT3-ITD | 7 (3.9) | 0 | 7 (22.6) | 0 | <.001 |
| Therapy-related AML, n (%) | 33 (20.8) | 13 (17.6) | 3 (9.7) | 17 (31.5) | .038 |
| Secondary AML, n (%) | 29 (18.2) | 15 (20.3) | 6 (19.4) | 8 (14.8) | .721 |
| Treatment protocol, n (%) | .130 | ||||
| Aza7Ven | 16 (10.1) | 9 (12.2) | 5 (16.1) | 2 (3.7) | |
| Dec10Ven | 86 (54.1) | 40 (54.1) | 12 (38.7) | 34 (63) | |
| Dec5Ven | 57 (35.8) | 25 (33.8) | 14 (45.2) | 18 (33.3) | |
KMT2Ar, KMT2A gene rearrangement; MECOMr, MECOM gene rearrangement; WBC, white blood cell.
Genetics and risk stratification. Oncoplot showing the mutational landscape (red) and cytogenetic abnormalities (blue) in the study patients. Untested mutations in the patients evaluated using the 28-gene NGS panel are shown in dark gray.
Genetics and risk stratification. Oncoplot showing the mutational landscape (red) and cytogenetic abnormalities (blue) in the study patients. Untested mutations in the patients evaluated using the 28-gene NGS panel are shown in dark gray.
According to the ELN22 classification, 30 (19%), 15 (9%), and 113 patients (71%) had favorable, intermediate, and adverse risk, respectively. When applying mPRS classification, it stratified 74 (47%), 31 (19%), and 54 patients (34%) into the higher-, intermediate-, and lower-benefit groups, respectively. In the intermediate-benefit group, 7 patients (23%) had FLT3-ITD mutations, with a median allelic ratio of 0.09 (range, 0.01-0.47), and 26 patients (84%) had RAS mutations (9 had KRAS mutations, whereas 21 had NRAS mutations). Patients with favorable risk according to the ELN22 system were allocated to the higher- (n = 22 [73%]) and intermediate-benefit (n = 8 [27%]) groups in the mPRS classification. In comparison, patients at intermediate risk by ELN22 were allocated to the higher- (n = 11 [73%]) and intermediate-benefit (n = 4 [27%]) groups in the mPRS classification. Of the patients at adverse risk by ELN22, 40 (35%), 19 (17%), and 54 (47%) were allocated to the higher-, intermediate-, and lower-benefit groups, respectively (supplemental Figure 1).
When comparing baseline characteristics among the mPRS classification groups, patients in the intermediate- and lower-benefit groups had lower hemoglobin levels and platelet counts than patients in the higher-benefit group. Furthermore, patients in the lower-benefit group had a lower incidence of diploid cytogenetics (3.7% vs 54.8% vs 50% for patients in the lower-, intermediate-, and higher-benefit groups, respectively; P < .001). A higher incidence of complex cytogenetics was detected in the lower-benefit group (89%), than in patients in the intermediate- (16.1%) and higher-benefit (13.5%) groups (P<.001). When analyzing mutation incidence (besides N/KRAS, FLT3-ITD, and TP53), we found that patients in the lower-benefit group had lower incidence of NPM1 (2%), ASXL1 (7%), RUNX1 (6%), and IDH1/2 (6%) mutations than patients in the higher and intermediate groups (P < .05). Furthermore, patients in the lower-benefit group had a higher incidence of therapy-related neoplasms (32%) than those in the higher- (18%) and intermediate-benefit (10%) groups (P = .038).
Treatment and response analysis
All patients received Ven on a 28-day schedule during the first cycle. Ven dose and duration could be modified in the subsequent cycles at the discretion of the treating physician based on patient tolerability or owing to protocol requirements. Regarding the HMAs, 86 patients (54%) received a 10-day course of decitabine (Dec10Ven), 57 (36%) received a 5-day course of decitabine (14 with an oral formulation; Dec5Ven), and 16 (10%) received a 7-day course of azacytidine (Aza7Ven).
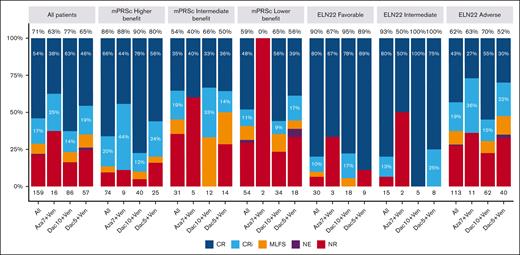
The median number of treatment courses received was 3 (range, 1-39), with a median of 2 (range, 1-29), 3 (range, 1-39), and 3 (range, 1-24) courses for Aza7Ven, Dec5Ven, and Dec10Ven, respectively. The ORR was 71%, with 54% and 17% of the patients experiencing CR and CRi, respectively. Moreover, the ORR was 86%, 54%, and 59% in the higher-, intermediate-, and lower-benefit groups in the mPRS classification, respectively (P < .001). When applying the ELN22 classification system, the ORR was 90%, 93%, and 62% for favorable, intermediate, and adverse risk, respectively (P = .001). A comparison of the ORRs for each treatment protocol in the 3 risk groups did not reveal any significant differences (Figure 2).
Response rates for all patients overall and according to mPRS and ELN 22 group. Percentages at the top of each bar represent the ORR. MLFS, morphologic leukemia-free status; mPRSc, mPRS classification NE, not evaluable; NR, no response.
Response rates for all patients overall and according to mPRS and ELN 22 group. Percentages at the top of each bar represent the ORR. MLFS, morphologic leukemia-free status; mPRSc, mPRS classification NE, not evaluable; NR, no response.
For patients who had treatment responses, the median number of cycles required to achieve best response was 1 (range, 1-8). The median cycles to response were 1 (range, 1-4), 1 (range, 1-8), and 1 (range, 1-6) for patients who received Aza7Ven, Dec10Ven, and Dec5Ven, respectively. Nineteen patients (12%) underwent allo-HSCT after achieving response (12, 6, and 1 patients receiving Dec10Ven, Dec5Ven, and Aza7Ven, respectively).
Survival analysis
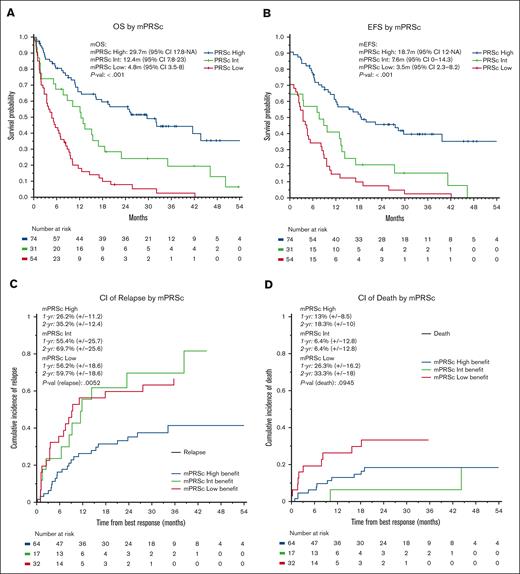
The median follow-up time for the entire cohort was 35 months (95% confidence interval [CI], 30-45 months). The median OS was 11 months (95% CI, 9-16 months), with 1- and 2-year OS rates of 48% and 34%, respectively. The median EFS was 9 months (95% CI, 7-12 months), with 1- and 2-year EFS rates of 43% and 29%, respectively. We observed no significant differences in OS or EFS according to treatment protocol. When stratifying patients using the mPRS, the median OS were 30, 12, and 5 months in the higher-, intermediate-, and lower-benefit groups, respectively (P < .001; C-index = 0.66). Furthermore, the median EFS times were 19, 8, and 4 months in the higher-, intermediate-, and lower-benefit groups, respectively (P < .001; C-index = 0.65, Figure 3). When applying the ELN22 system, the median OS times were 44, 13, and 8 months for favorable, intermediate, and adverse risk, respectively (P < .001; C-index = 0.60). Additionally, the median EFS time periods were 30, 8, and 6 months for favorable, intermediate, and adverse risk, respectively (P < .001; C-index = 0.62; supplemental Figures 2 and 3). When comparing the C-index obtained by applying the ELN22 and mPRS models, the mPRS had a better goodness of fit in predicting OS and EFS. We performed a multivariate OS analysis including cytogenetic abnormalities and mPRS classification, which demonstrated higher hazard ratios in the intermediate-benefit group (2.04 [95% CI, 1.20-3.40]; P = .005) and lower-benefit group (4.35 [95% CI, 2.80-6.80]; P < .001) than in the higher-benefit group.
Survival analysis. (A) OS, (B) EFS, (C) cumulative incidence of relapse, and (D) cumulative incidence of death according to mPRS classification. Adv, adverse; CI, cumulative incidence; Fav, favorable; Int, intermediate; mEFS, median EFS; mOS, median OS.
Survival analysis. (A) OS, (B) EFS, (C) cumulative incidence of relapse, and (D) cumulative incidence of death according to mPRS classification. Adv, adverse; CI, cumulative incidence; Fav, favorable; Int, intermediate; mEFS, median EFS; mOS, median OS.
For patients who had treatment responses, the 1- and 2-year cumulative incidence of relapse was 40% and 47%, respectively. Furthermore, the 1- and 2-year cumulative incidence of death as a competing event was 17% and 22%, respectively. When applying mPRS classification, the 2-year cumulative incidence of relapse was 35%, 70%, and 60% in the higher-, intermediate-, and lower-benefit groups, respectively (P = .005). The 2-year cumulative incidence of death was 18%, 6%, and 33% in the higher-, intermediate-, and lower-benefit groups, respectively (P = .09). When applying the ELN22 classification system, the 2-year cumulative incidence of relapse was 29%, 43%, and 55% for favorable, intermediate, and adverse risk, respectively, although the differences were not statistically significative (P = .1). Furthermore, the 2-year cumulative incidence of death was 12%, 21%, and 25% for favorable, intermediate, and adverse risk, respectively (P = .4); supplemental Figures 4 and 5).
Discussion
In this study, we analyzed a cohort of patients with newly diagnosed AML treated with HMA in combination with Ven, focusing on the differences in outcomes according to the ELN22 risk classification system and recently proposed mPRS classification. We included patients given treatment that included azacytidine or decitabine. When applying mPRS classification, patients were stratified into the higher- (47%), intermediate- (19%), and lower-benefit (34%) groups. Although using only a simple 4-gene classification system, these 3 groups had significant differences in terms of cytogenetic abnormalities as well as the overall mutational landscape. When analyzing response to therapy, mPRS classification discriminated 3 groups with different response rates, which was more evident when comparing patients in the higher-benefit group with those in the intermediate- and lower-benefit groups. Moreover, the outcome analysis demonstrated that mPRS classification performed well, segregating the 3 cohorts of patients with distinct survival as well as relapse risk.
The ELN classification systems are effective in stratifying patients according to risk, although investigators designed them based on patients receiving intensive treatments, most frequently with anthracyclines and cytarabine. However, because there are key differences in the patient population, leukemia biology, and mechanism of action between HMA plus Ven and intensive treatments, ELN classification does not optimally predict prognosis as well in the lower-intensity group. In our study, the ELN22 classification system stratified 71% of the patients as being at adverse risk, although only 47% were adverse (lower benefit) based on the mPRS classification. This paradoxical patient reallocation to nonadverse risk groups by the mPRS is explained by the presence of certain secondary-type mutations, such as those affecting splicing factors or chromatin modifiers, which are considered adverse by the ELN22 classification system but not by the mPRS. A recent report suggested that the addition of Ven to AML treatment could abrogate the adverse prognostic impact of splicing mutations in patients with AML.19 Moreover, recent publications have proposed that AML with ASXL1 mutation is especially sensitive to the combination of HMAs and Ven.20-22 Therefore, a reasonable position is that some mutations that are denoted as adverse by the ELN22 classification system may not actually be adverse in patients receiving HMAs and Ven.
When comparing the ability of ELN22 and mPRS classification to stratify patients according to outcome, both were able to segregate different cohorts of patients with significantly different outcomes. However, when assessing the performance of the 2 classification methods, the mPRS had a higher goodness of fit in predicting OS and EFS than the ELN22 system. The original cohort for which the mPRS classification method was developed had median OS times of 26.5, 12.1, and 5.5 months in the higher-, intermediate-, and lower-benefit groups, respectively.15 This is very similar to what we observed (29.7, 12.4, and 4.8 months in the higher-, intermediate-, and lower-benefit groups, respectively), providing external validation of the mPRS in an independent data set. Moreover, patients in the higher-benefit group according to mPRS classification had a markedly lower incidence of relapse (2-year cumulative incidence of relapse, 35%) than patients in the intermediate- and lower-benefit groups (2-year cumulative incidence of relapse, 60%-70%). Although not statistically significant, the 2-year cumulative incidence of relapse in the intermediate-benefit group was higher than that in the lower-benefit group. This could support the hypothesis that patients with N/KRAS and FLT3-ITD mutations are prone to present disease relapse when treated with HMA and Ven. Furthermore, the 2-year cumulative incidence of death in the intermediate-benefit group was lower than in the higher-benefit group, again nonstatistically significant. This might be explained because of fewer patients presenting relapse (therefore, being at risk of death as the competing event) and a higher incidence of these patients undergoing allo-HSCT, which exposes them to transplant-related mortality.
Notably, in the intermediate-benefit group in our study, more patients had RAS mutations than FLT3-ITD mutations. Furthermore, the proportion of patients in the intermediate-benefit group was lower in our cohort (19%) than in the original cohort that was used to design the mPRS classification (25%). RAS mutations are not considered to be unfavorable mutations according to the ELN17 and ELN22 classification systems,3,11 although both systems are optimized for patients receiving intensive chemotherapy. Several studies have suggested that patients with AML with RAS mutations may benefit from cytarabine-based regimens.23-25 Lower-intensity combinations using low-dose cytarabine and a second nucleoside analog such as cladribine may provide a tolerable approach to address RAS-mutated AML in an older, unfit population unable to tolerate intensive cytarabine-based approaches. We previously reviewed our own experience in RAS-mutated AML by therapy and observed improved outcomes with such an approach over HMAs.26FLT3-ITD was also a defining mutation in the intermediate-benefit group according to mPRS classification, although this group represented few patients in our study. Because the mPRS classification system was based on patients with AML on the VIALE-A study, treated only with azacytidine with or without Ven, it did not capture the impact of FLT3 inhibitors on response rate or survival. Novel, triplet combinations with an HMA, Ven, and a FLT3 inhibitor may abrogate the detrimental effect of FLT3-ITD mutations on response and survival.27-29
TP53 mutation was the defining mutation among patients classified into the lower-benefit group by the mPRS classification, and most had a complex karyotype. Of note, our cohort was enriched in these patients of the lower-benefit group (34%) compared with the original cohort, in which the mPRS classification was developed (23%). These AML cases are characterized by high genetic instability, a poor response rate (for both intensive and low-intensity therapy), and dismal outcomes.30,31 For this reason, TP53 mutations are considered adverse by both the ELN22 and mPRS classification methods. Because current standard AML treatments do not produce long-term remission of TP53-mutated disease, exploration of novel drugs or combinations in clinical trials is warranted.32 Moreover, in eligible patients, allo-HSCT may provide an additional graft-versus-leukemia effect, although outcomes of AML with TP53 mutations after allo-HSCT are also poor.33
The major limitation of this study is that it was a retrospective analysis from a single center with a limited number of patients, which is insufficient to definitely validate this prognostic tool. For this reason, a larger study including cooperative groups or a multi-institutional study would be necessary to support our data. Another important limitation is that this study included ∼50% of patients treated with a 10-day decitabine regimen, which is more intense than the usual HMA regimens.
In conclusion, in this study, we validated the mPRS classification method in a cohort of patients with AML treated with HMAs and Ven across different protocols. mPRS classification satisfactorily stratified patients into 3 groups with different response rates, survival times, and relapse rates. When compared with the ELN22 system, mPRS classification performs better in predicting survival. In the mPRS intermediate-benefit group, the impact of the addition of FLT3 inhibitors or the use of cytarabine-based regimens needs to be explored.
Acknowledgment
This study was partly funded by the Rama Guntupalli Endowment for Leukemia Research.
Authorship
Contribution: A. Bataller participated in study conception, data curation, formal data analysis, patient care, and writing, review, and editing of the manuscript. A. Bazinet participated in data curation, patient care, and review and editing of the manuscript. C.D.D., A.M., G.B., N.J.S., E.J.J., K.T., N.G.D., G.C.I., N.P., M.Y., G.M.-B., G.G.-M., F.R., and H.M.K. provided patient care and reviewed and edited the manuscript. S.L. provided molecular interpretation and reviewed and edited the manuscript. T.M.K. participated in study conception, patient care, and writing, review, and editing of the manuscript.
Conflict-of-interest disclosure: C.D.D. has been a board of directors or advisory committee member for Genmab, GlaxoSmithKline, Kura Oncology, and Notable Labs; has received honoraria from Kura, Astellas Pharma, bluebird bio, Bristol Myers Squibb, Foghorn Therapeutics, Immune-Onc Therapeutics, Novartis, Takeda Oncology, Gilead Sciences, and Jazz Pharmaceuticals; is a current holder of stock options for Notable Labs; has been a consultant for AbbVie and Servier; and has received research funding from Servier, Bristol Myers Squibb, Foghorn, Immune-Onc Therapeutics, Loxo Oncology, Astex Pharmaceuticals, Cleave, and Forma. A.M. reports support from BioSight, Sanofi, and Astex Pharmaceuticals. G.B. has received research funding from Astex Pharmaceuticals, Ryvu Therapeutics, and PTC Therapeutics; has been a board of directors or advisory committee member for Pacyclex Pharmaceuticals, Novartis, CytomX, and Bio Ascend; and has been a consultant for Catamaran Bio, AbbVie, PPD Development, Protagonist Therapeutics, and Janssen. N.S. has been a consultant for Takeda Oncology, AstraZeneca, Amgen, Novartis, and Pfizer; received research funding from Takeda Oncology, Astellas, and Stemline Therapeutics; and received honoraria from Amgen. K.T. has been a consultant for SymBio Pharmaceuticals; and received honoraria from Mission Bio, Illumina, and Otsuka Pharmaceutical. N.G.D. has received research funding from Astellas Pharma, AbbVie, Genentech, Daiichi Sankyo, Gilead Sciences, ImmunoGen, Pfizer, BristolMyers Squibb, Trovagene, Servier, Novimmune, Incyte, Hanmi Pharm, Fate Therapeutics, Amgen, Kite Pharma, Novartis, Astex Pharmaceuticals, KAHR, Shattuck, Sobi, GlycoMimetics, and Trillium; has been an adviser for Astellas Pharma, AbbVie, Genentech, Daiichi Sankyo, Novartis, Jazz Pharmaceuticals, Amgen, Servier, Karyopharm Therapeutics, Trovagene, Trillium, Syndax, Gilead Sciences, Pfizer, Bristol Myers Squibb, Kite Pharma, Actinium Pharmaceuticals, Arog Pharmaceuticals, ImmunoGen, Arcellx, and Shattuck; has been a data-monitoring committee member for Kartos Therapeutics and Jazz Pharmaceuticals; has been a consultant or board of directors or advisory committee member for Agios, Celgene, Sobi, and STAR Therapeutics; and has received research funding from Karyopham Therapeutics and Newave Pharmaceutical. G.I. has been a consultant for Novartis, Kura Oncology, and NuProbe and received research funding from Celgene, Kura Oncology, Syndax, Merck, Cullinan Oncology, and Novartis. N.P. has been a consultant for AbbVie, Astellas Pharma, Cimeio Therapeutics, CTI BioPharma, ImmunoGen, Intellisphere, Patient Power, PharmaEssentia, Protagonist Therapeutics, and TotalCME; has been a scientific/advisory committee member for Blueprint Medicines, Cancer.Net, CareDx, CTI BioPharma, Incyte, Menarini Group, and Pacylex Pharmaceuticals; has been a speaker or held a preceptorship for AbbVie, the Aplastic Anemia & MDS International Foundation, Curio Bioscience, DAVA Oncology, Intellisphere, Magdalen Medical Publishing, Medscape, Novartis, the Physician's Education Resource, Protagonist Therapeutics, and Targeted Oncology; and has received research support from the US Department of Defense. M.Y. has received research funding from Daiichi-Sankyo and Pfizer. G.G.-M. has received research funding from Astex Pharmaceuticals, Novartis, AbbVie, Bristol Myers Squibb, Genentech, Aprea Therapeutics, Curis, and Gilead Sciences; has been a consultant for Astex Pharmaceuticals, Acceleron Pharma, and Bristol Myers Squibb; and has received honoraria from Astex Pharmaceuticals, Acceleron Pharma, AbbVie, Novartis, Gilead Sciences, Curis, Genentech, and Bristol Myers Squibb. F.R. has received research funding from Amgen, Astex Pharmaceuticals/Taiho Oncology, Bristol Myers Squibb/Celgene, Syos, AbbVie, Prelude, Xencor, Astellas Pharma, and Biomea Fusion; honoraria from Amgen, Bristol Myers Squibb/Celgene, Syos, AbbVie, and Astellas Pharma; has been a board of directors or advisory committee member for Astex Pharmaceuticals/Taiho Oncology; and has been a consultant for Bristol Myers Squibb/Celgene, Syos, Novartis, AbbVie, AstraZeneca, and Astellas Pharma. H.M.K. has received research funding from AbbVie, Amgen, Ascentage Pharma, Bristol Myers Squibb, Daiichi Sankyo, ImmunoGen, Jazz Pharmaceuticals, and Novartis as well as honoraria from AbbVie, Amgen, Amphista Therapeutics, Ascentage Pharma, Astellas Pharma, Biologix, Curis, Ipsen, KAHR, Novartis, Pfizer, Precision BioSciences, Shenzhen TargetRx, and Takeda Oncology. T.M.K. has been a consultant for AbbVie, Agios, Bristol Myers Squibb, Genentech, Jazz Pharmaceuticals, Novartis, Servier, and PinotBio; has received research funding from AbbVie, Bristol Myers Squibb, Genentech, Jazz Pharmaceuticals, Pfizer, Cellenkos, Ascentage Pharma, Galectin Therapeutics, Astellas Pharma, AstraZeneca, Amgen, Cyclacel Pharmaceuticals, Delta-Fly Pharma, Iterion Therapeutics, GlycoMimetics, and Regeneron Pharmaceuticals; and has received honoraria from Astex Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Tapan M. Kadia, Department of Leukemia, Unit 428, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; email: tkadia@mdanderson.org.
References
Author notes
The data used for this study are not publicly available to protect patient confidentiality. Deidentified data are available on reasonable request from the corresponding author (tkadia@mdanderson.org).
The full-text version of this article contains a data supplement.