Key Points
Development of a genotyped and phenotyped cohort of patients with CH is required to establish clinical guidelines and translational research.
Initial data from a prospective registry and biorepository of patients with CH recapitulate findings derived from retrospective studies.
Visual Abstract
Clonal hematopoiesis (CH) is an age-associated phenomenon leading to an increased risk of both hematologic malignancy and nonmalignant organ dysfunction. Increasingly available genetic testing has made the incidental discovery of CH clinically common yet evidence-based guidelines and effective management strategies to prevent adverse CH health outcomes are lacking. To address this gap, the prospective CHIVE (clonal hematopoiesis and inflammation in the vasculature) registry and biorepository was created to identify and monitor individuals at risk, support multidisciplinary CH clinics, and refine taxonomy and standards of practice for CH risk mitigation. Data from the first 181 patients enrolled in this prospective registry recapitulate the molecular epidemiology of CH from biobank-scale retrospective studies, with DNMT3A, TET2, ASXL1, and TP53 as the most commonly mutated genes. Blood counts across all hematopoietic lineages trended lower in patients with CH. In addition, patients with CH had higher rates of end organ dysfunction, in particular chronic kidney disease. Among patients with CH, variant allele frequency was independently associated with the presence of cytopenias and progression to hematologic malignancy, whereas other common high-risk CH clone features were not clear. Notably, accumulation of multiple distinct high-risk clone features was also associated with cytopenias and hematologic malignancy progression, supporting a recently published CH risk score. Surprisingly, ∼30% of patients enrolled in CHIVE from CH clinics were adjudicated as not having clonal hematopoiesis of indeterminate potential, highlighting the need for molecular standards and purpose-built assays in this field. Maintenance of this well-annotated cohort and continued expansion of CHIVE to multiple institutions are underway and will be critical to understanding how to thoughtfully care for this patient population.
Introduction
Clonal hematopoiesis (CH) is an overrepresentation of mature blood cells derived from a single, genetically identical clone.1 CH is highly correlated with increasing age, with 15% of patients over the age of 65 estimated to have CH with a variant allele fraction (VAF) of at least 2%.1-4 CH is genetically heterogeneous, with most cases resulting from somatically derived mutations in leukemogenic driver genes within hematopoietic stem and progenitor cells.3 Variants have been reported from 72 CH driver genes, though more than two-thirds of CH mutations are found in 1 of 3 genes: DNMT3A, TET2, and ASXL1 (DTA mutations).3,5,6 Although CH-associated genes span a diverse set of cellular functions and processes, including epigenetic regulation, transcription, and RNA splicing,7,8 the resulting effect of a CH driver mutation is enhanced cellular fitness, leading to a selective advantage for the clone and subsequent clonal expansion.1,8,9
CH presents as clonal hematopoiesis of indeterminate potential (CHIP), an asymptomatic state with normal complete blood counts (CBC). Clonal cytopenia of uncertain significance (CCUS) connotes the presence of a clone and one or more associated cytopenias without a clear identifiable cause and a bone marrow biopsy lacking morphologic myelodysplasia.10 Numerous studies have demonstrated that CHIP and CCUS both increase the potential to progress to hematologic malignancy.1,8 Thus, CH is considered a premalignant state, and it is estimated that 0.5% to 1% of CHIP cases transform into an overt hematologic malignancy per year after acquiring additional somatic mutations.9 By definition, CCUS includes a hematologic phenotype and thus can be more pervasive in patients with multiple mutations, high VAFs, and/or those with non-DTA, myeloid neoplasm–type clones.11 The concept of high vs low risk of progression to myeloid neoplasm has been recently evaluated, and a risk score calculator has been developed for the evaluation of individuals for CH, though this has not been prospectively validated.12
In addition to malignancy risk, CH is associated with a high burden of organ dysfunction and confers a 40% increase in all-cause mortality.2 Recent reports of CH-associated organ dysfunction include increased risk of stroke and atherosclerotic vascular disease (ASCVD),13-16 inflammation and autoimmune disease,17-19 chronic obstructive pulmonary disease,20 infection,21 and chronic kidney disease,22-24 among others.25 Moreover, the mechanisms driving CH-associated adverse outcomes, in particular ASCVD, have recently been the subject of intense study.13,14,16,26 Given the diversity of genes involved in CH, myriad inflammatory and noninflammatory mechanisms are likely to exist for all downstream pathologies. As such, the prevailing immune dysregulation hypothesis as it currently exists does not completely reflect the complexity of CH across disease manifestations, and prospective assessments of collected patient samples and variant-specific mutational changes are needed.
Despite ample evidence supporting the association of CH with malignancy risk and end organ dysfunction, clinical guidelines for the identification, management, and surveillance of patients with CH are lacking, and treatment strategies for CH are nascent. Similarly, although recent progress has been made in our mechanistic understanding of CH progression and association with adverse outcomes, our clinical comprehension of factors mediating CH expansion and risk of progression remains poorly understood. This is due, in part, to the retrospective nature of available data and the paucity of serial evaluation of CH-related clones. Because CH represents a broad array of genes and clinical outcomes, research infrastructure must be built to study patients in a prospective manner to understand the mechanisms of progression at the level of the individual mutation in variable germ line contexts. Large-scale prospective cohorts will allow for the resolution needed to not only codify the natural history of CH but also give insight into possible therapeutic approaches. Ultimately, the development of clinical guidelines to risk stratify and treat patients with CH is needed to advance health care for a significant proportion of our aging population. To begin to address these challenges, we developed CHIVE (clonal hematopoiesis and inflammation in the vasculature), a collaborative registry and biorepository, and here, we describe a novel approach to the identification and enrollment of patients at risk for CH. We collate an early evaluation of clinical outcomes in patients with CH compared with those considered at risk for CH demonstrating the importance of development and maintenance of multidisciplinary, prospective CH clinics and biorepositories.
Methods
Identification and inclusion of study participants
Prospective CHIVE participants were identified by outpatient providers across Vanderbilt University Medical Center. Patients were eligible for study inclusion if they were either over 40 years of age or over 18 years of age with a known risk factor for CH, could give informed consent, and did not have an active hematologic malignancy (Figure 1). Our existing CH Clinic (established in 2018) includes patients with idiopathic cytopenia of uncertain significance (ICUS), CCUS, or CH as noted on clinical next-generation sequencing (NGS). These patients were included in arm A of CHIVE (n = 58). By contrast, we also sought out a population at risk for CH, which liberally included patients over 40 years of age or 18 years of age with a history of solid tumor, cardiovascular disease, renal disease, rheumatologic disease, or diabetes: arm B (n = 123). To identify patients at risk, we collaborated with specialists in hematology/oncology, cardiology, rheumatology, and genetic medicine. Given the known associations between CH and cardiovascular disease,13,14,16 autoimmune disease,17,19 history of chemotherapy/radiation treatment,27,28 and common hereditary cancer-related mutations,29 our initial collaborations were between these subspecialists.
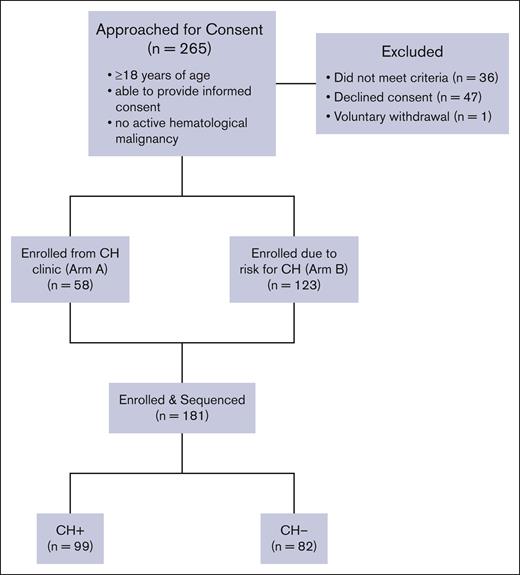
A total of 265 patients were approached for consent and 181 were enrolled; 47 patients were excluded because they declined consent. Thirty-six patients did not meet inclusion criteria. One patient voluntarily withdrew from the study. Of the enrolled patients, 63 were recruited from CH Clinic in the department of hematology (arm A) and 118 were recruited from elsewhere in the hospital system (arm B). Patients who were CH+ were defined as individuals with a CH variant present with a VAF of 2% or greater on the CHIVEseq assay (n = 99).
A total of 265 patients were approached for consent and 181 were enrolled; 47 patients were excluded because they declined consent. Thirty-six patients did not meet inclusion criteria. One patient voluntarily withdrew from the study. Of the enrolled patients, 63 were recruited from CH Clinic in the department of hematology (arm A) and 118 were recruited from elsewhere in the hospital system (arm B). Patients who were CH+ were defined as individuals with a CH variant present with a VAF of 2% or greater on the CHIVEseq assay (n = 99).
Patients in arm A were actively and voluntarily enrolled in CHIVE directly from CH Clinic. For those patients in arm B, enrollment occurred via initial collaborators, as noted above, or from direct patient interest in participation in research studies. In the case of the latter, our institution offers patients with an interest in research studies to be contacted when new opportunities arise, so, eligible patients with an interest in research studies were considered for CHIVE if deemed eligible via review of the electronic medical record (EMR). This subset of patients with upcoming appointments at the hospital system were contacted via telephone, introduced to the study, and, if meeting criteria, offered the opportunity to voluntarily participate in CHIVE.
Once enrolled, the research study was integrated with their regularly scheduled clinical care for all patients. Consensus guidelines are lacking regarding clinical and laboratory monitoring of patients with CH, though experts agree that patients with CH with high-risk features (multiple mutations, high VAF, high-risk mutations) should be monitored with greater frequency.12,30 Accordingly, follow-up samples were collected at 6- to 12-month intervals based on a rubric created by clinical experts in CH (supplemental Figure 1). Samples were collected from patients who were high-risk every 6 months, whereas collection was extended to 12 months for those without CH or with low-risk CH. This study was reviewed and approved by the Vanderbilt Institutional Review Board, and the enrollment is ongoing.
Development of a biorepository
After participants gave informed consent, clinical data available from standard clinical care were collected, including demographic data, medical histories, vital signs, laboratory studies, imaging studies, data from bone marrow biopsies, and clinically ordered genotyping. Genetic data included germ line testing, clinical NGS mutation analyses, and chromosome analyses. Additional cardiovascular studies, including electrocardiogram data and echocardiogram data were collected when available. Upon study enrollment, initial research samples from peripheral blood draws were obtained at the time of planned, routine, clinical sample collection. Similarly, aliquots from bone marrow aspirates and biopsies were procured when available.
Blood samples collected at 6- to 12-month intervals were stored within the CHIVE biorepository, a dedicated space with the capacity to store peripheral blood and bone marrow samples. Specimens were maintained in a liquid nitrogen freezer with a password-protected lock, which was housed in a dedicated laboratory space under supervision. Specimens were assigned a deidentified participant number for archiving to prevent subject identification; patient identifiers were separately stored in a REDcap database, accessible only with a secure ID and password.
CH status ascertainment
A low-cost custom oligonucleotide gene capture panel (Twist Bioscience) covering 95% of all CH gene mutations (supplemental Table 1) was used to perform targeted sequencing of prospective CHIVE participants as previously described.31 Briefly, DNA was extracted from whole blood using Qiagen Mini kits (Cat #27104) according to the manufacturer's recommendations. DNA library preparation was performed using a hybrid capture system to selectively amplify genes of interest (Twist Bioscience) before sequencing on an Illumina Novaseq 6000 targeting 1000× coverage. Mutect2, a publicly available somatic variant caller in the Genome Analysis Toolkit (GATK), was used to detect CH variants within the aligned sequencing data using the Tera biocomputing platform (http://Terra.bio).32 A putative variant list was created and filtered. Variants with a total low read depth (<100), a low variant allele read depth (<3), or a VAF below the threshold for CHIP (<2%) were removed from the data set. Manual filtering was performed to remove known sequencing artifacts, including variants that appear recurrently in multiple samples and/or across multiple projects but that are not known CHIP hotspots.2,6,33 We additionally sought to distinguish germ line from somatically occurring variants. We performed a binomial test to determine whether the measured allele depth for the variant is statistically different from half of the total allele depth at that site. We removed any variants that were not significant at P < .05. We refer to this sequencing assay henceforth as CHIVEseq.
For analysis, patients from arm A or arm B were included in the CH-positive (CH+) group if they met criteria for CH via the CHIVEseq assay at any time during their enrollment in the study (ie, on initial or follow-up sequencing). Patients with confirmed CH mutations, both with cytopenias (CCUS) and without cytopenias (CHIP), were included in the CH+ group. All consented, genotyped patients who were not found to have CH by the CHIVEseq assay at any point in time were included in the CH-negative (CH–) group. Notably, these patients with CH– were not true controls because all patients enrolled in CHIVE were at higher risk for CH than the general population based on clinical characteristics.
For any cases in which CH status determined via CHIVEseq assay differed from that determined via clinical genotyping, the discrepancy was rectified, and CH status was designated via manual evaluation of genotyping results. Common reasons for such discrepancies included misidentification of a germ line variant as CH, VAF below 2%, and mutations found via clinical sequencing in genetic regions not covered by the CHIVEseq assay. In the latter case, these variants were reviewed and classified as CH if they otherwise met criteria used for CHIP determination in whole genome– or exome-based methods.33
Clinical management
All patients followed in CH Clinic (arm A) were referred to collaborating cardio-oncology for individualized risk assessment for cardiovascular disease. Patients in arm A had blood pressure measurement, a physical examination, and basic laboratory testing, including CBC with differential, complete metabolic panel, and inflammatory markers. For patients in arm B who were not followed in CH clinic, laboratory and imaging data were recorded and maintained in the CHIVE database as available from routine clinical care.
Care for patients with CH demands a multidisciplinary approach, as input from investigators across clinical specialties enriches patient care. Our CH clinic, like most early CH clinics, emanated in hematology, but as experience grows, it is important to assure that patients with CH or at risk for CH can be initially managed at various points of entry. At our institution, patients can enter CHIVE from throughout the medical system, and providers across specialties can initiate evaluation and enroll patients.
Baseline clinical characteristics and laboratory results collected during routine care were extracted from the EMR and organized according to CH status. Laboratories were analyzed if they were collected within 6 months of the most recently sequenced sample. If multiple laboratories were collected within this 6-month range, the laboratory values collected nearest in time to the sequencing sample were used for analysis. Laboratory results from hemoglobin A1c (HbA1c) and brain natriuretic peptide (BNP) were analyzed if collected as part of routine care within 1 year of the most recently sequenced sample. Resting ejection fraction (EF) measurements were collected from a variety of tests of cardiovascular function, including echocardiograms, cardiac magnetic resonance imaging, myocardial perfusion imaging, and left heart catheterization.
Statistical analysis
χ2 tests were performed to determine statistical significance of categorical variables, and 2 sample t tests were used to compare the means of continuous variables. If the assumptions of these parametric tests were violated, nonparametric alternatives were used. To account for possible confounders, a multivariable regression was performed for each statistically significant outcome to assess the independent contribution from age, sex, and body mass index (BMI), in addition to CH status.
The largest VAF detected per individual with a CH variant was tested for an association with age using linear regression analysis and Pearson correlation coefficient. CH risk scores were derived from the initial blood count and sequencing information to categorize patient prognostic risk as high, intermediate or low. A Fisher exact test was used to determine if there was a statistically significant correlation with risk categorization. All analyses were performed using R statistical software (v4.1.1) with a statistical significance alpha value defined as 0.05. For visualization of the mutational landscape among patients with CH, an UpSet plot was generated using the UpSetR package.34,35
Results
Study population characteristics
From October 2020 to April 2023, 261 patients were approached for consent to enroll in CHIVE. Of whom, 34 patients did not meet inclusion criteria, 47 declined consent, and 1 patient voluntarily withdrew from the study. Therefore, genotyping results were obtained from 336 samples from 181 total patients: 99 patients were determined to have at least 1 CH mutation (CH+), and 82 patients had no CH mutations (CH–; Figure 1). Two hundred and 46 samples were serial samples collected from 89 patients at regular intervals as dictated by the testing rubric (supplemental Figure 1). Of the 181 patients included in the study, 63 were recruited from hematology, 69 from cardiology, 45 from genetics, and 4 from rheumatology (supplemental Table 2).
Males comprised 50.5% of the patients who were CH+ and 29.3% of the patients who were CH– (P = .006) (Table 1). Median age of patients who were CH+ was 71.91 years and that of patients who were CH– was 62.90 years (P < .001), and 74.2% of patients in the CH+ group were ≥65 years old compared with 46.3% of those in the CH– group. The median BMI was similar between groups (P = .256).
Baseline demographic characteristics of patients who were CH+ patients compared with those who were CH−
| . | CH− (n = 82) . | CH+ (n = 99) . | P value . |
|---|---|---|---|
| Sex | |||
| Male | 24 (29.3) | 50 (50.5) | .009 |
| Female | 58 (70.7) | 49 (49.5) | |
| Age | |||
| 18-29 | 3 (3.7) | 0 (0.0) | |
| 30-49 | 14 (17.1) | 1 (1.0) | |
| 50-64 | 27 (32.9) | 24 (24.8) | |
| 65+ | 38 (46.3) | 74 (74.2) | |
| Median ± IQR | 62.9 (51.5-72.7) | 71.9 (64.0-77.5) | <.001 |
| BMI | |||
| <18.5 | 3 (3.7) | 1 (1.0) | |
| 18.5-24.9 | 23 (28.0) | 22 (22.7) | |
| 25.0-29.9 | 28 (34.2) | 34 (35.1) | |
| 30.0-34.9 | 17 (20.7) | 23 (23.7) | |
| >35 | 11 (13.4) | 17 (17.5) | |
| Median ± IQR | 27.4 (24.1-32.4) | 28.9 (25.4-32.1) | .253 |
| Tobacco use | |||
| Current smoker | 2 (2.4) | 4 (4) | .691 |
| Former smoker | 22 (26.8) | 30 (30.3) | .624 |
| . | CH− (n = 82) . | CH+ (n = 99) . | P value . |
|---|---|---|---|
| Sex | |||
| Male | 24 (29.3) | 50 (50.5) | .009 |
| Female | 58 (70.7) | 49 (49.5) | |
| Age | |||
| 18-29 | 3 (3.7) | 0 (0.0) | |
| 30-49 | 14 (17.1) | 1 (1.0) | |
| 50-64 | 27 (32.9) | 24 (24.8) | |
| 65+ | 38 (46.3) | 74 (74.2) | |
| Median ± IQR | 62.9 (51.5-72.7) | 71.9 (64.0-77.5) | <.001 |
| BMI | |||
| <18.5 | 3 (3.7) | 1 (1.0) | |
| 18.5-24.9 | 23 (28.0) | 22 (22.7) | |
| 25.0-29.9 | 28 (34.2) | 34 (35.1) | |
| 30.0-34.9 | 17 (20.7) | 23 (23.7) | |
| >35 | 11 (13.4) | 17 (17.5) | |
| Median ± IQR | 27.4 (24.1-32.4) | 28.9 (25.4-32.1) | .253 |
| Tobacco use | |||
| Current smoker | 2 (2.4) | 4 (4) | .691 |
| Former smoker | 22 (26.8) | 30 (30.3) | .624 |
Values are presented as counts (percentage) for categorical variables or median (interquartile range) for continuous variables. Bold indicates significance P < 0.05.
IQR, interquartile range.
CH mutation analysis
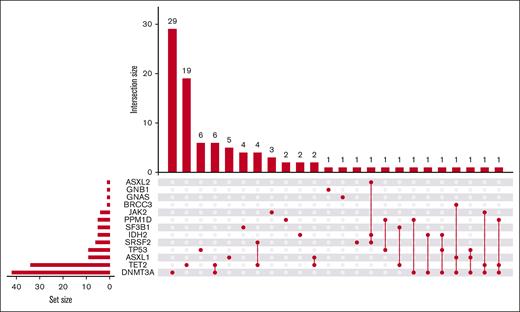
The most commonly mutated genes were DNMT3A (n = 44), TET2 (n = 40), ASXL1 (n = 10), and TP53 (n = 10; Figure 2). Other genes found to be mutated in this population were SRSF2, IDH2, SF3B1, PPM1D, JAK2, ASXL2, BRCC3, GNB1, and GNAS. Most patients who were CH+ (n = 99) had single mutations in either DNMT3A (n = 29) or TET2 (n = 21). Twenty-eight of the 99 total patients with CH (28.9%) had multiple mutations (Figure 2). VAF ranged from 2.1% to 79.8%. The highest VAF for any gene for each patient who was CH+ was plotted against age at genotyping, revealing a nominal positive correlation between age and VAF (Pearson correlation coefficient r = 0.18, P = .079, supplemental Figure 2). Bone marrow aspirates were collected during routine care for 15 patients. Of these, 8 had concordant mutations with similar overall VAFs between bone marrow and peripheral blood. Five samples lacked mutations in either the bone marrow or the peripheral blood. There was a sample with a mutation in the peripheral blood with no corresponding mutation in the bone marrow, and similarly, one mutation was found in a bone marrow aspirate without concordant findings in the peripheral blood. Putative germ line variants were also able to be identified within our CHIVEseq assay. Thirteen patients harbored 17 mutations in CH genes in germ line distribution. None were known pathologic variants (supplemental Table 3).
UpSet plot of CH-associated genes. Horizontal bars (set size) represent the number of individual mutations in each gene present within our cohort of patients who were CH+ (n = 99). Vertical bars (intersection size) represent the number of patients who were CH+ with a given mutational landscape. Connecting dot plot displays the specific gene or combination of genes that are mutated in each patient group.
UpSet plot of CH-associated genes. Horizontal bars (set size) represent the number of individual mutations in each gene present within our cohort of patients who were CH+ (n = 99). Vertical bars (intersection size) represent the number of patients who were CH+ with a given mutational landscape. Connecting dot plot displays the specific gene or combination of genes that are mutated in each patient group.
Of note, 57 patients recruited to CHIVE were included in arm A and were suspected to have a CH mutation based on a clinical sequencing panel. However, only 41 of these patients were found to have CH using the CHIVEseq assay. We investigated the 16 discrepant results and identified that all variants reported in clinical panels did not meet conventional CH criteria (Table 2). Most of these (n = 12 patients, n = 16 variants) were missense variants in CH genes at nonhotspot sites present at a VAF of roughly 50% and failing the binomial test, consistent with germ line variants. Two patients had a missense mutation in CUX1, a gene not included on the CHIVEseq assay. One individual had a JAK2 V617F variant at 0.2% VAF, which was below the current 2% VAF threshold for CH diagnosis. One individual had a SF3B1 K700E variant at a low VAF (3.1%) via clinical sequencing that was not subsequently identified via CHIVEseq, even at a VAF below 2%. The same patient had a DNMT3A L888Q variant, which does not meet conventional criteria for CH.13,33 Finally, one patient had a ASXL1 G646Wfs∗12 variant in a homopolymer region; by CHIVEseq, variants within this region with a VAF below 10% are considered artifactual.33 This patient’s ASXL1 G646Wfs∗12 VAF was 6% and therefore not considered to represent CH.
Patients who were referred with positive clinical CH determination found to be CH− on CHIVEseq assay
| Study ID . | Clinically identified mutation(s) . | Clinical panel . | Reason(s) . |
|---|---|---|---|
| 0006 | JAK2 p.V617F (VAF 0.2%) | Tempus (liquid biopsy) | Below 2% VAF threshold |
| 1020 | TET2 p.A915P (VAF 45%) | Myeloid neoplasm NGS Panel | Germ line mutation; mutation is outside of TET2 catalytic domain & therefore not considered a CH variant by CHIVEseq |
| 1071 | BCOR p.E1081K (VAF > 99%) | Myeloid neoplasm NGS panel | Germ line mutation; BCOR gene not covered on CHIVEseq assay |
| 1085 | CUX1 p.P1080∗ (VAF 5.18%) | Myeloid neoplasm NGS panel | CUX1 gene not covered on CHIVEseq assay |
| 1095 | JAK2 p.N691H (VAF 47.87%) | Myeloid neoplasm NGS panel | Germ line mutation; mutation not considered a CH variant on CHIVEseq assay |
| 1103 | TET2 p.C88R (VAF 45.94%) CSF3R p.R698C (VAF 48.92%) | Myeloid neoplasm NGS panel | TET2: germ line mutation; mutation is outside of TET2 catalytic domain and therefore not considered a CH variant by CHIVEseq CSF3R: germ line mutation; CSF3R gene not covered on CHIVEseq assay |
| 2001 | BCORL p.V881E (VAF 49.52%) | Myeloid neoplasm NGS panel | Germ line mutation; BCORL gene not covered on CHIVEseq assay |
| 2006 | SH2B3 p.L429V (VAF 48.58%) | Myeloid neoplasm NGS panel | Germ line mutation; SH2B3 gene not covered on CHIVEseq assay |
| 2011 | CALR p.E381del (VAF 48.97%) BCOR p.S209L (VAF 99.2%) | Myeloid neoplasm NGS panel | CALR: germ line mutation; CALR gene not covered on CHIVEseq assay BCOR germ line mutation; BCOR gene not covered on CHIVEseq assay |
| 2015 | JAK2 p.I724T (VAF 48.99%) SMC1A p.I1062M (VAF 50.47%) TET2 p.A289P (VAF 48.99%) | Myeloid neoplasm NGS panel | JAK2: germ line mutation; mutation not considered a CH variant by CHIVEseq SMC1A: germ line mutation; SMC1A gene not covered on CHIVEseq assay TET2: germ line mutation; mutation is outside of TET2 catalytic domain and therefore not considered a CH variant by CHIVEseq |
| 2016 | CUX1 p.E1373Q (VAF 48.62%) CUX1 p.R843K (VAF 50.35%) | Myeloid neoplasm NGS panel | Germ line mutations; CUX1 gene not covered on CHIVEseq assay |
| 2019 | DNMT3A p.A462V (VAF 57%) | Myeloid neoplasm NGS panel | Germ line mutation |
| 2041 | SF3B1 p.K700E (VAF 3.1%) DNMT3A p.L888Q (VAF 2.1%) | NeoTYPE MDS/CMML profile | SF3B1: this site was sequenced but the SF3B1 K700E variant was not identified, including at VAF <2%. DNMT3A: mutation is not considered CH by conventional definitions |
| 2046 | SH2B3 p.S213R (VAF 47.77%) | Myeloid neoplasm NGS panel | Germ line mutation; SH2B3 gene not covered on CHIVEseq assay |
| 2059 | KDM6A p.R621H (VAF 51%) | Advanced NGS myeloid report | Germ line mutation; KDM6A gene not covered on CHIVEseq assay |
| 2068 | ASXL1 p.G646Wfs∗12 (10%) | Advanced NGS myeloid report | Commonly encountered artifact in a homopolymer region of the CHIVEseq assay. Our practice is to categorize these variants as CH when VAF is > 10%. This variant was not considered CH because the VAF was 6%. |
| Study ID . | Clinically identified mutation(s) . | Clinical panel . | Reason(s) . |
|---|---|---|---|
| 0006 | JAK2 p.V617F (VAF 0.2%) | Tempus (liquid biopsy) | Below 2% VAF threshold |
| 1020 | TET2 p.A915P (VAF 45%) | Myeloid neoplasm NGS Panel | Germ line mutation; mutation is outside of TET2 catalytic domain & therefore not considered a CH variant by CHIVEseq |
| 1071 | BCOR p.E1081K (VAF > 99%) | Myeloid neoplasm NGS panel | Germ line mutation; BCOR gene not covered on CHIVEseq assay |
| 1085 | CUX1 p.P1080∗ (VAF 5.18%) | Myeloid neoplasm NGS panel | CUX1 gene not covered on CHIVEseq assay |
| 1095 | JAK2 p.N691H (VAF 47.87%) | Myeloid neoplasm NGS panel | Germ line mutation; mutation not considered a CH variant on CHIVEseq assay |
| 1103 | TET2 p.C88R (VAF 45.94%) CSF3R p.R698C (VAF 48.92%) | Myeloid neoplasm NGS panel | TET2: germ line mutation; mutation is outside of TET2 catalytic domain and therefore not considered a CH variant by CHIVEseq CSF3R: germ line mutation; CSF3R gene not covered on CHIVEseq assay |
| 2001 | BCORL p.V881E (VAF 49.52%) | Myeloid neoplasm NGS panel | Germ line mutation; BCORL gene not covered on CHIVEseq assay |
| 2006 | SH2B3 p.L429V (VAF 48.58%) | Myeloid neoplasm NGS panel | Germ line mutation; SH2B3 gene not covered on CHIVEseq assay |
| 2011 | CALR p.E381del (VAF 48.97%) BCOR p.S209L (VAF 99.2%) | Myeloid neoplasm NGS panel | CALR: germ line mutation; CALR gene not covered on CHIVEseq assay BCOR germ line mutation; BCOR gene not covered on CHIVEseq assay |
| 2015 | JAK2 p.I724T (VAF 48.99%) SMC1A p.I1062M (VAF 50.47%) TET2 p.A289P (VAF 48.99%) | Myeloid neoplasm NGS panel | JAK2: germ line mutation; mutation not considered a CH variant by CHIVEseq SMC1A: germ line mutation; SMC1A gene not covered on CHIVEseq assay TET2: germ line mutation; mutation is outside of TET2 catalytic domain and therefore not considered a CH variant by CHIVEseq |
| 2016 | CUX1 p.E1373Q (VAF 48.62%) CUX1 p.R843K (VAF 50.35%) | Myeloid neoplasm NGS panel | Germ line mutations; CUX1 gene not covered on CHIVEseq assay |
| 2019 | DNMT3A p.A462V (VAF 57%) | Myeloid neoplasm NGS panel | Germ line mutation |
| 2041 | SF3B1 p.K700E (VAF 3.1%) DNMT3A p.L888Q (VAF 2.1%) | NeoTYPE MDS/CMML profile | SF3B1: this site was sequenced but the SF3B1 K700E variant was not identified, including at VAF <2%. DNMT3A: mutation is not considered CH by conventional definitions |
| 2046 | SH2B3 p.S213R (VAF 47.77%) | Myeloid neoplasm NGS panel | Germ line mutation; SH2B3 gene not covered on CHIVEseq assay |
| 2059 | KDM6A p.R621H (VAF 51%) | Advanced NGS myeloid report | Germ line mutation; KDM6A gene not covered on CHIVEseq assay |
| 2068 | ASXL1 p.G646Wfs∗12 (10%) | Advanced NGS myeloid report | Commonly encountered artifact in a homopolymer region of the CHIVEseq assay. Our practice is to categorize these variants as CH when VAF is > 10%. This variant was not considered CH because the VAF was 6%. |
Mutation in question, clinical panel, and reasoning for non-CH diagnosis are reported.
BCOR, BCL6 corepressor.
Clinical findings
The median white blood cell count, hemoglobin, hematocrit, and platelets, and the depth of cytopenias were not statistically significant between patients who were CH– and CH+ (Table 3). Creatinine values were significantly elevated in patients who were CH+ than in those who were CH– (0.98 vs 0.90, P = .015; Table 3); however, multivariable regression analyses revealed that these differences were independently associated with patient BMI but not with CH status (supplemental Figure 3). Finally, 40 of 99 patients with CH+ (41.2%) met the criteria for chronic kidney disease (CKD) compared with 15 of 82 controls (18.3%) (P = .002; Table 3). In this case, multivariable regression analyses showed these differences were independently associated with both patient age (P = .012) and CH status (P = .039; supplemental Figure 3).
Selected laboratory values and clinical diagnoses of patients who were CH+ patients compared with those who were CH−
| . | CH− . | CH+ . | Unit . | P value . |
|---|---|---|---|---|
| Blood counts | ||||
| White blood cells | 6.8 (5.2-8.4) | 6.2 (4.5 8.1) | ×1000/mcL | .455 |
| Hemoglobin | 13.3 (12.3-14.6) | 12.9 (11.4-14.1) | gm.dL | .136 |
| Hematocrit | 42 (37.0-44.0) | 39.0 (35.0-43.0) | % | .134 |
| Platelet | 245.0 (198.0-284.0) | 203.0 (163.0-262.0) | ×1000/mcL | .093 |
| Kidney function | ||||
| BUN | 16.0 (12.0-21.0) | 18.0 (14.0-23.0) | mg/dL | .168 |
| Creatinine | 0.9 (0.76-1.11) | 0.97 (0.84-1.29) | mg/dL | .015 |
| CKD diagnosis, n (%) | 15 (18.3) | 41 (41.2) | .002 | |
| Blood glucose | ||||
| Glucose | 97.0 (88.0-117.0) | 106.0 (91.0-121.0) | mg/dL | .21 |
| HbA1c | 6.0 (5.4-6.8) | 5.8 (5.3-6.4) | % | .45 |
| Diabetes mellitus diagnosis, n (%) | 22 (26.8) | 31 (30.9) | .522 | |
| Inflammatory markers | ||||
| ESR | 16.0 (6.0-33.0) | 20.0 (15.0-32.0) | mm/h | .67 |
| CRP | 3.2 (1.2-13.1) | 8.2 (2.3-37.3) | mg/L | .183 |
| . | CH− . | CH+ . | Unit . | P value . |
|---|---|---|---|---|
| Blood counts | ||||
| White blood cells | 6.8 (5.2-8.4) | 6.2 (4.5 8.1) | ×1000/mcL | .455 |
| Hemoglobin | 13.3 (12.3-14.6) | 12.9 (11.4-14.1) | gm.dL | .136 |
| Hematocrit | 42 (37.0-44.0) | 39.0 (35.0-43.0) | % | .134 |
| Platelet | 245.0 (198.0-284.0) | 203.0 (163.0-262.0) | ×1000/mcL | .093 |
| Kidney function | ||||
| BUN | 16.0 (12.0-21.0) | 18.0 (14.0-23.0) | mg/dL | .168 |
| Creatinine | 0.9 (0.76-1.11) | 0.97 (0.84-1.29) | mg/dL | .015 |
| CKD diagnosis, n (%) | 15 (18.3) | 41 (41.2) | .002 | |
| Blood glucose | ||||
| Glucose | 97.0 (88.0-117.0) | 106.0 (91.0-121.0) | mg/dL | .21 |
| HbA1c | 6.0 (5.4-6.8) | 5.8 (5.3-6.4) | % | .45 |
| Diabetes mellitus diagnosis, n (%) | 22 (26.8) | 31 (30.9) | .522 | |
| Inflammatory markers | ||||
| ESR | 16.0 (6.0-33.0) | 20.0 (15.0-32.0) | mm/h | .67 |
| CRP | 3.2 (1.2-13.1) | 8.2 (2.3-37.3) | mg/L | .183 |
Values are presented as median (interquartile range) for continuous variables and counts (percentage) for categorical variables. For blood counts, n = 151 (86 CH+ and 65 CH−). For BUN and creatinine, n = 155 (88 CH+ and 67 CH−). For glucose, n = 158 (90 CH+ and 68 CH−). For HbA1c, n = 83 (53 CH+ and 30 CH−). For ESR, n = 31 (19 CH+ and 12 CH−). For CRP, n = 35 (21 CH+ and 14 CH−). For CKD and diabetes mellitus diagnoses, n = 181 (99 CH+ and 82 CH−). Bold indicates significance P < 0.05.
BUN, blood urea nitrogen; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate.
Significantly more patients who were CH+ had a history of coronary artery disease (CAD) compared with those who were CH– (55.7% vs 32.9%, P = .004). Similarly, patients who were CH+ in this cohort had increased rates of hypertension diagnosis (79.4% vs 52.4%, P < .001) and heart failure diagnosis (22.7% vs 9.8%, P = .035) compared with the CH– group (Table 4). However, multivariable regression revealed that these differences were not independently associated with CH status in this small cohort.
Selected measures of cardiovascular health in patients with CH compared with those without CH
| . | CH− . | CH+ . | Unit . | P value . |
|---|---|---|---|---|
| Cardiovascular measurements | ||||
| Systolic blood pressure | 125 (117-134) | 129 (118-139) | mmHg | .249 |
| Diastolic blood pressure | 75 (68-82) | 72 (66-78) | mmHg | .177 |
| CAD diagnosis, n (%) | 27 (32.9) | 55 (55.7) | .004 | |
| Hypertension diagnosis, n (%) | 43 (52.4) | 77 (79.4) | <.001 | |
| Clinical heart failure diagnosis, n (%) | 8 (9.8) | 24 (22.7) | .035 | |
| Brain natriuretic peptide | 79 (47-110) | 56 (34-184) | pg.mL | .908 |
| Ejection fraction | 60 (55-63) | 61 (54-68) | % | .424 |
| . | CH− . | CH+ . | Unit . | P value . |
|---|---|---|---|---|
| Cardiovascular measurements | ||||
| Systolic blood pressure | 125 (117-134) | 129 (118-139) | mmHg | .249 |
| Diastolic blood pressure | 75 (68-82) | 72 (66-78) | mmHg | .177 |
| CAD diagnosis, n (%) | 27 (32.9) | 55 (55.7) | .004 | |
| Hypertension diagnosis, n (%) | 43 (52.4) | 77 (79.4) | <.001 | |
| Clinical heart failure diagnosis, n (%) | 8 (9.8) | 24 (22.7) | .035 | |
| Brain natriuretic peptide | 79 (47-110) | 56 (34-184) | pg.mL | .908 |
| Ejection fraction | 60 (55-63) | 61 (54-68) | % | .424 |
Values are presented as median (interquartile range) for continuous variables and counts (percentage) for categorical variables. For BNP, n = 61 (46 CH+ and 15 CH−). For EF, n = 43 (15 CH+ and 28 CH−). For systolic blood pressure, diastolic blood pressure, CAD diagnosis, hypertension diagnosis, and heart failure diagnosis, n = 181 (99 CH+ and 82 CH−). Bold indicates significance P < 0.05.
Overall, nonhematologic cancers were prevalent in CHIVE patients because of recruitment from a hereditary cancer clinic. Among patients who were CH, 30 (30.9%) had other forms of malignancy, whereas patients without CH totaled 37 (45.1%). Breast cancer was the most frequent nonhematologic cancer in both subgroups (supplemental Table 4).
Patient outcomes
Of the 99 patients who were CH+, 6 (4.7%) progressed to frank hematologic malignancy over the course of the 2.5-year study. Two of these 6 patients (33%) harbored CH mutations in 2 or more genes, 4 (67%) had mutations in genes considered to be high risk, and 5 (83.3%) had one or more variants with a VAF > 10% (Table 5). The average VAF among these 8 patients was 35.7%, compared with 10.8% in those patients who were CH+ and had not progressed to malignancy. Interestingly, when also accounting for age, sex, and BMI, CH VAF was independently and significantly associated with a diagnosis of CCUS (P = .004) and with progression to hematologic malignancy (P = .009), whereas other high-risk clone features, presence of multiple mutations, and presence of mutations in high-risk genes, were not independently associated with either CCUS or malignancy progression in this initial cohort. However, the total number of high-risk clone features present was significantly associated with both the diagnosis of CCUS (P = .009) and progression to hematologic malignancy (P = .004, supplemental Figure 4). The Clonal Hematopoiesis Risk Score (CHRS) calculator was used to assign prognostic risk categories (low, intermediate, and high) across the CHIVE cohort.12 Fifty patients were determined to be low risk, with 1 patient (2%) progressing to malignancy. Intermediate risk included 32 patients, with 1 (3.1%) progressing to malignancy, whereas 14 patients were high risk and 4 (28%) progressed to malignancy. CHRS categorization in our cohort was predictive of malignancy progression (Fisher exact test, P = .007).
Clone features of patients with CH that developed hematologic malignancy over the course of the study
| Patient ID . | Number of mutations . | Mutation . | Maximum VAF . | High risk gene . | Average VAF . | Type of malignancy . |
|---|---|---|---|---|---|---|
| 0004 | 4 | TET2 R1516X | 0.398 | No | 0.217 | MDS |
| TET2 Q695X | 0.369 | No | ||||
| SRSF2 P95H | 0.177 | Yes | ||||
| JAK2 V617F | 0.02 | Yes | ||||
| 1060 | 1 | SF3B1 R625C | 0.241 | Yes | 0.236 | MDS |
| 1073 | 4 | TET2 Q742X | 0.422 | No | 0.237 | CMML |
| SRSF2 P95R | 0.330 | Yes | ||||
| TET2 Y1245Lfs∗22 | 0.270 | No | ||||
| TET2 N535Kfs∗6 | 0.033 | No | ||||
| 1096 | 1 | TET2 G1288D | 0.796 | No | 0.758 | CMML |
| 2026 | 1 | TP53 Y220C | 0.676 | Yes | 0.676 | AML |
| 2038 | 1 | TET2 Q749Rfs∗15 | 0.021 | No | 0.021 | MDS |
| Patient ID . | Number of mutations . | Mutation . | Maximum VAF . | High risk gene . | Average VAF . | Type of malignancy . |
|---|---|---|---|---|---|---|
| 0004 | 4 | TET2 R1516X | 0.398 | No | 0.217 | MDS |
| TET2 Q695X | 0.369 | No | ||||
| SRSF2 P95H | 0.177 | Yes | ||||
| JAK2 V617F | 0.02 | Yes | ||||
| 1060 | 1 | SF3B1 R625C | 0.241 | Yes | 0.236 | MDS |
| 1073 | 4 | TET2 Q742X | 0.422 | No | 0.237 | CMML |
| SRSF2 P95R | 0.330 | Yes | ||||
| TET2 Y1245Lfs∗22 | 0.270 | No | ||||
| TET2 N535Kfs∗6 | 0.033 | No | ||||
| 1096 | 1 | TET2 G1288D | 0.796 | No | 0.758 | CMML |
| 2026 | 1 | TP53 Y220C | 0.676 | Yes | 0.676 | AML |
| 2038 | 1 | TET2 Q749Rfs∗15 | 0.021 | No | 0.021 | MDS |
Clone features considered to be high-risk include total number of mutations, VAF, and mutation(s) in high-risk genes.
AML, acute myeloid leukemia; CMML, chronic myelomonocytic leukemia; MDS, myelodysplastic syndrome.
Eight out of 99 patients who were CH+ (8.1%) and 2 of 82 patients who were CH– (2.4%) died over the course of the study period resulting in a trend toward decreased survival in patients who were CH+ (P = .112). The most common causes of death among patients who were CH+ were malignancy (n = 5) and septic shock (n = 3), whereas the 2 patients who were CH– died of trauma and renal failure (supplemental Table 5).
Discussion
CH is an increasingly recognized condition with broad implications among aging populations. As NGS continues to evolve and become more widely adopted, additional patients will be identified with putative CH, underscoring the need for a greater understanding of the natural history of CH as well as the development of hematopathology and clinical guidelines. Registries, biorepositories, and prospective observational studies with well-described patients who were CH are critical to meeting this need. The CHIVE biorepository is a proof-of-principle, single institution–wide effort to define and catalog patients with CH with the intention of building a cohort of patients from which new clinical insights and treatment strategies can be derived. To our knowledge, this is the first prospective CH registry and biorepository, with preliminary outcomes to be described.
Analysis of several aspects of our data aligns with previous research from large-scale studies. Patients with CH variants followed the expected proportions of known CH genes, with DNMT3A, TET2, ASXL1, and TP53 being the most prominent. The median age of patients who were CH+ was a decade older than patients who were CH-, providing further data supporting CH as an age-associated process. Patients with CH also had higher mortality rates compared with those without CH, without a clear categorical trend. Patients who were CH+ in this cohort had decreased blood counts across all hematopoietic lineages as well as increased rates of CKD. Finally, the accumulation of multiple high-risk features of CH clones was associated with increased rates of CCUS diagnosis and the development of hematologic malignancy. Taken together, these findings mirror larger retrospective data sets, demonstrating enhanced risk of CH across a spectrum of pathologies.
Some elements of our data were not congruent with currently published studies. Although we observed higher rates of cardiovascular diagnoses in the CH+ group compared with the CH– group, these outcomes were independently associated with other patient factors, such as age, sex, and BMI, and not with CH status. These findings are attributable to a lack of power in our currently small cohort, a more stringent definition of CH driver genes, and/or additional patient factors that could serve as potential confounders. Notably, our CH– cohort is not a true control group, as all patients enrolled were deemed at risk for CH; many of them (29%) were enrolled via cardiology because of known cardiovascular dysfunction. Given the disproportionately high rates of cardiovascular disease in the CH– group compared with that in the general population, the emergence of a trend toward worse cardiovascular outcomes in the CH+ group suggests an association between CH and cardiovascular disease that may be partially masked within our cohort.
We also identified the largest VAF as positively associated with CCUS and the development of malignancy in our cohort, whereas other clone features commonly considered high-risk (multiple mutations, specific high-risk mutations) were not. This observation may point to clone features within the high-risk category that confer even higher risk. The CHRS categorization of CHIVE patients was highly congruent with malignancy outcomes in this small cohort with 1-2 years of follow-up. One-third of patients progressed to malignancy who were categorized as high-risk, whereas only 2% progressed to malignancy who were in the low-risk category. Although there is not enough longitudinal data to prospectively validate this score, it affirms the utility of such a score and represents an easy-to-use tool for clinicians. Continued refinement of this score through the accrual of prospective data will better inform and equip clinicians in serial monitoring and implementation of early prevention strategies, medical therapies, and treatment plan adjustments.
To our knowledge, this study is the first to actively recruit patients from outside of hematology, and 56 of the 117 patients recruited from cardiology, genetics, and rheumatology were found to have CH variants not previously identified. It is important to continue to find and recruit these patients to CH clinics to advance clinical and translational research into the natural history of CH, to identify potential high-risk features of clones that may warrant CLIA-approved sequencing and close monitoring, and to guide specialty-specific follow-up of these newly identified patients with CH.
This study also highlights the current challenges associated with increasing rates of clinical use of NGS and subsequent referral practices. Although there are specific genes that account for most of the reported CH, there is not a universally accepted list of CH driver genes.33 Furthermore, the various NGS platforms and panels differ considerably in which genomic regions are sequenced and if and how variants are reported out.33 Predictably, these factors cause confusion among health care providers regarding what constitutes a potential pathologic clone and ultimately lead to referrals to clinics that may not be necessary. In this real-world cohort, 16 patients were referred to the CH Clinic and enrolled in CHIVE based on mutations identified via a clinical NGS panel but who did not have a bona fide CH mutation. Though these patients did not meet current criteria for CH, prospective data such as those from CHIVE can be used to clarify and validate definitions of CH, so following these patients for future analyses is critical. This will serve as a basis for buttressed CH definitions, which will provide guidelines for surveillance and referral practices and be useful in the evolving CH clinical trials.
There are certain limitations in this study that should be addressed and will improve with the enrollment from more patients from multiple sites. First, CHIVE is a single-center study with limited ethnic diversity. Our novel recruitment strategy with the inclusion of patients seen in hereditary cancer clinic (25% of the cohort) likely contributed to the female skew and limited ethnic diversity of the patients in our cohort,36 which limits generalizability to other populations. We have opened CHIVE and are beginning enrollment at other sites to address this directly. As a growing biorepository, our study currently lacks a sufficient sample size, which limits the ability to discern all but the strongest relationships within the data. For instance, some measures of cardiovascular and renal function demonstrate decreased function in patients with CH and will be expected to meet statistical significance with larger sampling. Enrollment in CHIVE is ongoing, and we anticipate that our ability to perform complex analyses will increase as we accumulate a more robust data set. The expansion of participant recruitment intra- and inter-institutionally will both increase sample size and further diversify our patient population.
In conclusion, we demonstrate the feasibility of a prospective, observational study of patients with CH using a robust referral network to support both clinical care and translational research. A well-genotyped and -phenotyped cohort of patients with CH will be critical to future translational research efforts and clinical trials, a key function of our study design. Early clinical findings from our cohort recapitulate large-scale retrospective data sets, in which patients with CH are at an increased risk of developing hematologic malignancy, end organ damage, and all-cause mortality. Scaling this resource in collaboration with other centers is underway and will enable the development of clinical guidelines and treatment strategies for this increasingly recognized patient population.
Authorship
Contribution: M.L.S., J.B.H., and S.O. collected and analyzed data, recruited patients, drafted the original manuscript, and revised the manuscript; A.D., Z.C., C.S., A.J.S., D.B., T.S., M.B., K.P., A.K., S.S.S., and J.U. collected data, recruited patients, and revised the manuscript; C.V. collected and analyzed data and revised the manuscript; Y.X., M.B., J.M., E.J., P.B.F., and D.S. conceived the study, collected and analyzed data, and revised the manuscript; and A.G.B. and M.R.S. conceived of the study, supervised the study, and revised the manuscript.
Conflict-of-interest disclosure: M.R.S. has served on advisory boards for AbbVie, Bristol Myers Squibb, CTI, Geron, GlaxoSmithKline, Novartis, Rigel, and Treadwell; received personal fees and other support from Geron; and reports personal fees and equity from Empath Biosciences, Karyopharm and Ryvu, outside of the submitted work. J.M. has served on advisory boards for Bristol Myers Squibb, AstraZeneca, Myovant, Cytokinetics, Takeda, BeiGene, Kiniksa, Kurome Therapeutics, BitterRoot Bio, Deciphera, Regeneron, Repare Therapeutics, Antev, Daiichi Sankyo, Prelude Therapeutics, and Voyager Therapeutics, outside of the submitted work. P.B.F. reports grants from Incyte, outside of the submitted work. A.G.B. is a scientific cofounder and has equity in TenSixteen Bio, outside of the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Michael R. Savona, Vanderbilt-Ingram Cancer Center, Vanderbilt University School of Medicine, 2200 Pierce Ave, Preston Resea, Nashville, TN 37232; email: michael.savona@vanderbilt.edu.
References
Author notes
M.L.S., J.B.H., and S.O. contributed equally to this study.
Deidentified data set will be made available upon publication. Access to data can be made at https://doi.org/10.5281/zenodo.10413401.
The full-text version of this article contains a data supplement.