tCMML has lower incidence of high-risk genomic features characteristic of other therapy-related myeloid neoplasms.
tCMML does not exhibit the poor outcomes of therapy-related myeloid neoplasms.
Visual Abstract
Therapy-related myeloid neoplasms (t-MNs) arise after exposure to cytotoxic therapies and are associated with high-risk genetic features and poor outcomes. We analyzed a cohort of patients with therapy-related chronic myelomonocytic leukemia (tCMML; n = 71) and compared its features to that of de novo CMML (dnCMML; n = 461). Median time from cytotoxic therapy to tCMML diagnosis was 6.5 years. Compared with dnCMML, chromosome-7 abnormalities (4% vs 13%; P = .005) but not complex karyotype (3% vs 7%; P = .15), were more frequent in tCMML. tCMML was characterized by higher TP53 mutation frequency (4% vs 12%; P = .04) and lower NRAS (6% vs 22%, P = .007) and CBL (4% vs 12%, P = .04) mutation frequency. Prior therapy with antimetabolites (odd ratio [OR], 1.22; 95% confidence interval [CI], 1.05-1.42; P = .01) and mitotic inhibitors (OR, 1.24; 95% CI, 1.06-1.44; P = .009) was associated with NF1 and SETBP1 mutations whereas prior mitotic inhibitor therapy was associated with lower TET2 mutation frequency (OR, 0.71; 95% CI, 0.55-0.92; P = .01). Although no differences in median overall survival (OS) were observed among tCMML and dnCMML (34.7 months vs 35.9 months, P = .26), multivariate analysis for OS revealed that prior chemotherapy was associated with increased risk of death (hazard ratio, 1.76; 95% CI, 1.07-2.89; P = .026). Compared with a cohort of therapy-related myelodysplastic syndrome, tCMML had lower TP53 mutation frequency (12% vs 44.4%, P < .001) and less unfavorable outcomes. In summary, tCMML does not exhibit the high-risk features and poor outcomes of t-MNs.
Introduction
Therapy-related myeloid neoplasms (t-MNs) are defined by the World Health Organization (WHO) 2016 classification as acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), and myelodysplastic/myeloproliferative neoplasms that occur as late complications of cytotoxic chemotherapy and/or radiotherapy.1 More recently, the WHO 2022 classification modified the definition to MNs after cytotoxic therapy but essentially maintained the original criteria.2 The incidence rate for t-MNs is ∼10% to 20% of all MNs and is expected to increase in parallel with the increase in the number of cancer survivors.3-5 In the past, t-MNs were proposed to be direct consequences of induction of DNA damage in hematopoietic progenitors by cytotoxic therapy. However, ensuing studies have demonstrated that t-MNs result from a combination of factors that include genetic damage to the hematopoietic precursor, inherited predisposition to genetic damage, clonal selection of previously mutated precursors, and bone marrow microenvironment disruption.3,6,7 Furthermore, t-MNs are associated with high-risk genetic features (eg, complex karyotype and TP53 mutations) and have dismal outcomes.8-11
Chronic myelomonocytic leukemia (CMML) is a hematopoietic stem cell neoplasm characterized by sustained monocytosis together with myelodysplastic and myeloproliferative features.1,2 Compared with AML and MDS, cytogenetic abnormalities are not as frequent in CMML cases (∼30%).12,13 Also, CMML has a unique genetic landscape, with TET2, SRSF2, and ASXL1 being the most commonly mutated genes.14,15 Despite increasing knowledge about the biology of CMML, effective therapies are limited, and patients are at risk of transformation to AML, which is associated with poor survival.14 Although prior studies suggest that therapy-related CMML (tCMML) accounts for 9% to 11% of all CMMLs,16-18 data on this entity remain scarce owing to its rarity and lack of specific reporting. In addition, although some studies have demonstrated increased incidence of high-risk genetic features in tCMML cases, data remain contradictory and few studies performed detailed mutational profile analysis. Therefore, there is a need to evaluate the clinical and genetic features of tCMML in large cohorts of patients.
To study further the natural history and features of tCMML, we evaluated the largest cohort of such patients reported to date, and compared their baseline cytogenetic and molecular characteristics with those of de novo CMML (dnCMML). We also describe the results of a comprehensive survival analysis of patients with tCMML compared with patients with dnCMML and other t-MNs.
Methods
Study design and patient inclusion
This was a single-center retrospective analysis of all patients diagnosed with CMML from 2005 to 2022 at The University of Texas MD Anderson Cancer Center. Patients were diagnosed with CMML according to the 2016 WHO classification criteria.1 Patients with tCMML were defined as those with a documented history of exposure to cytotoxic therapy (chemotherapy and radiotherapy). Types of cytotoxic therapy were grouped by mechanism of action (supplemental Table 1). Responses to treatment were assessed using the 2006 modified International Working Group response criteria for MDS.19 An MD Anderson database of patients diagnosed with MDS spanning 2017 to 2022 was used for comparison with the patients with tCMML. This study was approved by the MD Anderson institutional review board and was conducted in accordance with the Declaration of Helsinki.
Genetic assessment
Cytogenetic analysis was performed using conventional chromosome banding and, in some cases, fluorescence in situ hybridization. Mutational analysis was performed using 28- and 81-gene next-generation sequencing (NGS) panels. The NGS gene coverage for these panels is described in supplemental Tables 2 and 3. TP53 multihit status was defined as the presence of ≥2 TP53 mutations, 1 TP53 mutation with a variant allele frequency (VAF) of >50%, or 1 TP53 mutation with a deletion of chromosome 17/17p.
Statistical methods
Baseline patient characteristics were analyzed using descriptive statistics. The Student t test and Mann-Whitney U test were used for comparison of continuous variables with normal and nonnormal distribution, respectively. The χ2 and Fisher exact test were used to compare categorical variables. Logistic regression was used to test for associations between exposure to therapy and tCMML characteristics. Median follow-up times were calculated using the Kaplan-Meier estimate of potential follow-up.20 Overall survival (OS) was calculated from diagnosis to death. Leukemia-free survival (LFS) was calculated from diagnosis to AML transformation or death. OS and LFS distributions were estimated using the Kaplan-Meier method and compared using a log-rank test. Cumulative incidence analyses were performed using death as the primary event and AML transformation as a competing event. Comparison of cumulative incidence was performed using the Gray test. Univariate and multivariate analyses were performed using Cox proportional hazards regression. Cox proportional hazard assumption was evaluated with the Schoenfeld residuals. All statistical analyses were performed using the R computing language (version 4.2.2).
Results
Baseline characteristics
We included a total of 532 patients with CMML in this study, 71 (13%) of whom had tCMML. Baseline patient characteristics are listed in Table 1. The median age was 70 years (range, 25-94) for the entire cohort, 70 years (range, 25-94 years) for patients with dnCMML, and 73 years (range, 30-89) for those with tCMML (P = .054). Of 532 patients, 358 patients (67%) were male. The median white blood cell count was 12.6 × 109/L (range, 2.4 × 109/L to 214.0 × 109/L) overall, 13.3 × 109/L (range, 2.6 × 109/L to 214.0 × 109/L) in patients with dnCMML, and 10.7 × 109/L (range, 2.40 × 109/L to 132.90 × 109/L) in those with tCMML (P = .03). According to the French-American-British classification,21 228 (43%) patients were classified as having dysplastic CMML (n = 193 [43%] and n = 35 [49%], for dnCMML and tCMML, respectively), and 299 (56%) were classified as having proliferative CMML (n = 263 [57%] and n = 36 [41%], for dnCMML and tCMML, respectively; P = .33). According to the WHO 2016 classification,1 206 patients (39%) were classified as having CMML-0 (n = 176 [38%] and n = 30 [42%], for dnCMML and tCMML, respectively), 205 (39%) were classified as having CMML-1 (n = 179 [39%] and n = 26 [36%], for dnCMML and tCMML, respectively), and 116 (22%) were classified as having CMML-2 (n = 101 [22%] and n = 15 [21%], for dnCMML and tCMML, respectively; P = .839). According to the WHO 2022 classification,2 401 patients (76%) were classified as having CMML-1 (n = 346 [75%] and n = 55 [77%], for dnCMML and tCMML, respectively), with the same proportion of patients in the CMML-2 subgroup of the previous WHO classification.
Baseline characteristics of the study patients
| Characteristic . | All patients (N = 532) . | Patients with dnCMML (n = 461) . | Patients with tCMML (n = 71) . | P value . |
|---|---|---|---|---|
| Median age, y (range) | 70 (25-94) | 70 (25-94) | 73 (30-89) | .055 |
| Male sex, n (%) | 358 (67) | 309 (67) | 49 (69) | .844 |
| Median hemoglobin level, g/dL (range) | 10.8 (5.3-17.3) | 10.8 (5.3-17.3) | 10.9 (6.4-15.6) | .530 |
| Median WBC count, ×109 cells/L (range) | 12.6 (2.4-214.0) | 13.3 (2.6-214.0) | 10.7 (2.4-132.9) | .030 |
| Median absolute neutrophil count, ×109 cells/L (range) | 6.5 (0-108.0) | 6.8 (0-108.0) | 5.3 (0.5-50.6) | .100 |
| Median absolute monocyte count, ×109 cells/L (range) | 2.4 (1-74.7) | 2.4 (1-74.7) | 1.9 (1-73.1) | .698 |
| Median platelet count, ×109 cells/L (range) | 101 (7-1386) | 101 (7-1386) | 115 (16-638) | .629 |
| Median bone marrow blast percentage (range) | 6 (0-18) | 6 (0-18) | 5 (1-18) | .879 |
| WHO 2016 classification, n (%) | ||||
| CMML-0 | 206 (39) | 176 (38) | 30 (42) | .840 |
| CMML-1 | 205 (39) | 179 (39) | 26 (37) | |
| CMML-2 | 116 (22) | 101 (22) | 15 (21) | |
| Not classifiable∗ | 5 (1) | 5 (1) | 0 | |
| FAB CMML classification, n (%) | ||||
| Dysplastic | 228 (43) | 193 (42) | 35 (49) | .330 |
| Proliferative | 299 (56) | 263 (57) | 36 (50) | |
| Not classifiable∗ | 5 (1) | 20 (1) | 0 | |
| Cytogenetics, n (%) | ||||
| Diploid | 334 (63) | 293 (64) | 41 (58) | .235 |
| Chr 5/5q abnormality | 12 (2) | 9 (2) | 3 (4) | .464 |
| Chr 7 abnormality | 27 (5) | 18 (4) | 9 (13) | .005 |
| Trisomy 8 | 40 (8) | 36 (8) | 4 (6) | .642 |
| del(13q) | 9 (2) | 7 (2) | 2 (3) | .794 |
| del(20q) | 19 (4) | 16 (3) | 3 (4) | .999 |
| Complex | 18 (3) | 13 (3) | 5 (7) | .154 |
| CPSS CG classification, n (%) | ||||
| Favorable | 328 (62) | 284 (62) | 44 (62) | .917 |
| Intermediate | 89 (17) | 77 (17) | 12 (17) | |
| Adverse | 79 (15) | 67 (15) | 12 (17) | |
| Not classifiable∗ | 36 (6) | 33 (6) | 3 (4) | |
| CPSS-Mol risk group, n (%) | ||||
| Low | 26 (5) | 24 (5) | 2 (3) | .792 |
| Intermediate-1 | 70 (13) | 59 (13) | 11 (15) | |
| Intermediate-2 | 150 (28) | 129 (28) | 21 (30) | |
| High | 113 (21) | 97 (21) | 16 (23) | |
| Not classifiable∗ | 173 (33) | 153 (33) | 21 (29) | |
| Characteristic . | All patients (N = 532) . | Patients with dnCMML (n = 461) . | Patients with tCMML (n = 71) . | P value . |
|---|---|---|---|---|
| Median age, y (range) | 70 (25-94) | 70 (25-94) | 73 (30-89) | .055 |
| Male sex, n (%) | 358 (67) | 309 (67) | 49 (69) | .844 |
| Median hemoglobin level, g/dL (range) | 10.8 (5.3-17.3) | 10.8 (5.3-17.3) | 10.9 (6.4-15.6) | .530 |
| Median WBC count, ×109 cells/L (range) | 12.6 (2.4-214.0) | 13.3 (2.6-214.0) | 10.7 (2.4-132.9) | .030 |
| Median absolute neutrophil count, ×109 cells/L (range) | 6.5 (0-108.0) | 6.8 (0-108.0) | 5.3 (0.5-50.6) | .100 |
| Median absolute monocyte count, ×109 cells/L (range) | 2.4 (1-74.7) | 2.4 (1-74.7) | 1.9 (1-73.1) | .698 |
| Median platelet count, ×109 cells/L (range) | 101 (7-1386) | 101 (7-1386) | 115 (16-638) | .629 |
| Median bone marrow blast percentage (range) | 6 (0-18) | 6 (0-18) | 5 (1-18) | .879 |
| WHO 2016 classification, n (%) | ||||
| CMML-0 | 206 (39) | 176 (38) | 30 (42) | .840 |
| CMML-1 | 205 (39) | 179 (39) | 26 (37) | |
| CMML-2 | 116 (22) | 101 (22) | 15 (21) | |
| Not classifiable∗ | 5 (1) | 5 (1) | 0 | |
| FAB CMML classification, n (%) | ||||
| Dysplastic | 228 (43) | 193 (42) | 35 (49) | .330 |
| Proliferative | 299 (56) | 263 (57) | 36 (50) | |
| Not classifiable∗ | 5 (1) | 20 (1) | 0 | |
| Cytogenetics, n (%) | ||||
| Diploid | 334 (63) | 293 (64) | 41 (58) | .235 |
| Chr 5/5q abnormality | 12 (2) | 9 (2) | 3 (4) | .464 |
| Chr 7 abnormality | 27 (5) | 18 (4) | 9 (13) | .005 |
| Trisomy 8 | 40 (8) | 36 (8) | 4 (6) | .642 |
| del(13q) | 9 (2) | 7 (2) | 2 (3) | .794 |
| del(20q) | 19 (4) | 16 (3) | 3 (4) | .999 |
| Complex | 18 (3) | 13 (3) | 5 (7) | .154 |
| CPSS CG classification, n (%) | ||||
| Favorable | 328 (62) | 284 (62) | 44 (62) | .917 |
| Intermediate | 89 (17) | 77 (17) | 12 (17) | |
| Adverse | 79 (15) | 67 (15) | 12 (17) | |
| Not classifiable∗ | 36 (6) | 33 (6) | 3 (4) | |
| CPSS-Mol risk group, n (%) | ||||
| Low | 26 (5) | 24 (5) | 2 (3) | .792 |
| Intermediate-1 | 70 (13) | 59 (13) | 11 (15) | |
| Intermediate-2 | 150 (28) | 129 (28) | 21 (30) | |
| High | 113 (21) | 97 (21) | 16 (23) | |
| Not classifiable∗ | 173 (33) | 153 (33) | 21 (29) | |
CG, cytogenetic; Chr, chromosome; FAB, French-American-British; WBC, white blood cell.
Patients not classifiable because of unavailable laboratory, cytogenetic, or genetic data.
The median time from cytotoxic therapy to tCMML diagnosis was 6.5 years (range, 0.5-24.0 years). At the moment of tCMML diagnosis, 11 patients (15.5%) had an active neoplasm. The most frequent neoplasms preceding tCMML diagnosis (Table 2) were prostate cancer (n = 18 [25%]), lymphoproliferative disease (n = 16 [23%]), breast cancer (n = 10 [14%]), head and neck cancer (n = 8 [11%]), and colorectal cancer (n = 8 [11%]). Patients received radiotherapy (n = 25 [35%]), chemotherapy (n = 22 [31%]), or a combination of radiotherapy and chemotherapy (n = 24 [34%]). Among those who received chemotherapy, 32 patients (45%) received alkylating agents, 26 (37%) received mitotic inhibitors, 25 (35%) received antimetabolite drugs, 17 (24%) received cytotoxic antibiotics, and 7 (10%) received type II topoisomerase inhibitors. We found no differences in the median time from cancer therapy to tCMML diagnosis when comparing different types of therapy.
Prior neoplasms and therapy in patients with tCMML
| Variable . | n (%) . |
|---|---|
| Median time from previous cancer to tCMML diagnosis, y (range) | 6.5 (0.1-24.4) |
| Prior neoplasms | |
| Prostate | 18 (25) |
| Lymphoid | 16 (23) |
| Breast | 10 (14) |
| Head and neck | 8 (11) |
| Colorectal | 8 (11) |
| Lung | 5 (7) |
| Ovarian | 3 (4) |
| Other | 7 (10) |
| Therapy | |
| RT only | 25 (35) |
| CT only | 22 (31) |
| CT and RT | 24 (34) |
| Type of chemotherapy | |
| Alkylating agents | 32 (45) |
| Mitotic inhibitors | 26 (37) |
| Antimetabolites | 25 (35) |
| Antitumor antibiotics | 17 (24) |
| Type II topoisomerase inhibitors | 7 (10) |
| Other | 2 (3) |
| Unknown | 8 (11) |
| Variable . | n (%) . |
|---|---|
| Median time from previous cancer to tCMML diagnosis, y (range) | 6.5 (0.1-24.4) |
| Prior neoplasms | |
| Prostate | 18 (25) |
| Lymphoid | 16 (23) |
| Breast | 10 (14) |
| Head and neck | 8 (11) |
| Colorectal | 8 (11) |
| Lung | 5 (7) |
| Ovarian | 3 (4) |
| Other | 7 (10) |
| Therapy | |
| RT only | 25 (35) |
| CT only | 22 (31) |
| CT and RT | 24 (34) |
| Type of chemotherapy | |
| Alkylating agents | 32 (45) |
| Mitotic inhibitors | 26 (37) |
| Antimetabolites | 25 (35) |
| Antitumor antibiotics | 17 (24) |
| Type II topoisomerase inhibitors | 7 (10) |
| Other | 2 (3) |
| Unknown | 8 (11) |
Data from n = 71 patients.
CT, chemotherapy; RT, radiotherapy.
Cytogenetic and molecular findings
At diagnosis, 334 patients (63%) had normal karyotype (n = 293 [64%] and n = 41 [58%] in patients with dnCMML and tCMML, respectively; P = .23]). Chromosome-7 abnormalities occurred in 27 patients (5%) and were more frequent in patients with tCMML (n = 9 [13%]) than in those with dnCMML (n = 18 [4%]; P = .005). Complex karyotype, defined as ≥3 chromosomal abnormalities, was more frequent in patients with tCMML (n = 5 [7%]) than in those with dnCMML (n = 13 [3%]), although the difference was not significant (P = .15). According to the CMML prognostic scoring system (CPSS) cytogenetic risk classification,13 328 patients (62%) were in the low-risk group (n = 284 [62%] and n = 44 [62%] in patients with dnCMML and tCMML, respectively), 89 patients (17%) were in the intermediate-risk group (n = 77 [17%] and n = 12 [17%] in patients with dnCMML and tCMML, respectively), and 79 patients (15%) were in the high-risk group (n = 67 [15%] and n = 12 [17%] in patients with dnCMML and tCMML, respectively; P = .98]).
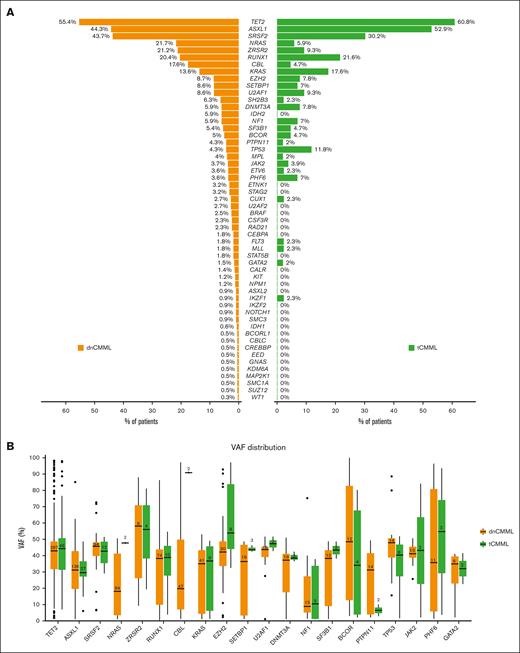
Among all patients, 374 (70%) had available NGS testing data (51 patients with tCMML [72%], and 323 with dnCMML [70%]) obtained by evaluating a panel of 28 or 81 genes in 109 (29%) and 265 (71%) patients, respectively. The most commonly observed mutations included TET2 (n = 210 [56%]), ASXL1 (n = 170 [45%]), SRSF2 (n = 109 [29%]), and RUNX1 (n = 77 [21%]), with no significant differences in the incidence of these mutations between dnCMML and tCMML. Frequencies and VAFs of identified mutations are shown in Figure 1. Compared with patients with dnCMML, those with tCMML had a lower incidence of NRAS mutations (n = 70 [22%] vs n = 3 [6%]; P = .007) and CBL mutations (n = 39 [12%] vs n = 2 [4%]; P = .04) and a higher incidence of TP53 mutations (n = 14 [4%] vs n = 6 [12%]; P = .04). TP53 multihit status was present in 7 patients with dnCMML (50% of patients with dnCMML with TP53 mutations) and 4 patients with tCMML (67% of with tCMML with TP53 mutations; P = .84). When comparing VAF differences between patients with dnCMML and those with tCMML, those with tCMML had a higher median VAF for CBL (91% vs 19%; P = .01) and NRAS (48% vs 18%; P = .04) mutations.
Mutations in patients with dnCMML and tCMML. (A) Frequency of each mutation in dnCMML and tCMML. (B) VAF for each of the most frequently mutated genes in dnCMML and tCMML.
Mutations in patients with dnCMML and tCMML. (A) Frequency of each mutation in dnCMML and tCMML. (B) VAF for each of the most frequently mutated genes in dnCMML and tCMML.
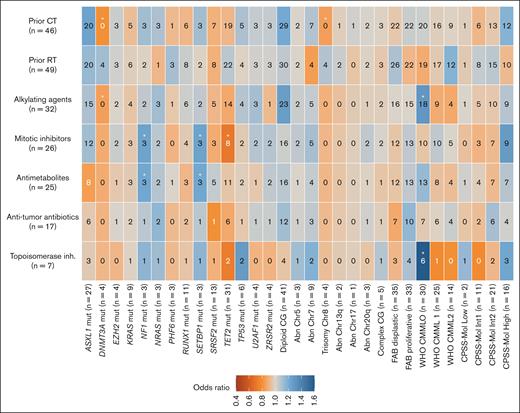
We also did an exploratory analysis of the association between prior types of treatment received for the antecedent malignancy and biological variables at tCMML diagnosis (Figure 2). No patients with trisomy of chromosome 8 (odds ratio [OR], 0.85; 95% confidence interval [CI], 0.75-0.95; P = .006) or DNMT3A mutation (OR, 0.80 [95% CI, 0.69-0.93]; P = .004) received prior chemotherapy. Patients who received alkylating agents or type II topoisomerase inhibitors were associated with tCMML classified as CMML-0 according to the WHO classification (OR, 1.30 [95% CI, 1.04-1.64]; P = .03, for alkylating agents; OR, 1.60 [95% CI, 1.10-2.33]; P = .02, for type II topoisomerase inhibitors). All patients with NF1 or SETBP1 mutations had received antimetabolites and mitotic inhibitors (OR, 1.22 [95% CI, 1.05-1.42]; P = .01, for antimetabolites; OR, 1.24 [95% CI, 1.06-1.44]; P = .009, for mitotic inhibitors). Also, patients who received mitotic inhibitors were associated with a lower frequency of TET2 mutations (OR, 0.71 [95% CI, 0.55-0.92]; P = .01).
Plot of the association between cytotoxic therapies and baseline and biological characteristics of patients with tCMML. The numbers in the cells represent the absolute numbers of patients with each association. The color gradient represents the OR. Cells with an asterisk represent significant associations (unadjusted P value < .05).
Plot of the association between cytotoxic therapies and baseline and biological characteristics of patients with tCMML. The numbers in the cells represent the absolute numbers of patients with each association. The color gradient represents the OR. Cells with an asterisk represent significant associations (unadjusted P value < .05).
Among 359 patients in whom molecular-CPSS (CPSS-Mol) scores were evaluable, 26 (5%) were classified as being at low risk (n = 24 [5%] vs n = 2 [3%] for patients with dnCMML and tCMML, respectively), 70 (13%) as being at intermediate-1 risk (n = 59 [13%] vs n = 11 [15%] for patients with dnCMML and tCMML, respectively), 150 (28%) as being at intermediate-2 risk (n = 129 [28%] vs n = 21 [30%] for patients with dnCMML and tCMML, respectively), and 113 (21%) as being at high risk (n = 97 [21%] vs n = 16 [23%] for patients with dnCMML and tCMML, respectively; P = .79). An additional classification using the Molecular International Prognostic Score System for MDS22 is available in supplemental Material (supplemental Analysis 1).
Treatment responses and survival analysis
Among all patients in the cohort, 351 (66%) received treatment for CMML. Hypomethylating agents were most frequently used (n = 328 [93%]), followed by intensive chemotherapy (n = 10 [3%]) and low-dose chemotherapy (n = 5 [1%]). Among patients treated with hypomethylating agents, the overall response rate (complete remission or marrow complete remission) was 59% (195 of 328), with no differences in rate between patients with dnCMML (167 of 281 [59%]) and those with tCMML (28 of 47 [60%]; P = 1). Overall, 6 patients with tCMML (7.4%) and 72 patients with dnCMML (15.6%) underwent allogeneic stem cell transplantation (P = .12).
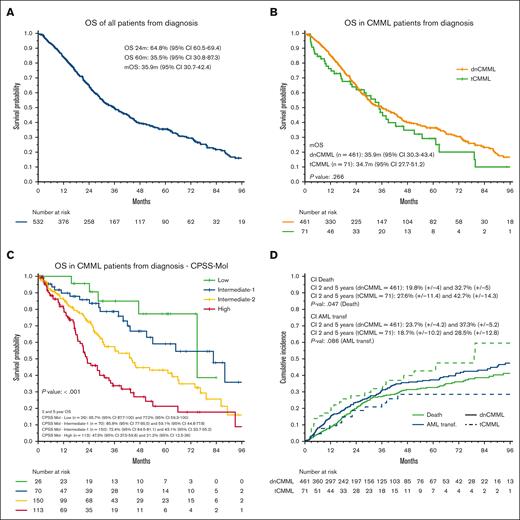
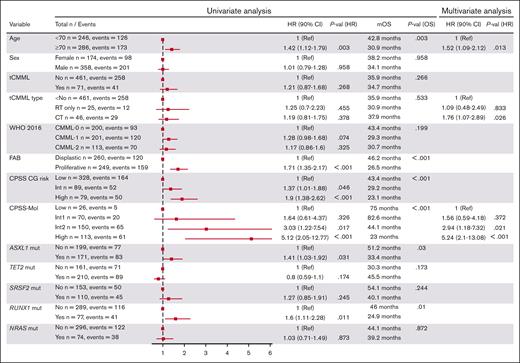
The median follow-up time was 58.3 months (95% CI, 49.5-62.8). The median OS (mOS) and median LFS times were 35.9 months (95% CI, 30.7-42.4) and 28.6 months (95% CI, 25.0-34.1), respectively (Figure 3). The 2-year OS and LFS rates were 64.8% and 56.1%, respectively. The mOS and median LFS times were 35.9 months and 28.9 months, respectively, in patients with dnCMML, and 34.7 months and 28.2 months, respectively, in patients with tCMML (P = .26 for OS, P = .8 for LFS). We found no OS differences between patients with tCMML who had previously received only radiotherapy and those who had received chemotherapy for their antecedent malignancy (mOS time, 37.9 months vs 30.9 months; P = .811). The mOS times were 75.0, 82.6, 44.1, and 22.9 months for patients in the low, intermediate-1, intermediate-2, and high CPSS-Mol risk groups, respectively (P < .001). The mOS time was 75.0 months (dnCMML) vs not reached (tCMML; P = .605), 88.0 months vs 37.9 months (P = .016), 44.1 months vs 36.3 months (P = .515), and 23.1 months vs 21.4 months (P = .399) for patients in the low, intermediate-1, intermediate-2, and high CPSS-Mol risk groups, respectively. Additional OS and LFS analysis results are detailed in supplemental Figure 2 and 3. Results of a univariate analysis for OS for all patients with CMML are detailed in Figure 4. Multivariate analysis for survival identified age of >70 years (hazard ratio [HR], 1.52 [95% CI, 1.09-2.12]; P = .013), previous chemotherapy for tCMML (HR, 1.76 [95% CI, 1.07-2.89]; P = .026), intermediate-2 CPSS-Mol risk group (HR, 2.94 [95% CI, 1.18-7.32]; P = .021), and high CPSS-Mol risk group (HR, 5.24 [95% CI, 2.10-13.10]; P < .001) as independent risk factors for OS. Univariate analysis for patients with tCMML is detailed in the supplemental Table 4.
Survival analysis of patients with tCMML. (A) OS of all patients with CMML. (B) OS of patients with dnCMML and tCMML. (C) OS of all CMML by CPSS-Mol risk classification. (D) Cumulative incidence of relapse and AML transformation in patients with dnCMML and tCMML.
Survival analysis of patients with tCMML. (A) OS of all patients with CMML. (B) OS of patients with dnCMML and tCMML. (C) OS of all CMML by CPSS-Mol risk classification. (D) Cumulative incidence of relapse and AML transformation in patients with dnCMML and tCMML.
Results of univariate and multivariate analysis of patients with CMML.
The 2- and 5-year cumulative incidence rates for AML transformation were 23.0% and 36.1%, respectively. The 2- and 5-year cumulative incidence rates for death without AML transformation were 20.8% and 33.9%, respectively. When comparing patients with dnCMML and with patients with tCMML, the 2-year cumulative incidence rates for AML transformation were 23.7% and 18.7%, respectively (P = .086), and the 2-year cumulative incidence rates for death without transformation were 19.8% and 27.6%, respectively (P = .047). We also compared the cumulative incidence of AML transformation and death without transformation in each CPSS-Mol risk group (supplemental Figure 4). We found a significant difference in the 2-year cumulative incidence rate for death without transformation in patients with dnCMML (6.9%) and patients with tCMML (39.4%) in the intermediate-1 group (P = .002).
Therapy-related MDS vs tCMML
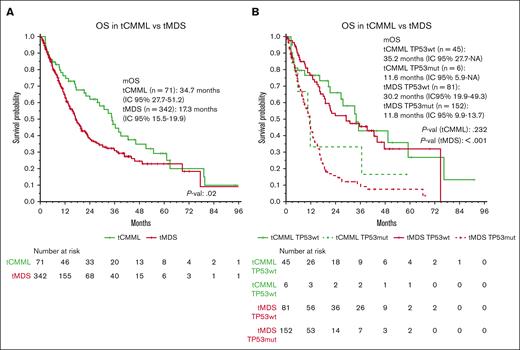
To determine whether the clinical outcomes of t-MNs are driven by underlying phenotypic and genomic features, we compared the characteristics of tCMML with those of a cohort of therapy-related MDS (tMDS). We analyzed 998 patients with MDS with a median follow-up of 23.9 months: 342 (34%) with tMDS and 656 (66%) with de novo MDS. Patients with tMDS had higher incidence rates for chromosome-7 abnormalities (30.1% vs 9.1%; P < .001), complex karyotype (39.8% vs 19.4%; P < .001), and TP53 mutations (44.4% vs 20.1%; P < .001), compared with those with de novo MDS. TP53 multihit status was present in 64% and 59% patients with TP53 mutations in the tMDS and de novo MDS groups, respectively (P = .49). When compared with tCMML, tMDS exhibited significantly higher incidence rates of complex karyotype (39.8% vs 7%, P < .001) and TP53 mutations (44.4% vs 12%, P < .001). Normal karyotype was less common in patients with tMDS than in those with de novo MDS (17.3% vs 42.8%; P < .001). The mOS time for tMDS was 17.3 months (vs 34.7 for tCMML; P = .02). When stratifying patients by disease and TP53 status, patients with tCMML and tMDS with TP53 mutations had worse mOS times (11.6 months and 11.8 months, respectively) than did patients with tCMML and tMDS without TP53 mutations (35.2 months and 30.2 months, respectively; P = .2 [tCMML] and P < .001 [tMDS]; Figure 5; supplemental Figure 5).A simulation using the new WHO 20222 and International Consensus Classification23 diagnostic criteria is detailed in the supplemental Material (supplemental Analysis 2).
Survival analysis comparing tCMML and tMDS. (A) OS of patients with tCMML and tMDS. (B) OS of patients with tCMML and tMDS according to TP53 mutation status.
Survival analysis comparing tCMML and tMDS. (A) OS of patients with tCMML and tMDS. (B) OS of patients with tCMML and tMDS according to TP53 mutation status.
Discussion
In this study, to our knowledge, we performed a comprehensive analysis of the largest published cohort of patients with tCMML to date. The incidence rate for tCMML was 13%, which is consistent with previous reports of incidence rates of 9% to 11%.16-18 Also, the median time from exposure to therapy to diagnosis of 6.5 years is consistent with previous tCMML reports and similar to reported times from cytotoxic drug exposure to AML and MDS diagnosis (5-7 years after alkylating agent use and/or radiotherapy).9,24 Authors have described reduced time from therapy to t-MN development (2-3 years) and increased risk of balanced translocations in patients receiving type II topoisomerase inhibitors.25 We did not find this phenomenon in this study, although most patients received these drugs in combination with other cytotoxic agents. Moreover, balanced translocations are rare in CMML cases, suggesting that they are specific to the development of therapy-related AML. The neoplasms most frequently preceding tCMML were prostate, lymphoid, and breast tumors, which coincides with the most frequent prior tumors in patients with t-MNs.3 This is likely a result of the wide use of chemotherapeutic agents in the treatment of these neoplasms, together with increased long-term survival of patients with cancer, which increases the chances of late development of t-MN.
The baseline characteristics of our patients with dnCMML and tCMML were similar, with no significant differences according to CMML subtype (dysplastic vs proliferative) or blast percentage. When analyzing cytogenetic characteristics, only chromosome-7 abnormalities were markedly more common in patients with tCMML than in those with dnCMML (3.9% vs 12.7%); the incidence of chromosome-5 abnormalities (4.2% in tCMML) and complex karyotype (7% in tCMML) did not differ. Accordingly, we saw no differences in cytogenetic risk according to CPSS classification. In contrast, t-MN cases have a high incidence of chromosome-5, -7, and -17 abnormalities as well as complex karyotype (40%-50% of all t-MNs).3 Moreover, Patnaik et al16 found a significantly increased incidence of complex karyotype in tCMML cases (12%), but, again, this was far below the incidence in cases of other t-MNs. Complex karyotype usually occurs in the context of TP53 mutations, likely as a result of inactivation of this gene, which drives cells into a highly unstable genome.26 In our study, the overall incidence of TP53 mutations was 5.5%, which is higher than the 2.4% reported in a recent publication, likely because a higher incidence of tCMML in our cohort together with a referral bias given the study was performed in an academic referral cancer center.27 Specifically, the incidence of TP53 mutations in patients with tCMML was 11.8%, which was considerably higher than that in patients with dnCMML (4.3%) but again below the reported incidence rates for patients with other t-MNs, which range from 23% to 37%.3 Consistent with these data, our cohort of patients with tMDS revealed TP53 mutation and complex karyotype incidence rates of 44.4% and 39.8%, respectively. This low incidence of TP53 mutations and complex karyotype suggests that the leukemogenic mechanisms in the development of tCMML differ from those reported for other t-MNs. Another hypothesis is that in some of these tCMML cases, previous exposure to therapy may not have been directly involved in the development or progression of the disease, thus behaving as dnCMML. Although this hypothesis is supported by our observation that tCMML does not have significantly worse survival than dnCMML unless corrected for other high-risk biologic features, it requires further experimental validation. The WHO definition of t-MNs is very broad because it includes all MNs occurring after exposure to cytotoxic therapy without requiring a proven causality. With better knowledge of the specific leukemogenic mechanisms of cytotoxic therapy, future classifications may refine the definition of t-MNs. We observed a low incidence of NRAS and CBL mutations with a high VAF in individuals with tCMML. However, it is important to interpret this finding cautiously given the limited number of patients.
In our study, outcomes of tCMML were not distinct from those of dnCMML. The response rates for hypomethylating agent–based therapy in the 2 groups were similar, and the mOS times were 35.9 months and 34.7 months for dnCMML and tCMML, respectively. This differs from previously published outcomes in patients with t-MN, including a mOS time of <1 year.8,11 Also, when specifically comparing tCMML and tMDS, the mOS time was substantially lower in the latter (17.3 months). As discussed above, the incidence of TP53 mutations and complex karyotype is low in patients with tCMML, which is associated with treatment resistance and poor outcomes. This may be the main reason why tCMML does not have more dismal outcomes than other t-MNs. A hypothesis is that exposure to cytotoxic therapy selects resistant hematopoietic stem cell subclones, likely with TP53 mutations, that are more prone to later develop tMDS or therapy-related AML instead of tCMML. Clonal selection of TP53 subclones induced by treatments has been described in AML and MDS.28,29 This could mean that perhaps the majority of tCMMLs are not truly therapy related and that the exposure to these therapies did not induce clonal selection leading to disease evolution. A deeper survival analysis demonstrated that OS was significantly reduced in patients with tCMML in the intermediate-1 CPSS-Mol risk group, with an increased cumulative incidence of death but not of AML transformation. Indeed, tCMML patients who received previous chemotherapy had an increased risk of death in the multivariate analysis (HR, 1.76). Moreover, the cumulative incidence of AML transformation was lower in patients with tCMML, possibly attributed to a higher risk of nontransformation death, which was assessed as a competing event. This increased risk of death in this non–high-risk group of patients with tCMML may result from potential comorbidities related to a previous neoplasm or an active neoplasm co-occurring with tCMML.
Exposure to specific antineoplastic agents is associated to recurrent genetic abnormalities in patients with t-MNs. Indeed, the most known associations are exposure to type II topoisomerase inhibitors with 11q23 translocations, and exposure to alkylating agents with chromosome-5 and -7 abnormalities.30,31 Guerra et al analyzed a large cohort of patients with t-MN to look for associations between chemotherapy exposure and specific mutations and found some interesting associations between EZH2 mutation and vinca alkaloid use as well as TP53 mutation with immunomodulatory imide drug use.29 In this study, we conducted a hypothesis-generating analysis to look for associations between therapies received and tCMML characteristics. We found some interesting preliminary associations, such as a low incidence of TET2 mutations in patients exposed to mitotic inhibitors, which was described by Sperling et al specifically in the context of vinca alkaloids. However, our analysis was exploratory using a small cohort of patients with tCMML and therefore it is premature to make assumptions based on the associations found. The associations and the potential mechanisms by different chemotherapeutic agents to induce specific genetic abnormalities need to be confirmed in the context of tCMML.
A limitation of this study is that it was a retrospective analysis of patients given treatment over a wide interval of time and in a single institution. Moreover, such patients commonly receive multiple combinations of chemotherapy and radiotherapy for their neoplasms, some of whom can receive multiple lines if their disease relapses. This treatment heterogenicity makes it difficult to establish associations between specific exposures and t-MN characteristics.
In conclusion, tCMML differs biologically from other t-MNs, with fewer high-risk features such as complex karyotype and TP53 mutations. These variations may stem from unique leukemogenic mechanisms in tCMML when compared with those of tMDS or therapy-related AML. Therefore, the outcomes of tCMML are not as dismal as those of other t-MNs. Our findings support the eligibility of patients with tCMML to be potential candidates for clinical trials, given the clinical and biological similarity to dnCMML. Related to that, a definition based on clinical and genetic characteristics in t-MNs would be beneficial, encompassing previous exposure to cytotoxic agents together with specific genetic abnormalities implicated in their leukemogenic mechanisms, such as TP53 mutations or chromosome abnormalities.
Authorship
Contribution: A. Bataller was responsible for study conception, data curation, formal data analysis and interpretation, patient care, and writing, review, and editing of the manuscript; G.G.-R., S.U., E.A.-H., K.S.C., and A. Bazinet were responsible for data curation, patient care, and review and editing of the manuscript; J.-J.R.-S. and D.H. were responsible for data curation, and review and editing of the manuscript; K.S. was responsible for data curation, patient care, and review and editing of the manuscript; K.T., C.D.D., F.R., G.B., and T.M.K. were responsible for patient care and review and editing of the manuscript; R.K.-S. was responsible for diagnostic analysis and molecular interpretation, and review and editing of the manuscript; H.M.K. was responsible for patient care and review and editing of the manuscript; G.G.-M. was responsible for study conception, patient care, writing of the manuscript, and review and editing of the manuscript; and G.M.-B. was responsible for study conception, patient care, data interpretation, writing of the manuscript, and review and editing of the manuscript.
Conflict-of-interest disclosure: K.T. has been a consultant for SymBio Pharmaceuticals and received honoraria from Mission Bio, Illumina, and Otsuka Pharmaceutical. C.D.D. has been a board of directors or advisory committee member for Genmab, GlaxoSmithKline, Kura Oncology, and Notable Labs; has received honoraria from Kura, Astellas Pharma, bluebird bio, Bristol Myers Squibb, Foghorn Therapeutics, Immune-Onc Therapeutics, Novartis, Takeda Oncology, Gilead Sciences, and Jazz Pharmaceuticals; is a current holder of stock options for Notable Labs; has been a consultant for AbbVie and Servier; and has received research funding from Servier, Bristol Myers Squibb, Foghorn, Immune-Onc Therapeutics, Loxo Oncology, Astex Pharmaceuticals, Cleave, and Forma. F.R. has received research funding from Amgen, Astex Pharmaceuticals/Taiho Oncology, Bristol Myers Squibb/Celgene, Syos, AbbVie, Prelude, Xencor, Astellas Pharma, and Biomea Fusion; has received honoraria from Amgen, Bristol Myers Squibb/Celgene, Syos, AbbVie, and Astellas Pharma; has been a board of directors or advisory committee member for Astex Pharmaceuticals/Taiho Oncology; and has been a consultant for Bristol Myers Squibb/Celgene, Syos, Novartis, AbbVie, AstraZeneca, and Astellas Pharma. G.B. has received research funding from Astex Pharmaceuticals, Ryvu Therapeutics, and PTC Therapeutics; has been a board of directors or advisory committee member for Pacyclex Pharmaceuticals, Novartis, CytomX, and Bio Ascend; and has been a consultant for Catamaran Bio, AbbVie, PPD Development, Protagonist Therapeutics, and Janssen. T.M.K. has been a consultant for AbbVie, Agios, Bristol Myers Squibb, Genentech, Jazz Pharmaceuticals, Novartis, Servier, and PinotBio; has received research funding from AbbVie, Bristol Myers Squibb, Genentech, Jazz Pharmaceuticals, Pfizer, Cellenkos, Ascentage Pharma, GenFleet Therapeutics, Astellas Pharma, AstraZeneca, Amgen, Cyclacel Pharmaceuticals, Delta-Fly Pharma, Iterion Therapeutics, GlycoMimetics, and Regeneron Pharmaceuticals; and has received honoraria from Astex Pharmaceuticals. H.M.K. has received research funding from AbbVie, Amgen, Ascentage Pharma, Bristol Myers Squibb, Daiichi Sankyo, ImmunoGen, Jazz Pharmaceuticals, and Novartis; and has received honoraria from AbbVie, Amgen, Amphista Therapeutics, Ascentage Pharma, Astellas Pharma, Biologix, Curis, Ipsen, KAHR, Novartis, Pfizer, Precision Biosciences, Shenzhen TargetRx, and Takeda Oncology. G.G.-M. has received research funding from Astex Pharmaceuticals, Novartis, AbbVie, Bristol Myers Squibb, Genentech, Aprea Therapeutics, Curis, and Gilead Sciences; has been a consultant for Astex Pharmaceuticals, Acceleron Pharma, and Bristol Myers Squibb; and has received honoraria from Astex Pharmaceuticals, Acceleron Pharma, AbbVie, Novartis, Gilead Sciences, Curis, Genentech, and Bristol Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Guillermo Montalban-Bravo, Department of Leukemia, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 428, Houston, TX 77030; email: gmontalban1@mdanderson.org.
References
Author notes
The study data (including genetic information) are not publicly available to respect patient confidentiality. Requests for sharing of deidentified data should be directed to the corresponding author, Guillermo Montalban-Bravo (gmontalban1@mdanderson.org).
The full-text version of this article contains a data supplement.