Cord blood–derived CBFA2T3::GLIS2 high-fidelity AMKL models contribute to capture the cellular and molecular heterogeneity of the disease.
BCL-XL inhibition is revealed as a therapeutic vulnerability in CG2 and NUP98r AMKL.
Visual Abstract
Acute megakaryoblastic leukemia (AMKL) is a rare, developmentally restricted, and highly lethal cancer of early childhood. The paucity and hypocellularity (due to myelofibrosis) of primary patient samples hamper the discovery of cell- and genotype-specific treatments. AMKL is driven by mutually exclusive chimeric fusion oncogenes in two-thirds of the cases, with CBFA2T3::GLIS2 (CG2) and NUP98 fusions (NUP98r) representing the highest-fatality subgroups. We established CD34+ cord blood–derived CG2 models (n = 6) that sustain serial transplantation and recapitulate human leukemia regarding immunophenotype, leukemia-initiating cell frequencies, comutational landscape, and gene expression signature, with distinct upregulation of the prosurvival factor B-cell lymphoma 2 (BCL2). Cell membrane proteomic analyses highlighted CG2 surface markers preferentially expressed on leukemic cells compared with CD34+ cells (eg, NCAM1 and CD151). AMKL differentiation block in the mega-erythroid progenitor space was confirmed by single-cell profiling. Although CG2 cells were rather resistant to BCL2 genetic knockdown or selective pharmacological inhibition with venetoclax, they were vulnerable to strategies that target the megakaryocytic prosurvival factor BCL-XL (BCL2L1), including in vitro and in vivo treatment with BCL2/BCL-XL/BCL-W inhibitor navitoclax and DT2216, a selective BCL-XL proteolysis-targeting chimera degrader developed to limit thrombocytopenia in patients. NUP98r AMKL were also sensitive to BCL-XL inhibition but not the NUP98r monocytic leukemia, pointing to a lineage-specific dependency. Navitoclax or DT2216 treatment in combination with low-dose cytarabine further reduced leukemic burden in mice. This work extends the cellular and molecular diversity set of human AMKL models and uncovers BCL-XL as a therapeutic vulnerability in CG2 and NUP98r AMKL.
Introduction
Acute megakaryoblastic leukemia (AMKL) is a developmentally restricted and early-onset cancer affecting children aged 0 to 4 years, including neonates,1-4 pointing to a stem or progenitor cell of origin that is only transiently present in time.5 It represents ∼10% of pediatric acute myeloid leukemia (AML) cases and is a molecularly heterogenous and highly lethal leukemia, with dismal cure rates (<50%).6 Two-thirds of AMKL cases are driven by mutually exclusive chimeric fusion oncogenes including RBM15::MKL1,7,8KMT2A translocations,9 and cytogenetically cryptic rearrangements involving either NUP9810,11 (NUP98r, specifically NUP98::KDM5A [N5A]12 and NUP98::BPTF [NTF]13), HOX genes,1,2,9 or the CBFA2T3::GLIS2 (CG2) fusion.14-18 The mapping of functional dependencies of de novo pediatric AMKL is emerging,19,20 and a repertoire of relevant preclinical models is needed to develop cell- and genotype-specific treatments for pediatric patients with leukemia.
CG2 and NUP98r AMKL are recognized as the highest relapse risk subgroups.6,16,21 The CG2 fusion results from a cryptic inversion on chromosome 16, joining the N-terminal nervy homology regions (protein-interacting domains) of corepressor CBFA2T3 (ETO2) to the C-terminal zinc finger domains of transcription factor GLIS2, which allows for binding to GLI DNA consensus sequences.14,15 CBFA2T3 is involved in hematopoietic stem cell maintenance and differentiation as well as in the regulation of erythro-megakaryocytic progenitors.22-24 GLIS2 was primarily identified in adult kidneys, maintaining normal tissue architecture and function.25 CG2-targeting treatments are actively explored, including Aurora A kinase inhibitors14,26; interference with CG2 fusion oligomerization27; and targeting of Neural Cell Adhesion Molecule 1 (NCAM1),17 the Folate Receptor Alpha (FOLR1),18,28 or the JAK-STAT pathway with ruxolitinib.12,29
Resisting cell death is a long-recognized hallmark of cancer,30 and agents that promote apoptosis entered the therapeutic arena for AML.31 The subsequent development of pharmacological inhibitors (BH3 mimetics) that target single or multiple prosurvival proteins32,33 has sparked significant interest in oncology because these agents can induce mitochondrial apoptosis in multiple hematological malignancies,34,35 with significant clinical responses,36 including in children.37,38 Evidence suggests that prosurvival factor dependency is both lineage-39-42 and mutation–specific43,44 in leukemia, and the integration of BH3 mimetics into current treatment protocols is under active investigation.
To define AMKL functional dependencies and overcome the paucity of pediatric patient samples, we have engineered CG2 AMKL models derived from cord blood (CB) hematopoietic stem and progenitor cells (HSPCs) that phenocopy the human disease. Across AMKL genotypes, CG2 leukemia distinctively upregulate the prosurvival factor B-cell lymphoma 2 (BCL2) yet are resistant to BCL2-specific inhibition with venetoclax or genetic knockdown (KD). In contrast, targeting the megakaryocytic lineage prosurvival factor BCL-XL45,46 with navitoclax47 (binding to BCL2, BCL-XL, and BCL-W) or BCL-XL proteasomal degrader DT221648 induced apoptosis of CG2 and NUP98r AMKL cells, with minimal cross toxicity on normal CB CD34+ HSPCs. Our work highlights prosurvival protein BCL-XL as a therapeutic vulnerability in pediatric AMKL and is timely aligned with a recent study suggesting that BCL-XL is also a potential target of adult megaerythroid leukemia.49 Addition of anti–BCL-XL agents to cytarabine-based chemotherapy regimens in pediatric AMKL clinical trials is within reach and not exploited.
Methods
Extended methods are provided in supplemental Methods.
Patient samples
Pediatric AMKL samples (supplemental Table 1; and as previously reported12) were collected with approval from the research ethics boards of Centre Hospitalier Universitaire (CHU) Sainte-Justine, and informed patient consent in agreement with the Declaration of Helsinki. Biobanking was overseen by the Quebec Leukemia Cell Bank (BCLQ) (Montréal, Canada).
Generation of CG2 models
Xenotransplantation
Mouse experimental procedures were performed in accordance with the Canadian Council of Animal Care and with approval from the CHU Sainte-Justine research ethics board and animal institutional ethic committee (approval number 2020-2666). Xenotransplantation and characterization protocols are provided in the supplemental Methods.
Flow cytometry
Immunophenotyping antibodies are listed in supplemental Table 3. Apoptotic assays and intracellular staining are detailed in supplemental Methods.
Molecular studies
Detailed protocols and sample descriptions are presented in supplemental Methods and supplemental Table 4.
RNA sequencing
RNA sequencing and data processing were performed at the Institute for Research in Immunology and Cancer’s Genomics Platform (Montréal, Canada) using the Illumina Nextseq500 with 150-cycle paired-end runs. Sequences were aligned to the reference human genome version GRCh38.
Single-cell RNA sequencing
Single-cell suspensions were processed on the Chromium Controller according to the manufacturer’s protocol, and sequenced at the CHU Sainte-Justine Integrated Centre for pediatric clinical genomics (Montréal, Canada).
In vitro pharmacological inhibition assays
Synthetic models were maintained in optimized serum-free media. Navitoclax (ABT-263), venetoclax (ABT-199), DT2216, cytarabine and staurosporine were purchased from MedChemExpress (Monmouth Junction, NJ). Media composition, dose response curves, and functional assays are described in supplemental Methods.
In vivo drug studies
Mice that underwent AMKL xenotransplantation were treated either daily via oral gavage with navitoclax (100 mg/kg; ABT-26347), DT2216 intraperitoneal every 4 days (15 mg/kg),48 or vehicle only. For drug combinations, mice were additionally treated with cytarabine (20 mg/kg, intraperitoneal) for the first 5 days. Detailed protocols are described in supplemental Methods.
Results
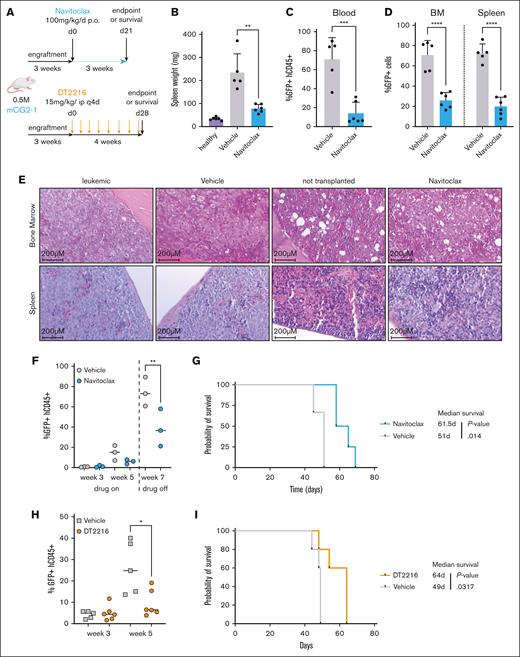
CG2 gene fusion transforms CD34+ CB cells
To generate human leukemia models, we overexpressed CG2 in pooled CB CD34+ purified cells by transduction with a lentiviral construct carrying the fusion oncogene and a green fluorescent protein (GFP) reporter (Figure 1A). Gene transfer rates averaged 12.5% (Figure 1B), and 80% of each well was separately transplanted into 1 recipient NSG mouse. Leukemia penetrance was of 60%, with latencies between 9.9 and 36.1 weeks (Figure 1B). Expression of the CG2 fusion transcript was confirmed by reverse transcription polymerase chain reaction analysis (Figure 1C) and RNA sequencing of leukemic cells. Expression levels of the CG2 fusion transcript and wild-type CBFA2T3 were comparable between models and patient samples (supplemental Figure 8). At necropsy of leukemic mice, infiltration of hematopoietic organs by leukemic blasts was confirmed in the bone marrow (BM) and spleen (hCD45+GFP+ cells; Figure 1D-F; supplemental Figure 1; supplemental Table 5). No adverse outcomes were documented in the empty-vector control group (n = 10; Figure 1F-G). All 6 engineered leukemic models were able to robustly sustain serial transplantation with consistent disease phenotype and latencies (∼8-12 weeks), tested for up to 4 passages in vivo (Figure 1G; supplemental Figure 1; supplemental Table 5). As seen in patients with CG2, some mice exhibited neurological deficits of variable intensities indicative of leukemic paraspinal infiltration, which was confirmed by bioluminescent imaging of luciferase-transduced grafts (supplemental Figure 2).
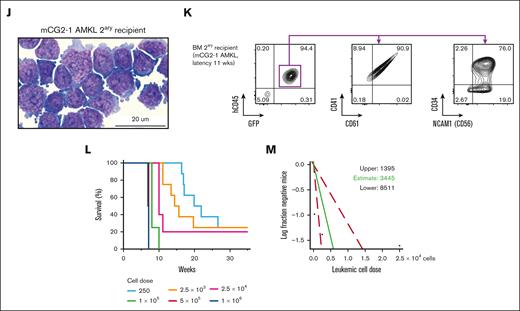
Generation of human models of CG2 leukemia. (A) Experimental procedure used to establish xenograft models of CG2 AMKL (mCG2 AMKL) using independent lentiviral transduction in CB CD34+ cells (CB CD34+, pool of 6 CB units) and transplantation in recipient NSG mice. (B) Schematic representation of 6 CG2 leukemia models (mCG2) describing initial gene transfer (GT; %GFP) at the time of transplantation and leukemia latency in primary recipient mice. Mouse identification shown in brackets; 1 of 10 mice was not available for analysis. (C) Detection of CG2 fusion transcript expression by reverse transcription polymerase chain reaction (RT-PCR) with RNA isolated from leukemic blasts, as indicated. M07e and normal lineage-depleted CB (CB LIN−) cells were used as the positive and negative control, respectively. ABL1 was used as housekeeping gene. (D) Percentage of infiltrating human blasts (hCD45+ GFP+) and (E) CD41+ cells (of hCD45+ GFP+ population) in the BM and spleen of CG2 primary recipient mice (color code indicates distinct primary mice). (F) Hematoxylin phloxine saffron–stained longitudinal sections of tibia bones harvested from primary recipient mice that received transplantation with control CB CD34+ cells transduced with empty vector (top panel, 47 weeks after transplantation) and from a CG2-1 AMKL model (secondary recipient mice that received transplantation with 1 × 106 CG2-1 AMKL xenograft cells and were euthanized 11.1 weeks after transplantation) (bottom). (G) Survival curves of primary recipient mice that received transplantation with CB CD34+ cells transduced with CG2 (black line) or empty vector (gray dashes), and mice that serially received transplantation up to 3 times with CG2-1 AMKL (red lines). Giemsa-stained cytospin and flow cytometry profiles of leukemic BM cells from representative (H-I) primary (1ary) and (J-K) secondary (2ary) CG2-1 AMKL recipient mice (2ary recipient mice that received transplantation with 1.4 × 106 CG2-1 AMKL xenograft cells and that were euthanized 6.9 weeks after transplantation). Detailed characteristics of other CG2 leukemia xenografts in recipient mice that serially received transplantation are described in supplemental Table 5 and supplemental Figure 1. (L) Survival curves of recipient NSG mice that received transplantation with CG2-1 AMKL xenograft cells in a limiting-dilution cell transplantation assay and (M) estimation of LIC frequency and 95% confidence interval (red dashes) using the extreme limiting dilution analysis software (http://bioinf.wehi.edu.au/software/elda/).
Generation of human models of CG2 leukemia. (A) Experimental procedure used to establish xenograft models of CG2 AMKL (mCG2 AMKL) using independent lentiviral transduction in CB CD34+ cells (CB CD34+, pool of 6 CB units) and transplantation in recipient NSG mice. (B) Schematic representation of 6 CG2 leukemia models (mCG2) describing initial gene transfer (GT; %GFP) at the time of transplantation and leukemia latency in primary recipient mice. Mouse identification shown in brackets; 1 of 10 mice was not available for analysis. (C) Detection of CG2 fusion transcript expression by reverse transcription polymerase chain reaction (RT-PCR) with RNA isolated from leukemic blasts, as indicated. M07e and normal lineage-depleted CB (CB LIN−) cells were used as the positive and negative control, respectively. ABL1 was used as housekeeping gene. (D) Percentage of infiltrating human blasts (hCD45+ GFP+) and (E) CD41+ cells (of hCD45+ GFP+ population) in the BM and spleen of CG2 primary recipient mice (color code indicates distinct primary mice). (F) Hematoxylin phloxine saffron–stained longitudinal sections of tibia bones harvested from primary recipient mice that received transplantation with control CB CD34+ cells transduced with empty vector (top panel, 47 weeks after transplantation) and from a CG2-1 AMKL model (secondary recipient mice that received transplantation with 1 × 106 CG2-1 AMKL xenograft cells and were euthanized 11.1 weeks after transplantation) (bottom). (G) Survival curves of primary recipient mice that received transplantation with CB CD34+ cells transduced with CG2 (black line) or empty vector (gray dashes), and mice that serially received transplantation up to 3 times with CG2-1 AMKL (red lines). Giemsa-stained cytospin and flow cytometry profiles of leukemic BM cells from representative (H-I) primary (1ary) and (J-K) secondary (2ary) CG2-1 AMKL recipient mice (2ary recipient mice that received transplantation with 1.4 × 106 CG2-1 AMKL xenograft cells and that were euthanized 6.9 weeks after transplantation). Detailed characteristics of other CG2 leukemia xenografts in recipient mice that serially received transplantation are described in supplemental Table 5 and supplemental Figure 1. (L) Survival curves of recipient NSG mice that received transplantation with CG2-1 AMKL xenograft cells in a limiting-dilution cell transplantation assay and (M) estimation of LIC frequency and 95% confidence interval (red dashes) using the extreme limiting dilution analysis software (http://bioinf.wehi.edu.au/software/elda/).
The generated leukemia showed megakaryocytic morphology and lineage markers (hCD45+CD34+/−CD41+CD61+) with significant CD56 (NCAM1) expression, a hallmark of CG2 AMKL14,15,52 (Figure 1H-K; supplemental Figure 4; supplemental Table 5). The CG2-6 model was hCD45+CD71+CD117+ by immunophenotyping, with lower surface expression of CD41 (Figure 1E), CD34, CD33, and CD56 and absence of myelomonocytic markers (myeloperoxidase [MPO], CD68, lysozyme [LYZ], fucosyltransferase 4 [FUT4/CD15] and integrin subunit alpha M [ITGAM/CD11b]) by transcriptomic and flow cytometry (supplemental Table 5; supplemental Figures 3 and 4), pointing to an early megaerythroid progenitor state. AMKL development in vivo was retained after in vitro culture for 6 days, providing physiologically relevant cellular substrate for biochemical analyses (supplemental Figure 5).
AMKL leukemia initiating cell (LIC) frequency was assessed by limiting dilution assay using cells isolated from synthetic model of CG2 (mCG2-1, mCG2-2, and mCG2-6) recipients and approximated to 1 in 3445 cells, 1 in 14 938 cells, and 1 in 15 286 cells, respectively, close to the 1 in 10 300 LIC frequency assessed in a sample from a patient-derived NUP98r sample (Figure 1L-M; supplemental Table 6). The time to overt leukemia was proportional to the cell dose, with 4 of 8 mice of mCG2-1 succumbing to AMKL 20 to 30 weeks after injection of 250 cells (Figure 1L; supplemental Table 6). Overall, the engineered CG2 AMKL faithfully recapitulate human disease, reside in the megaerythroid differentiation space and harbor high LIC frequencies, as reported for patient samples.14
AMKL CG2 models phenocopy human disease at the molecular level
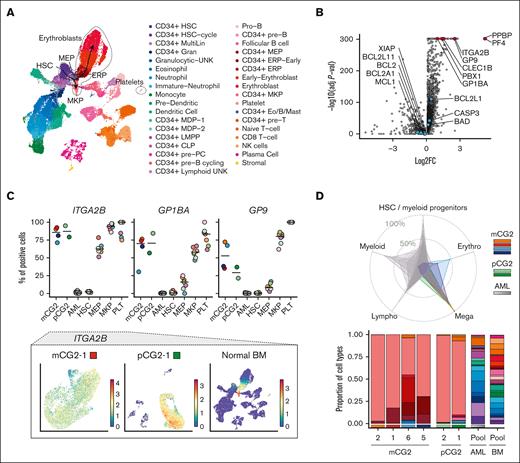
Transcriptomic profiling of the CG2 models closely correlate with genotype-matched patient samples (r = 0.64; n = 2, institutional cohort; n = 12, validation cohort9; Figure 2A), with high expression of NCAM1, BMP2 and ERG, and low levels of GATA1.5,14,15,27 A CG2-specific gene signature of upregulated and downregulated markers was derived by comparing the expression profiles of CG2 models and patients (n = 10) against institutional NUP98r AMKL (n = 7) and CB CD34+ cells (n = 4), crossvalidated in a patient cohort comprising different AMKL genotypes9 (n = 73 total, with CG2 n = 12; Figure 2A; supplemental Table 7). CG2 AMKL is a non-HOXA/B, non-MEIS1/2 leukemia, which distinctively expresses GLIS2, whereas CBFA2T3 is expressed across AMKL genotypes (supplemental Figure 6B). Remarkably, CG2 AMKL upregulated prosurvival factor BCL2 (Figure 2A), pointing to a potential subtype-specific vulnerability. Furthermore, the engineered CG2 AMKL models cluster with patient-derived CG2 samples (n = 14) by expression profiling (Figure 2B) and apart from other AMKL genotypes and CB CD34+ cells. Principal component analysis revealed clustering of primary and tertiary xenografts based on expression profile (supplemental Figure 6A).
Gene expression in CG2 leukemia models correlate with pediatric disease. (A) (Left) Correlation of differential gene expression (log2 fold change [L2FC]) in CG2 AMKL models and patients from our institutional data set (Centre hospitalier universitaire Sainte-Justine [CHUSJ]) compared with a validation data set of pediatric CG2 AMKL (St Jude). Differentially expressed genes (DEGs), defined as |L2FC > 1| and false discovery rate (FDR) q value < .05, common to both data sets are indicated in blue or red and define the CG2 signature (supplemental Table 7). Institutional (Inst) data set: CG2 AMKL models (n = 10) and patient-derived CG2 samples (n = 2) compared with N5A AMKL models (n = 5), patient-derived N5A samples (n = 2), and normal CB CD34+ cells (n = 4). Validation (Val) data set: CG2 AMKL (n = 12) vs other genetic subtypes of AMKL (n = 61) from pediatric patients at diagnosis.9 (Right) Upset plots showing DEGs that are jointly overexpressed (n = 399) or underexpressed (n = 330) in CG2 leukemias, corresponding to blue and red dots in panel A. (B) Hierarchical clustering using the 729 DEGs of the CG2 gene expression signature. (C) Heat map showing protein (log2[mass spectrometry [MS] values, left panels) and messenger RNA (RNAseq; fragments per kilobase of transcript per million mapped reads [FPKM] values, right panels) expression of cell surface markers associated to CG2 leukemia (high expression in mCG2-1 and mCG2-2 leukemia models log2[MS values] ≥ 16 and RNAseq FPKM ≥ 5) and weak/no expression in normal CB CD34+ cells (log2[MS values] < 16 and RNAseq FPKM < 5). Samples were analyzed in triplicates and represented as mean expression (biological triplicates for RNAseq and technical triplicates for proteomic data). For comparison, values for mN5A and pdxNTF are shown alongside mCG2 samples. The St Jude Val cohort RNAseq expression is presented as a separate column. (D) Star plot presenting the adjusted P values of DEGs and differentially expressed proteins (FDR < 0.05) (transcriptome: CHUSJ CG2 vs N5A AMKL and normal CB CD34+; surfaceome: CG2 vs NUP98r and CB CD34+). The scores were calculated by multiplying the algebraic sign (+ or −) of the log2FC, surfaceome or transcriptome, by the corresponding log10(adjusted P value). Significantly upregulated CG2 AMKL–specific surface markers intersecting both data sets are labeled. (E) Validation by flow cytometry of PCDH19 surface expression on CG2 AMKL cells from patient (pCG2-1), model mCG2-1 and M07e cell line. PCDH19 is not expressed on lineage-depleted human CB cells (CB CD34+). Samples were costained with NCAM1 (CD56). (F) Scatterplot representations showing the best pairwise correlations of the 2 sets of cell surface marker genes that select for CG2 genotype in a validation data set of pediatric AMKL (of 8 cell surface marker combinations, see supplemental Figure 13). Values, from top to bottom, represent the global and subtype-specific Kendall rank correlation coefficients. P values: ∗P < .05 and ∗∗P < .005.
Gene expression in CG2 leukemia models correlate with pediatric disease. (A) (Left) Correlation of differential gene expression (log2 fold change [L2FC]) in CG2 AMKL models and patients from our institutional data set (Centre hospitalier universitaire Sainte-Justine [CHUSJ]) compared with a validation data set of pediatric CG2 AMKL (St Jude). Differentially expressed genes (DEGs), defined as |L2FC > 1| and false discovery rate (FDR) q value < .05, common to both data sets are indicated in blue or red and define the CG2 signature (supplemental Table 7). Institutional (Inst) data set: CG2 AMKL models (n = 10) and patient-derived CG2 samples (n = 2) compared with N5A AMKL models (n = 5), patient-derived N5A samples (n = 2), and normal CB CD34+ cells (n = 4). Validation (Val) data set: CG2 AMKL (n = 12) vs other genetic subtypes of AMKL (n = 61) from pediatric patients at diagnosis.9 (Right) Upset plots showing DEGs that are jointly overexpressed (n = 399) or underexpressed (n = 330) in CG2 leukemias, corresponding to blue and red dots in panel A. (B) Hierarchical clustering using the 729 DEGs of the CG2 gene expression signature. (C) Heat map showing protein (log2[mass spectrometry [MS] values, left panels) and messenger RNA (RNAseq; fragments per kilobase of transcript per million mapped reads [FPKM] values, right panels) expression of cell surface markers associated to CG2 leukemia (high expression in mCG2-1 and mCG2-2 leukemia models log2[MS values] ≥ 16 and RNAseq FPKM ≥ 5) and weak/no expression in normal CB CD34+ cells (log2[MS values] < 16 and RNAseq FPKM < 5). Samples were analyzed in triplicates and represented as mean expression (biological triplicates for RNAseq and technical triplicates for proteomic data). For comparison, values for mN5A and pdxNTF are shown alongside mCG2 samples. The St Jude Val cohort RNAseq expression is presented as a separate column. (D) Star plot presenting the adjusted P values of DEGs and differentially expressed proteins (FDR < 0.05) (transcriptome: CHUSJ CG2 vs N5A AMKL and normal CB CD34+; surfaceome: CG2 vs NUP98r and CB CD34+). The scores were calculated by multiplying the algebraic sign (+ or −) of the log2FC, surfaceome or transcriptome, by the corresponding log10(adjusted P value). Significantly upregulated CG2 AMKL–specific surface markers intersecting both data sets are labeled. (E) Validation by flow cytometry of PCDH19 surface expression on CG2 AMKL cells from patient (pCG2-1), model mCG2-1 and M07e cell line. PCDH19 is not expressed on lineage-depleted human CB cells (CB CD34+). Samples were costained with NCAM1 (CD56). (F) Scatterplot representations showing the best pairwise correlations of the 2 sets of cell surface marker genes that select for CG2 genotype in a validation data set of pediatric AMKL (of 8 cell surface marker combinations, see supplemental Figure 13). Values, from top to bottom, represent the global and subtype-specific Kendall rank correlation coefficients. P values: ∗P < .05 and ∗∗P < .005.
Akin to patient samples,2,9,15 the genomic landscape of the engineered CG2 models was rather silent as assessed by whole-exome sequencing and comparative genomic hybridization (supplemental Figure 7), with no recurrent cooperating mutations, pathogenic single-nucleotide variants, or indels. Comparable with the reported genomic landscape of human disease,15 copy number variations were identified in 4 of 6 models, and chromosomal alterations remained stable over serial transplantation for 2 models analyzed (supplemental Figure 7). We excluded insertional mutagenesis events by long-read DNA sequencing (n = 4), and confirmed the (oligo)-clonal character of the engineered AMKL models (supplemental Figure 9). The CG2 AMKL models thus transcriptionally phenocopy human disease and appear genetically stable through serial transplantation.
Proteomic analyses of human CG2 models identify genotype-specific surface markers
We used a surface proteome (surfaceome) approach, as previously described,12,53-55 to identify CG2-specific vs normal HSPC membrane proteins (CB CD34+, n = 11; CG2 AMKL, n = 2; and NUP98r AMKL, n = 212,13; Figure 2C; supplemental Figure 10; supplemental Table 9). Comparing proteomic data sets of CG2 and CB CD34+ cells confirmed that CG2 models express AMKL-associated markers CD151,56 ITGA2B, PECAM1 (CD31)57 and NCAM1 (CD56)14,15,17 (supplemental Figure 10E). The 25 most abundant and differentially expressed proteins in CG2 vs CB CD34+ were crossvalidated against an external transcriptomic data set from patients with AMKL9 (Figure 2C; supplemental Figure 11B). Eight of these markers (CSF2RA, DPP10, NCAM1, NRP1, PCDH10, PCDH19, RECK, and SCN9A) were significantly associated with the CG2 genotype (highlighted in Figure 2D; supplemental Figure 11B). Five of the 25 surfaceome proteins could be validated using available flow cytometry antibodies, showing that PCDH19 and JAG1 were detected on a subset of cells across all patients and models, whereas CSF2RA, NRP1, and INSR were only detected in a subset of samples (Figure 2E; supplemental Figure 12). Similarly, the FOLR1 receptor, recently reported to be associated with CG2 AMKL,18 was found to be expressed on a significant proportion of cells across models, except for mCG2-6. Of note, the CD41low mCG2-6 cells express CD31/CD151 but upregulate CD41/CD61 in culture (supplemental Figure 4), pointing to immature leukemic cells that are primed toward the megakaryocytic lineage. Pairwise correlation analyses of cell surface markers most associated with CG2 uncovered that the combination of NCAM1 with either PCDH10 or PCDH19 was most predictive of the CG2 genotype (Figure 2F; supplemental Figure 13; supplemental Table 10; P < .05). These markers can contribute to refine the panel of markers used for CG2 AMKL diagnosis and tracking of minimal residual disease in patients.58
CG2 leukemia lies in the megaerythroid stem and progenitor differentiation space
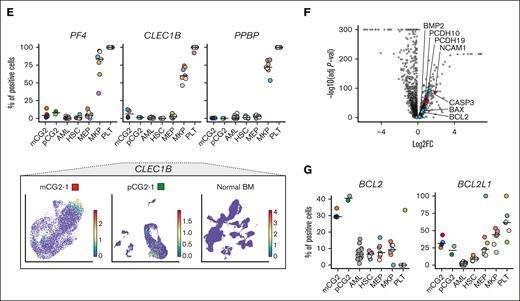
Engineered CG2 models (n = 4) and patient-derived samples (n = 2) were profiled by single-cell RNA sequencing to determine their cellular composition (Figure 3; supplemental Figures 14-17). Key upregulated genes of normal megakaryocyte-erythroid progenitors (MEPs), megakaryocyte progenitors (MKPs), and platelets were inferred from normal BM cell populations (Human Cell Atlas [HCA]59; Figure 3A-B; supplemental Table 11). MEP/MKP/platelet-specific genes, for example ITGA2B (CD41), are universally expressed in CG2 AMKL cells but not in adult AML cells60 (Figure 3C; supplemental Figures 15 and 16). The MKP-specific genes GP1BA (glycoprotein 1B platelet subunit α or CD42b) and GP9 (glycoprotein IX) are more expressed in CG2 than in healthy MEP cells, indicating that CG2 leukemic cells lie toward the primitive megakaryocytic differentiation space. Transferring the cell type assignment from the HCA to AMKL and AML data sets, confirmed the dominant MKP bias of CG2 models and patients compared with other AML samples (gray shaded samples in Figure 3D, top; supplemental Figure 14B-C). CG2 AMKL samples contain varying MEP proportions (Figure 3D, bottom), with the CG2-6 model partially projecting into the erythroid space (Figure 3D, blue shading in radar plot and CD34+ erythroid progenitors [ERP] fractions in the bottom panel). Although MKP genes are homogeneously expressed in CG2 samples, only small subsets express genes suggesting terminal differentiation such as CLEC1B (Figure 3E; supplemental Figures 15 and 16). These data indicate that CG2 models reside predominantly in the MKP differentiation space and recapitulate the lineage commitment in patients with CG2 AMKL.
CG2 leukemia consists of immature megaerythroid stem and progenitor lineages. (A) Uniform manifold approximation and projection (UMAP) of normal BM lineages. The stem-megaerythroid compartment is circled, relevant populations are labeled, and differentiation trajectories are highlighted with arrows. (B) DEGs between cells of the megakaryocytic lineage (CD34+ MEPs, CD34+ MKPs, and platelets) and all other cell types in normal BM (supplemental Table 11). Genes relevant to the megakaryocytic differentiation are highlighted in red, and genes of the mitochondrial apoptotic pathway are highlighted in blue. (C, top) Percentage positive cells for each gene in mCG2s, pCG2s, other subtypes of AML and in specific populations of the normal BM (HSCs, MEPs, MKPs, and PLTs). The horizontal line represents the median of the group, the patient and models are color coded as in panel D, and the normal samples are color coded based on donor as in supplemental Figure 14D. (Bottom) ITGA2B (CD41) expression per cell on a UMAP representation of mCG2-1, pCG2-1, and normal BM (extended samples in supplemental Figures 15 and 16). (D) Radar plot (top) of lineage composition assessed by single-cell RNAseq in CG2 AMKL models and pediatric patients, as compared with diverse phenotypic and genetic subtypes of adult AML. The detailed proportion of each cell type is presented as stacked bar plots (bottom). Color coding of populations is as depicted in panel A. (E) Representation as in panel C of terminal differentiation gene expression (extended samples in supplemental Figures 15 and 16). (F) DEGs in the CD34+ MKPs from the mCG2 and pCG2s compared with those in normal CD34+ MKPs (supplemental Table 12). Selected markers are highlighted in red, and genes of the mitochondrial apoptotic pathway are highlighted in blue. (G) Representation as in panel C of BCL2 and BCL2L1 (BCL-XL) expression. ERP, erythroid progenitors; HSCs, hematopoietic stem cells; PLT, platelet.
CG2 leukemia consists of immature megaerythroid stem and progenitor lineages. (A) Uniform manifold approximation and projection (UMAP) of normal BM lineages. The stem-megaerythroid compartment is circled, relevant populations are labeled, and differentiation trajectories are highlighted with arrows. (B) DEGs between cells of the megakaryocytic lineage (CD34+ MEPs, CD34+ MKPs, and platelets) and all other cell types in normal BM (supplemental Table 11). Genes relevant to the megakaryocytic differentiation are highlighted in red, and genes of the mitochondrial apoptotic pathway are highlighted in blue. (C, top) Percentage positive cells for each gene in mCG2s, pCG2s, other subtypes of AML and in specific populations of the normal BM (HSCs, MEPs, MKPs, and PLTs). The horizontal line represents the median of the group, the patient and models are color coded as in panel D, and the normal samples are color coded based on donor as in supplemental Figure 14D. (Bottom) ITGA2B (CD41) expression per cell on a UMAP representation of mCG2-1, pCG2-1, and normal BM (extended samples in supplemental Figures 15 and 16). (D) Radar plot (top) of lineage composition assessed by single-cell RNAseq in CG2 AMKL models and pediatric patients, as compared with diverse phenotypic and genetic subtypes of adult AML. The detailed proportion of each cell type is presented as stacked bar plots (bottom). Color coding of populations is as depicted in panel A. (E) Representation as in panel C of terminal differentiation gene expression (extended samples in supplemental Figures 15 and 16). (F) DEGs in the CD34+ MKPs from the mCG2 and pCG2s compared with those in normal CD34+ MKPs (supplemental Table 12). Selected markers are highlighted in red, and genes of the mitochondrial apoptotic pathway are highlighted in blue. (G) Representation as in panel C of BCL2 and BCL2L1 (BCL-XL) expression. ERP, erythroid progenitors; HSCs, hematopoietic stem cells; PLT, platelet.
Coembedding the data sets in the same uniform manifold approximation and projection (UMAP) revealed that CG2 models and patient samples lie in an area of the UMAP representation also populated by healthy BM cells (HCA) of the megakaryocytic lineage (supplemental Figure 17A), highlighting the low intersample heterogeneity, in contrast to the large cell heterogeneity of normal BM, as seen by the distribution overlap of the CG2 models and patient-derived samples (supplemental Figure 17B).
Furthermore, gene expression of leukemic CG2 MKP was compared with that of healthy MKPs to identify aberrantly expressed genes, such as NCAM1 and BCL2 (Figure 3F-G; supplemental Table 12), mirroring the CG2 bulk transcriptomic signature (Figure 2A). Although no other BCL2 family member was aberrantly expressed in CG2 MKP, BCL2L1 (BCL-XL) is specifically expressed in normal MEP/MKP/platelet subsets as compared with other cell types (adjusted P = 7.5 × 10−111; Figure 3B; supplemental Figure 14E), and BCL2L1 expression is maintained across CG2 models and patient-derived samples (Figure 3G; supplemental Figures 15 and 16). This provides a rationale to assess the functional dependency of CG2 cells to BCL-XL, based on lineage ontogeny, and to BCL2, given its aberrant expression in CG2 AMKL.
AMKL cells are resistant to venetoclax but sensitive to navitoclax
We tested the sensitivity of AMKL cells to BCL2 inhibitor venetoclax (CG2, n = 6; N5A, n = 212; and patient-derived xenograft [pdx]NTF, n = 113) as well as in AML samples (KMT2A::MLLT3 [MLLr], n = 1; NUP98::NSD1 [NNSD1], n = 1; and N5A monocytic [CD68+LYZ+] AML models, n = 212). All AMKL samples tested were resistant to venetoclax, with half-maximal inhibitory concentrations (IC50s) of >10 μM, whereas AML samples were sensitive to venetoclax (Figure 4A; supplemental Table 13). In contrast, both CG2 and NUP98r AMKL were sensitive to navitoclax, which has a broader affinity to BCL2, BCL-XL, and BCL-W as well as AML samples (Figure 4B; supplemental Table 13), whereas genotype-matched monocytic N5A AML cells were resistant, suggesting a lineage-specific vulnerability. AMKL cells exposed to navitoclax for 72 hours at concentrations spanning the leukemia-specific IC50 values exhibited higher levels of apoptosis than control CB CD34+ cells (Figure 4C). In contrast, exposure to venetoclax did not induce apoptosis in AMKL cells at doses up to 10 μM (Figure 4D). Levels of mitochondrial superoxide radicals and loss of mitochondrial potential were higher in AMKL cells treated with navitoclax than in cells treated with venetoclax (Figure 4E-F; supplemental Figure 18A), suggesting mitochondrial dysfunction. These results imply that AMKL cells are sensitive to BCL2/BCLXL/BCL-W inhibitor navitoclax with subsequent induction of mitochondrial apoptosis yet resistant to venetoclax that uniquely targets BCL2.
CG2 and NUP98r AMKL xenografts are sensitive to the induction of the intrinsic apoptotic pathway. (A-B) Dose-response curves and IC50s (supplemental Table 13) determined for each indicated sample of AMKL or AML, submitted to a viability assay in presence of venetoclax or navitoclax. (Cell-Titer Glo, 6-day incubation, 4 replicates). Viability readout was normalized to dimethyl sulfoxide (DMSO) controls for each sample. (C-D) Apoptosis assessed by annexin V staining and flow cytometry of AKML xenografts (mCG2, pdxNTF, and mN5A; n = 2 biological replicates for each cell type), ML2 (AML cell line, n = 2), or normal CB CD34+ cells (n = 3) treated for 72 hours in culture as triplicates with the indicated BH3 mimetics (123.5 nM, 370 nM, and 10 μM) or DMSO. P values: ∗∗P < .005; ∗∗∗P < .001; ∗∗∗∗P < .0001. (E-F) Mitochondrial superoxide production was assessed by MitoSox staining and flow cytometry of AMKL xenografts or normal CB CD34+ cells, treated with the indicated BH3 mimetics or DMSO for 72 hours in culture in duplicates. Staurosporine (STS) was used as positive control at 1 μM. mAMKL, synthetic xenograft of AMKL; mAML, synthetic xenograft of AML; pdx, patient-derived xenograft; CB CD34+, CB CD34+ cells.
CG2 and NUP98r AMKL xenografts are sensitive to the induction of the intrinsic apoptotic pathway. (A-B) Dose-response curves and IC50s (supplemental Table 13) determined for each indicated sample of AMKL or AML, submitted to a viability assay in presence of venetoclax or navitoclax. (Cell-Titer Glo, 6-day incubation, 4 replicates). Viability readout was normalized to dimethyl sulfoxide (DMSO) controls for each sample. (C-D) Apoptosis assessed by annexin V staining and flow cytometry of AKML xenografts (mCG2, pdxNTF, and mN5A; n = 2 biological replicates for each cell type), ML2 (AML cell line, n = 2), or normal CB CD34+ cells (n = 3) treated for 72 hours in culture as triplicates with the indicated BH3 mimetics (123.5 nM, 370 nM, and 10 μM) or DMSO. P values: ∗∗P < .005; ∗∗∗P < .001; ∗∗∗∗P < .0001. (E-F) Mitochondrial superoxide production was assessed by MitoSox staining and flow cytometry of AMKL xenografts or normal CB CD34+ cells, treated with the indicated BH3 mimetics or DMSO for 72 hours in culture in duplicates. Staurosporine (STS) was used as positive control at 1 μM. mAMKL, synthetic xenograft of AMKL; mAML, synthetic xenograft of AML; pdx, patient-derived xenograft; CB CD34+, CB CD34+ cells.
Inhibition of BCL-XL induces mitochondrial apoptosis of CG2 and NUP98r AMKL
To dissect whether the induction of apoptosis in AMKL cells by navitoclax is mediated via BCL2, BCL-XL, or BCL-W inhibition, expression levels of each gene were assessed using transcriptomic data sets. In accordance with the CG2 signature (Figure 2A), BCL2 was most abundant in CG2 compared with NUP98r AMKL cells (institutional cohort, Figure 5A) and all other AML genotypes (CG2, n = 7; patients total, validation cohort, n = 739; Figure 5B). In contrast, BCL2L1 (BCL-XL) is homogenously expressed across AMKL genotypes and at much lower levels in the N5A monocytic AML (n = 3). Intracellular protein levels of BCL-XL were correspondingly increased in CG2 and NUP98r AMKL as compared with normal CB CD34+ cells, whereas the BCL2 protein was only detected in the CG2 genotype, matching gene expression (Figure 5C). Short hairpin(sh)–mediated KD against BCL2, BCL2L1 or BCL2L2 (BCL-W) was used to assess the functional impact of reduced prosurvival factors in 2 distinct CG2 models (gene transfer of >95% GFP+mCherry+ cells; Figure 5D-E; supplemental Figure 18B-C), with confirmation of efficient protein reduction of BCL2 and BCL-XL (Figure 5F; supplemental Figure 18D). Increased apoptosis of AMKL cells (annexin V–positive cells; Figure 5D; supplemental Figure 18E) was observed only with KD of BCL-XL, but not BCL2 or BCL-W. Additionally, AMKL and monocytic N5A AML models were treated with DT2216,48 a selective BCL-XL proteolysis-targeting chimera that targets BCL-XL to the Von Hippel-Lindau E3 ligase (VHL) for degradation. VHL is abundantly expressed in our models of AMKL (Figure 5A) but not expressed in platelets, which are most vulnerable to BCL-XL inhibition.48 In a dose-response experiment, AMKL models were sensitive to DT2216, with IC50s < 200 nM (Figure 5G; supplemental Table 13). In contrast, monocytic N5A AML models demonstrated resistance to DT2216 treatment, with IC50 values > 10 μM (Figure 5G; supplemental Table 13). AMKL cells exposed to 100 nM and 1 μM of DT2216 showed significantly higher levels of apoptosis compared with normal CB CD34+ cells (Figure 5H) and higher mitochondrial stress, shown by increased superoxide levels and loss of mitochondrial membrane potential (supplemental Figure 18F-G). Furthermore, testing navitoclax, venetoclax, and DT2216 on several independent CB CD34+ pools revealed sample-dependent variability in drug response but overall lower toxicity with DT2216 in comparison to navitoclax (supplemental Figure 18H; supplemental Table 13). These results suggest that the sensitivity of AMKL cells toward navitoclax is due to the inhibition of BCL-XL, as demonstrated by genetic and pharmacological inhibition.
AMKL xenografts depend on prosurvival protein BCL-XL for survival. (A) RNAseq expression values (log2[FPKM]) of selected genes implicated in intrinsic apoptosis pathway in model and patient leukemias from our institutional data set. (B) Crossvalidation of gene expression in a large and published data set of pediatric AMKL.9 (C) Protein levels of BCL-XL (left) and BCL2 (right) in AMKL xenograft cells or normal CB CD34+ cells assessed by intracellular flow cytometry. (D) Apoptosis, assessed by annexin V staining and flow cytometry, (E) relative gene expression detected by quantitative reverse transcription polymerase chain reaction, and (F) prosurvival protein levels quantified by flow cytometry, after short hairpin RNA (shRNA)-mediated KD of prosurvival BCL2 proteins (BCL-XL, BCL2, and BCL-W) in CG2 xenografts (72 hours after infection, 2 selected shRNA per gene). Samples were compared with nontransduced (NT) or cells infected with control shRNA against Renilla (shRenilla). See supplemental Figure 18 for KD studies in mCG2-2 cells. (G) Dose response curves and IC50 values were determined after incubation of AMKL or AML xenografts with DT2216 (BCL-XL proteolysis-targeting chimera) or DMSO for 6 days, followed by viability readout with Cell-Titer Glo. Viability readout was normalized against DMSO for each sample. (H) Amount of apoptosis assessed by annexin V staining and flow cytometry in AMKL xenografts or normal CB CD34+ cells after 72 hours of incubation with DT2216 at 100 nM or 1 μM in comparison with cells treated with DMSO. GATA1s, GATA1 truncation; HOXr, HOX rearrangement; KMT2Ar, KMT2A rearrangement; MFI, mean fluorescence intensity; RBM-MKL, RBM15::MKL1; VHL, von Hippel-Lindau; xAMKL, synthetic model of AMKL; xAML, synthetic model of AML. P values: ∗P < .05; ∗∗P < .005; ∗∗∗∗P < .0001.
AMKL xenografts depend on prosurvival protein BCL-XL for survival. (A) RNAseq expression values (log2[FPKM]) of selected genes implicated in intrinsic apoptosis pathway in model and patient leukemias from our institutional data set. (B) Crossvalidation of gene expression in a large and published data set of pediatric AMKL.9 (C) Protein levels of BCL-XL (left) and BCL2 (right) in AMKL xenograft cells or normal CB CD34+ cells assessed by intracellular flow cytometry. (D) Apoptosis, assessed by annexin V staining and flow cytometry, (E) relative gene expression detected by quantitative reverse transcription polymerase chain reaction, and (F) prosurvival protein levels quantified by flow cytometry, after short hairpin RNA (shRNA)-mediated KD of prosurvival BCL2 proteins (BCL-XL, BCL2, and BCL-W) in CG2 xenografts (72 hours after infection, 2 selected shRNA per gene). Samples were compared with nontransduced (NT) or cells infected with control shRNA against Renilla (shRenilla). See supplemental Figure 18 for KD studies in mCG2-2 cells. (G) Dose response curves and IC50 values were determined after incubation of AMKL or AML xenografts with DT2216 (BCL-XL proteolysis-targeting chimera) or DMSO for 6 days, followed by viability readout with Cell-Titer Glo. Viability readout was normalized against DMSO for each sample. (H) Amount of apoptosis assessed by annexin V staining and flow cytometry in AMKL xenografts or normal CB CD34+ cells after 72 hours of incubation with DT2216 at 100 nM or 1 μM in comparison with cells treated with DMSO. GATA1s, GATA1 truncation; HOXr, HOX rearrangement; KMT2Ar, KMT2A rearrangement; MFI, mean fluorescence intensity; RBM-MKL, RBM15::MKL1; VHL, von Hippel-Lindau; xAMKL, synthetic model of AMKL; xAML, synthetic model of AML. P values: ∗P < .05; ∗∗P < .005; ∗∗∗∗P < .0001.
Monotherapy with navitoclax or DT2216 reduces leukemic burden in high-fatality AMKL in vivo
To investigate the in vivo impact of BCL-XL inhibition of navitoclax or DT2216 as single agent, 3 independent models of AMKL were tested (Figure 6A; supplemental Figures 19 and 20) as well as a chemotherapy refractory PDX of NTF13 (supplemental Figures 19 and 20). After a 3-week treatment cycle with navitoclax, a significant reduction of leukemic infiltration was noted in mCG2-1, as assessed based on spleen weight (Figure 6B) and abundance of GFP+hCD45+ AMKL cells in the blood, BM, and spleen (Figure 6C-D). Normal hematopoietic cell distribution and tissue architecture was restored in the BM and spleen of navitoclax-treated mice vs control mice (Figure 6E). Furthermore, navitoclax-treated mice showed reduced circulating blasts during treatment (week 5) as well as 1 week after treatment (week 7; Figure 6F), conferring a survival benefit over vehicle-treated mice (Figure 6G). For mCG2-2, treatment was stopped early because of rapid onset of hindleg paralysis. Nevertheless, a significant reduction of overall leukemic burden was detected after 2 weeks of treatment, measured by luciferase intensity (supplemental Figure 19A-B) as well as reduction of GFP+Ametrine+ cells in the spleen in comparison with vehicle controls (supplemental Figure 19C). Using in vivo imaging, reduced leukemic progression was seen in mice with pdxNTF xenografts treated with navitoclax, which was significantly lower than in vehicle-treated control mice (week 8; supplemental Figure 19D-E).
CG2 AMKL is impaired by navitoclax and DT2216 treatment in vivo. (A) Workflow of experimental procedures to assess the in vivo activity of navitoclax or DT2216 in CBFA2T3::GLS2 (mCG2-1) AMKL xenografts. On the day of euthanizing, (B) spleen weights, (C) percentage of leukemic blasts in the peripheral blood (% GFP+hCD45+), and (D) infiltration of the BM and spleen (% GFP+ cells) were assessed in mice that received transplantation after treatment with either vehicle only (n = 5) or navitoclax (n = 6) for 3 weeks. For comparison, spleen weights of healthy mice that did not receive transplantation (n = 6) were recorded for data shown in panel B. (E) Hematoxylin and eosin–stained longitudinal sections of the tibia and spleen collected from mice on the day of euthanizing. Conditions were as follows: transplantation with CG2 but not treated (leukemic), transplantation and treated with vehicle only (Vehicle), no transplantation and untreated, age-matched littermates of treatment groups (not transplanted), or transplantation and treated with navitoclax (Navitoclax). (F) Leukemic burden (% GFP hCD45+) was monitored during treatment in the blood of Vehicle- and navitoclax–treated mice. (G) Kaplan-Meier survival curves of mice that received transplantation with mCG2-1 treated either with vehicle or navitoclax. Log-rank Mantel-Cox test was used to determine survival benefit. (H) Leukemic burden (GFP+hCD45+) was monitored by bleeding in mCG2-1 vehicle-treated mice vs DT2216-treated mice. (I) Kaplan-Meier survival curves of mCG2-1 AMKL model treated either with vehicle or DT2216. Survival benefit was determined with log-rank Mantel-Cox test. P values: ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; ∗∗∗∗P < .0001.
CG2 AMKL is impaired by navitoclax and DT2216 treatment in vivo. (A) Workflow of experimental procedures to assess the in vivo activity of navitoclax or DT2216 in CBFA2T3::GLS2 (mCG2-1) AMKL xenografts. On the day of euthanizing, (B) spleen weights, (C) percentage of leukemic blasts in the peripheral blood (% GFP+hCD45+), and (D) infiltration of the BM and spleen (% GFP+ cells) were assessed in mice that received transplantation after treatment with either vehicle only (n = 5) or navitoclax (n = 6) for 3 weeks. For comparison, spleen weights of healthy mice that did not receive transplantation (n = 6) were recorded for data shown in panel B. (E) Hematoxylin and eosin–stained longitudinal sections of the tibia and spleen collected from mice on the day of euthanizing. Conditions were as follows: transplantation with CG2 but not treated (leukemic), transplantation and treated with vehicle only (Vehicle), no transplantation and untreated, age-matched littermates of treatment groups (not transplanted), or transplantation and treated with navitoclax (Navitoclax). (F) Leukemic burden (% GFP hCD45+) was monitored during treatment in the blood of Vehicle- and navitoclax–treated mice. (G) Kaplan-Meier survival curves of mice that received transplantation with mCG2-1 treated either with vehicle or navitoclax. Log-rank Mantel-Cox test was used to determine survival benefit. (H) Leukemic burden (GFP+hCD45+) was monitored by bleeding in mCG2-1 vehicle-treated mice vs DT2216-treated mice. (I) Kaplan-Meier survival curves of mCG2-1 AMKL model treated either with vehicle or DT2216. Survival benefit was determined with log-rank Mantel-Cox test. P values: ∗P < .05; ∗∗P < .005; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Furthermore, we tested DT2216 in 2 distinct models of CG2 (mCG2-1 and mCG2-6), presenting either as megakaryopoietic or megaerythroid leukemia, and in pdxNTF. Mice with xenografts were treated with DT2216 every 4 days for a total of 4 to 6 weeks. Treatment with DT2216 demonstrated reduced leukemic blasts in the blood (GFP+hCD45+ cells; Figure 6H) and significantly prolonged the survival of mice that received mCG2-1 transplantation in comparison with vehicle controls (Figure 6I). Mice that received CG2-6 transplantation were investigated by BM aspiration after 6 weeks of treatment and showed significant reduction of hCD45+GFP+CD31+CD151+ leukemic cells in the BM in comparison with vehicle controls (supplemental Figure 20A-B). Additionally, a 4-week treatment cycle of DT2216 significantly reduced the leukemic burden in mice with pdxNTF xenografts (supplemental Figure 20C). This work demonstrates that BCL-XL degradation either with navitoclax or DT2216 could be exploited as a strategy in the treatment of high-fatality CG2 and NUP98r AMKL.
Combinatorial use of navitoclax or DT2216 with cytarabine has potent anti-AMKL activity in vivo
Cytarabine is a standard-of-care component of chemotherapy induction and intensification cycles for patients with AML. In this context, combinatorial treatment with BCL-XL inhibitors and cytarabine was tested in CG2 AMKL. First, all CG2 models showed sensitivity toward cytarabine in vitro with IC50s ranging from 1 to 7 nM (Figure 7A; supplemental Table 13). To assess the in vivo antileukemia activity of either navitoclax or DT2216 combined with cytarabine, NSG mice received a xenograft of mCG2-1 and were treated with low-dose cytarabine (20 mg/kg) for 5 consecutive days alongside navitoclax or DT2216 (Figure 7B; supplemental Figure 21A). Engraftment was verified for all groups before treatment by subjecting them to bleeding 3 weeks after transplantation (supplemental Figure 21B-C). At the end point, mice treated with the combination of cytarabine and navitoclax showed significantly lower leukemic infiltration in the blood, BM, and spleen as compared with mice treated with single agent alone or vehicle control (Figure 7C). In monotherapy, navitoclax or cytarabine performed equivalently in terms of leukemic burden reduction (Figure 7C). The same in vivo response was observed after combinatorial treatment with DT2216 and cytarabine (supplemental Figure 21D). Of note, although navitoclax shows a steeper reduction of leukemic burden as a single agent in comparison with DT2216, treatment with either navitoclax or DT2216 combined with cytarabine was equally effective against CG2 AMKL (Figure 7D-E; supplemental Figure 21F-E). These results highlight the translational potential of combining standard-of-care cytarabine with BCL-XL inhibitors in the treatment of high-fatality pediatric AMKL.
Combinatorial use of BCL-XL inhibitors and cytarabine demonstrates greater reduction of leukemic burden in vivo than single-agent treatment. (A) Dose-response curves and IC50 values (supplemental Table 13) determined for all 6 samples of CG2 AMKL, submitted to a viability assay in presence of cytarabine. (Cell-Titer Glo, 6-day incubation, 4 replicates). Viability readout was normalized to DMSO controls for each sample. (B) Schematic overview of experimental design of combinatory treatments with navitoclax and cytarabine (AraC) of mice that received xenotransplantation. (C) Percentage of leukemic blasts in the peripheral blood (% GFP+hCD45+, left graph), infiltration of the BM (middle graph) and spleen (right graph) was assessed in mice that received transplantation, after 3 weeks of indicated treatments. (D) Percentage infiltration (% GFP+hCD45+) in the BM of mice that received xenotransplantation and that were treated was compared at the end point between matched Vehicle controls and mice either treated with DT2216 or navitoclax (Navito) (E) as well as their respective combinations with cytarabine (AraC).
Combinatorial use of BCL-XL inhibitors and cytarabine demonstrates greater reduction of leukemic burden in vivo than single-agent treatment. (A) Dose-response curves and IC50 values (supplemental Table 13) determined for all 6 samples of CG2 AMKL, submitted to a viability assay in presence of cytarabine. (Cell-Titer Glo, 6-day incubation, 4 replicates). Viability readout was normalized to DMSO controls for each sample. (B) Schematic overview of experimental design of combinatory treatments with navitoclax and cytarabine (AraC) of mice that received xenotransplantation. (C) Percentage of leukemic blasts in the peripheral blood (% GFP+hCD45+, left graph), infiltration of the BM (middle graph) and spleen (right graph) was assessed in mice that received transplantation, after 3 weeks of indicated treatments. (D) Percentage infiltration (% GFP+hCD45+) in the BM of mice that received xenotransplantation and that were treated was compared at the end point between matched Vehicle controls and mice either treated with DT2216 or navitoclax (Navito) (E) as well as their respective combinations with cytarabine (AraC).
Discussion
We present 6 human models of CG2 AMKL that can sustain serial transplantation and phenocopy the clinical features, expression signature, genomic landscape, cellular ontogeny, and high LIC frequencies of the disease. Progress in the field is hampered by limited patient samples currently available for preclinical testing, a restricted pool of representative cell lines, and limited human models. Synthetic CG2 models contribute to map functional dependencies and capture the cellular and molecular heterogeneity of the leukemia, maturation block, concomitant genetic alterations, and surface marker expression. They represent faithful and scalable biomasses for biochemical studies, such as proteomics, to uncover new CG2 biomarkers that are either megaerythroid-lineage specific (CD151 and PECAM1), distinct from normal CB progenitors (JAG1), or most predictive of the CG2 AMKL genotype (NCAM1, PCDH10, and PCDH19), which can contribute to the optimization of flow cytometry antibody panels for CG2 AMKL diagnosis or minimal residual disease tracking.
Cell- and genotype-specific therapies are needed for pediatric AMKL to improve patient survival. Our models of CG2 highlight prosurvival factor BCL-XL as a novel therapeutic target of high-fatality AMKL. In vitro pharmacological inhibition of BCL-XL with navitoclax or DT2216 induces apoptosis in all CG2 AMKL models, accompanied by mitochondrial dysfunction. This was recapitulated by genetic KD of BCL-XL but not BCL2 or BCL-W, suggesting that CG2 AMKL depend preferentially on BCL-XL to resist apoptosis. Similarly, NUP98r AMKL cells (n = 3) were also sensitive to navitoclax or DT2216 treatment, whereas genotype-matched monocytic AML cells were resistant, pointing to a lineage-specific dependency. Indeed, prosurvival factor dependencies are cell context–specific, with the megakaryocytic lineage relying predominantly on BCL-XL45,61,62 for proper maturation.
Importantly, after in vivo treatment with either navitoclax or DT2216, we note a reduction in tumor burden, and a survival benefit in leukemic mice, using 3 CG2 models (mCG2-1, -2, and -6) and a PDX of NUP98r AMKL, the 2 highest fatality AMKL subgroups. The mCG2-6 model resides in a distinct megaerythroid differentiation space (CD41lowCD31+CD151+), reflecting the cellular heterogeneity of CG2 AMKL, yet demonstrates a common BCL-XL dependency across this cellular context. Our findings are aligned with a recent publication reporting that adult leukemia of the megaerythrocytic lineage is sensitive to BCL-XL–specific inhibitor A-1331852, including the HEL erythrocytic line in vivo.49
Expression of BCL2 on its own is not considered a predictor of drug response to venetoclax63 and may explain the relative resistance of CG2 leukemic cells to BCL2 inhibition, as suggested by our work and that of others.64,65 This is an important finding, because venetoclax (potent BCL2 inhibitor) is gaining unprecedented momentum for leukemia treatment,37,38 including in children. Given that CG2 cells do express higher baseline BCL2 levels than NUP98r AMKL and normal MEP/MKPs, combinatorial strategies that trigger apoptosis such as cytotoxic or targeted chemotherapy, in addition to BCL2 inhibition, may ultimately induce apoptosis in CG2 cells that express BCL2, as recently suggested with either azacytidine66 or prosurvival protein myeloid cell leukemia 1 (MCL1) inhibitor S63845.64
Moving forward, our work demonstrates that combining BCL-XL inhibitors (navitoclax or DT2216) and low-dose cytarabine is more potent against AMKL in vivo than either single agent alone, with significant translational potential given the current use of venetoclax and high-dose cytarabine in the treatment of relapsed or refractory pediatric AML.37,66,67 However, thrombocytopenia is a dose-limiting toxicity of navitoclax due to BCL-XL inhibition in the megakaryocytic lineage.68 Platelets express BCL-XL at higher levels than MEP/MKPs (supplemental Figure 16), in line with the rapid drop in platelets observed in patients exposed to navitoclax.69 The BCL-XL degrader DT2216, developed to mitigate the on-target thrombocytopenia of navitoclax, interacts with the Von Hippel-Lindau E3 ligase, which is weakly expressed in platelets. When gauging treatment toxicity in normal cells, we found that DT2216 was less toxic than navitoclax to CB CD34+ cells, underscoring the translational potential of BCL-XL degraders for treatment of hematological malignancies of the megaerythrocytic lineages. Although monotherapy with navitoclax may seem slightly more effective than DT2216 against AMKL in vivo (Figure 7D), the DT2216 intermittent dosing scheme can be further optimized in future studies, and, importantly, both agents are equally effective in combination with cytarabine. Altogether, these preclinical studies suggest that integration of selective and potent BCL-XL inhibitors to existing treatment regiments may improve disease eradication in pediatric AMKL.
Acknowledgments
The authors thank the patients and their families and members of the pediatric Hematology-Oncology Team of CHU Sainte-Justine for their outstanding support; the members of the Quebec Leukemia Cell Bank (BCLQ) and the Hematology-Oncology Division of CHU Sainte-Justine for their contribution to the biobanking initiative; Elie Haddad, Kathie Béland, and the members of the humanized mouse core facility (CHU Sainte-Justine); Ines Boufaied of the flow cytometry facility (CHU Sainte-Justine) for cell sorting; Virginie Saillour, Charles Privé, and René Allard of the Integrated Centre for pediatric clinical genomics at CHU Sainte-Justine for whole-exome sequencing and single-cell RNA sequencing studies; France Léveillé, Frédérique Tihy, Géraldine Mathonnet, and Emmanuelle Lemyre of the Medical Genetics Service at CHU Sainte-Justine for comparative genomic hybridization analysis; Elke Küster-Schöck of the microscopy facility at CHU Sainte-Justine; Raphaëlle Lambert from the Genomics Core Facility (Institute for Research in Immunology and Cancer) for quantitative reverse transcription polymerase chain reaction analysis and RNA sequencing analysis; Simon Mathien and Jean Duchaine from the High Throughput Screening Core Facility (Institute for Research in Immunology and Cancer) for dose-response experiments; Marianne Issac from the Histology Platform (Institute for Research in Immunology and Cancer) for processing of histological samples; Carolina Marmolejo, Anne-Cécile Soufflet, Emma Rose Cheetham, Nadia Emely Chauca Torres, and Françoise Couture for their help with experiments; Sami Ayachi for bioinformatic analysis; Keith Humphries, Donald B. Kohn, and Connie Eaves for providing lentiviral vectors; and Michael A. Lieberman for offering the CHRF-288-11 cell line.
This study was supported by seminal Cole Foundation transition awards (S. Cellot and V.-P.L.), Terry Fox Research Institute new investigator grants, a Canadian Cancer Society Research Institute /Cole Foundation impact grant (S. Cellot and B.T.W.), an operating grant from the Leukemia and Lymphoma Society of Canada (S. Cellot), an operating grant from the Richard and Edith Strauss Foundation (S. Cellot and B.T.W.), an operating grant from the Canadian Institutes of Health Research (S. Cellot and H.B.) (FRN 178326), funding from the CHU Sainte-Justine Foundation (Fonds d’Innovation Thérapeutique), the Fondation Charles-Bruneau (operating grants), and the Canada Foundation for Innovation (John R. Evans Leaders Fund) (S. Cellot). This research work was made possible thanks to the financial support of the Oncopole, which receives funding from Merck Canada Inc, the Fonds de recherche du Québec-Santé (FRQS), Génome Québec, the Cancer Research Society, and the Institute for Research in Immunology and Cancer-Commercialization of Research (B.T.W., F.B., and S. Cellot). V.P.L. is supported by a FRQS Junior 1 Clinical Research Scholarship and by the CHU Sainte-Justine and Charles-Bruneau Foundations. S. Cellot is recipient of a Clinician scientist senior award from the FRQS. V.G., M.R., B.K., and L.T. are recipients of Cole Foundation fellowships. N.E.-H. is supported by a postdoctoral scholarship from the Institute for Data Valorization. The Quebec Leukemia Cell Bank is supported by the Cancer Research Network of the FRQS. The humanized mouse core and the flow cytometry facilities at CHU Sainte-Justine are supported by the Fondation Charles-Bruneau.
Authorship
Contribution: V.G. and M.R. designed and performed the experiments, analyzed the data, generated the figures, and wrote the manuscript; M.B. and S. Cardin led projects, performed experiments, analyzed data, and wrote the manuscript; L.B., L.L., and F.F. assisted with experiments and data collection; N.E.-H. performed bioinformatic analysis, generated figures, and wrote the manuscript; A.R., E.A., P.G., L.J., and S.J. performed bioinformatic analysis; V.L., B.K., and A.F. performed and analyzed single-cell RNA sequencing and wrote the manuscript; V.-P.L. supervised bioinformatics and single-cell experiments and analyses and wrote the corresponding manuscript sections; L.T., L.A., and P.P.R. performed and analyzed the cell surface proteomic experiments; E.B. performed mass spectrometry analysis; B.P. and S.M.S. performed nanopore genome sequencing and bioinformatic analysis under the supervision of M.A.S.; T.A.G. shared the AMKL validation cohort data set; T.H.T., R.S., M.D., H.B., P. Teira, V.-P.L., and S. Cellot oversaw patients and contributed to the biobanking; J.H. supervised the biobanking of patient specimens at the BCLQ; J.C., G.S., F.B., B.T.W., P. Thibault, and H.B. provided guidance in designing the experiments and analyses; S. Cellot conceived the experiments, supervised the biobanking of patient specimens at the BCLQ, and wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sonia Cellot, Pediatric Hematology-Oncology Division, Centre Hospitalier Universitaire (CHU) Sainte-Justine Research Center, 3175 Côte Sainte-Catherine, Room 6-17-007, Montréal, QC H3T 1C5, Canada; email: sonia.cellot@umontreal.ca.
References
Author notes
∗V.G. and M.R. contributed equally to this work.
†B.T.W., V.-P.L., and S.C. are joint senior authors.
The RNA-sequencing data set was deposited in the Gene Expression Omnibus database (accession number GSE209628).
Other data are available on request from the corresponding author, Sonia Cellot (sonia.cellot@umontreal.ca).
The full-text version of this article contains a data supplement.



![Gene expression in CG2 leukemia models correlate with pediatric disease. (A) (Left) Correlation of differential gene expression (log2 fold change [L2FC]) in CG2 AMKL models and patients from our institutional data set (Centre hospitalier universitaire Sainte-Justine [CHUSJ]) compared with a validation data set of pediatric CG2 AMKL (St Jude). Differentially expressed genes (DEGs), defined as |L2FC > 1| and false discovery rate (FDR) q value < .05, common to both data sets are indicated in blue or red and define the CG2 signature (supplemental Table 7). Institutional (Inst) data set: CG2 AMKL models (n = 10) and patient-derived CG2 samples (n = 2) compared with N5A AMKL models (n = 5), patient-derived N5A samples (n = 2), and normal CB CD34+ cells (n = 4). Validation (Val) data set: CG2 AMKL (n = 12) vs other genetic subtypes of AMKL (n = 61) from pediatric patients at diagnosis.9 (Right) Upset plots showing DEGs that are jointly overexpressed (n = 399) or underexpressed (n = 330) in CG2 leukemias, corresponding to blue and red dots in panel A. (B) Hierarchical clustering using the 729 DEGs of the CG2 gene expression signature. (C) Heat map showing protein (log2[mass spectrometry [MS] values, left panels) and messenger RNA (RNAseq; fragments per kilobase of transcript per million mapped reads [FPKM] values, right panels) expression of cell surface markers associated to CG2 leukemia (high expression in mCG2-1 and mCG2-2 leukemia models log2[MS values] ≥ 16 and RNAseq FPKM ≥ 5) and weak/no expression in normal CB CD34+ cells (log2[MS values] < 16 and RNAseq FPKM < 5). Samples were analyzed in triplicates and represented as mean expression (biological triplicates for RNAseq and technical triplicates for proteomic data). For comparison, values for mN5A and pdxNTF are shown alongside mCG2 samples. The St Jude Val cohort RNAseq expression is presented as a separate column. (D) Star plot presenting the adjusted P values of DEGs and differentially expressed proteins (FDR < 0.05) (transcriptome: CHUSJ CG2 vs N5A AMKL and normal CB CD34+; surfaceome: CG2 vs NUP98r and CB CD34+). The scores were calculated by multiplying the algebraic sign (+ or −) of the log2FC, surfaceome or transcriptome, by the corresponding log10(adjusted P value). Significantly upregulated CG2 AMKL–specific surface markers intersecting both data sets are labeled. (E) Validation by flow cytometry of PCDH19 surface expression on CG2 AMKL cells from patient (pCG2-1), model mCG2-1 and M07e cell line. PCDH19 is not expressed on lineage-depleted human CB cells (CB CD34+). Samples were costained with NCAM1 (CD56). (F) Scatterplot representations showing the best pairwise correlations of the 2 sets of cell surface marker genes that select for CG2 genotype in a validation data set of pediatric AMKL (of 8 cell surface marker combinations, see supplemental Figure 13). Values, from top to bottom, represent the global and subtype-specific Kendall rank correlation coefficients. P values: ∗P < .05 and ∗∗P < .005.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/1/10.1182_bloodadvances.2022008899/2/m_blooda_adv-2022-008899-gr2.jpeg?Expires=1765896648&Signature=qEIbPjSnaTBq4Tuy0VDvM8V3tAYSUOXSnR-ZkCjRA4AWWoIl2KqPsVNbYreDw-3wtDYzmSoXBH1orehEzTDhk13nZnEAwQu4WH7~MLjd76DFH-dq263zhSa0VtUXtu2dElwPPE5XQ976MfLIdNB1WdbBtbtWkiEk4Ft2jKWEhEWw5dxporMoDMs0b-QI-SRLDlzLmaqF-EwrOcIhligbio2XzGHnCjzg85BuJbfIzeY3Gd0kypdfHc6VzjFj2-lDMckOUygeVlCRa0QPe-IiZgM6DvpcE33tcSPQ0TD3Y9lLUs4DxOgWcq07Jxj37PoL47V4Xv44Qnwod0NBLYxTyw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![AMKL xenografts depend on prosurvival protein BCL-XL for survival. (A) RNAseq expression values (log2[FPKM]) of selected genes implicated in intrinsic apoptosis pathway in model and patient leukemias from our institutional data set. (B) Crossvalidation of gene expression in a large and published data set of pediatric AMKL.9 (C) Protein levels of BCL-XL (left) and BCL2 (right) in AMKL xenograft cells or normal CB CD34+ cells assessed by intracellular flow cytometry. (D) Apoptosis, assessed by annexin V staining and flow cytometry, (E) relative gene expression detected by quantitative reverse transcription polymerase chain reaction, and (F) prosurvival protein levels quantified by flow cytometry, after short hairpin RNA (shRNA)-mediated KD of prosurvival BCL2 proteins (BCL-XL, BCL2, and BCL-W) in CG2 xenografts (72 hours after infection, 2 selected shRNA per gene). Samples were compared with nontransduced (NT) or cells infected with control shRNA against Renilla (shRenilla). See supplemental Figure 18 for KD studies in mCG2-2 cells. (G) Dose response curves and IC50 values were determined after incubation of AMKL or AML xenografts with DT2216 (BCL-XL proteolysis-targeting chimera) or DMSO for 6 days, followed by viability readout with Cell-Titer Glo. Viability readout was normalized against DMSO for each sample. (H) Amount of apoptosis assessed by annexin V staining and flow cytometry in AMKL xenografts or normal CB CD34+ cells after 72 hours of incubation with DT2216 at 100 nM or 1 μM in comparison with cells treated with DMSO. GATA1s, GATA1 truncation; HOXr, HOX rearrangement; KMT2Ar, KMT2A rearrangement; MFI, mean fluorescence intensity; RBM-MKL, RBM15::MKL1; VHL, von Hippel-Lindau; xAMKL, synthetic model of AMKL; xAML, synthetic model of AML. P values: ∗P < .05; ∗∗P < .005; ∗∗∗∗P < .0001.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/1/10.1182_bloodadvances.2022008899/2/m_blooda_adv-2022-008899-gr5.jpeg?Expires=1765896648&Signature=3Y6dx9Yl3aswNNUmMoJV8LhHAZNZOXJivIMD~qDqWAffbfaploV4InJWtdQo9dAvSrxbpehzBO5LTJr3HtVMJXFsGRF53HkVbJtVsjk4ky6fot9jZlozWtmqlvL2duRc20nFuRjwF-Z~DYbk7i6tzmxpHAsg5HNHMb6ek71J1QGKMqvLwuLL7r5hZgsb33vpPiu9bMMfaiaZxDxSh79QsVRnrLVtQPvJAdvdzgQ-4-rQJUMtr6eXr7yov~NDDqyGO-cWy9vt6LdkomYWPl7p2MnGPJwaPosevSEPrOFclnvxhWdnp1IziHIObo57GRY7J7tnsn-q-u9LT2IYuqCpcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)