Key Points
Clonal heterogeneity occurs especially in patients with extramedullary MM.
Residual disease, secondary cancers, site-specific high-risk clones, and drug susceptibilities are detected by guided biopsies.
Abstract
Spatial heterogeneity is a common phenomenon in metastatic solid tumors and an evolving concept in multiple myeloma (MM). The interplay between malignant plasma cells (PCs) and the microenvironment has not yet been analyzed in MM. For this purpose, we performed bone marrow aspirates and imaging-guided biopsies of corresponding lesions in newly diagnosed MM (NDMM) and relapsed/refractory MM (RRMM) patients. PCs were isolated and subjected to whole-exome sequencing (WES). Non-PCs were studied with next-generation flow (NGF) and T-cell receptor sequencing (TCRseq) to analyze the connection between malignant and nonmalignant cells in the bone marrow and in lesions. Although we observed a strong overlap from WES, NGF, and TCRseq in patients with intramedullary disease, WES revealed significant spatial heterogeneity in patients with extramedullary disease. NGF showed significant immunosuppression in RRMM compared with NDMM as indicated by fewer myeloid dendritic cells, unswitched memory B cells, Th9 cells, and CD8 effector memory T cells but more natural killer and regulatory T cells. Additionally, fewer T-cell receptor (TCR) sequences were detected in RRMM compared with NDMM and healthy individuals. After induction therapy, TCR repertoire richness increased to levels of healthy individuals, and NGF showed more regulatory T cells and myeloid-derived suppressor cells, regardless of depth of response. Clinical significance of imaging-guided biopsies of lesions was demonstrated by detection of monoclonal PCs in patients without measurable residual disease (MRD) in aspirates from the iliac crest as well as identification of secondary primary malignancies in MRD− patients. Furthermore, site-specific clones with different drug susceptibilities and genetically defined high-risk features were detected by our workflow.
Introduction
Chromosomal aberrations and somatic mutations drive the progression from benign plasma cells (PCs) over asymptomatic precursor diseases to symptomatic, and ultimately refractory, multiple myeloma (MM).1,2 Some of the respective genetic alterations are already present at early stages of monoclonal gammopathy of undetermined significance and smoldering MM, suggesting that nonmalignant cells of the tumor microenvironment also contribute to the malignant transformation of benign PCs. This hypothesis was confirmed by a recent single-cell RNA sequencing study showing early immune dysregulation in precursor stages of MM.3 Additionally, targeting the immune system is a cornerstone of MM therapy, and modulating cellular immune response with T-cell engagers or chimeric antigen receptor T cells are emerging concepts in the treatment of MM.4
In solid tumors, there is a well-established interplay between somatic mutations in the coding genome, neoantigen expression, and antitumor immunity.5,6 Furthermore, metastastic solid tumors are comprised of spatially divergent ecosystems, and it has been shown that genomic heterogeneity drives coevolution of site-specific T-cell immune responses.7
Spatial genomic heterogeneity is also an emerging concept in MM.8,9 However, so far, no study examined the interactions between spatially divergent clones and host immune cells in MM. In the current study, we performed bone marrow biopsies from the iliac crest and imaging-guided biopsies of corresponding lesions in patients with newly diagnosed MM (NDMM) and relapsed/refractory MM (RRMM). To decipher genomic heterogeneity of the PC compartment, we performed whole-exome sequencing (WES) on purified PCs from both locations. The non-PC compartment was investigated with next-generation flow cytometry (NGF) to analyze the cellular composition of the bone marrow and lesion microenvironments. T-cell immune response was furthermore studied with T-cell receptor sequencing (TCRseq). Beyond the assessment of spatially divergent PC clones and immune repertoires, analyses were repeated after induction therapy with immunomodulatory drugs (IMiD) in combination with proteasome inhibitors (PI) and steroids.
We demonstrate spatial heterogeneity in patients with extramedullary disease, compromised immunity in relapsed MM, and immunological effects of combination induction therapy. The site-specific detection of emerging heterogeneous clones through imaging-guided biopsies in patients without measurable residual disease (MRD) from regular bone marrow biopsies underlines the prognostic significance of our study.
Patients and methods
Imaging-guided biopsies of osteolytic lesions
Patients with NDMM or RRMM and new positron emission tomography (PET)-positive lesions were discussed in a weekly, multidisciplinary tumorboard. If interventional radiologists (A.B. and R.A.) identified lesions accessible for imaging-guided biopsy, patients were consented for a biopsy of the respective lesion and a regular bone marrow biopsy of the iliac crest. Both interventions were performed under computed tomography (CT) or fluoroscopyguidance in the same session under conscious sedation. The study was approved by the local ethics review board and performed according to the Declaration of Helsinki. The entire study protocol can be found in supplemental Material.
As described previously,9 bone marrow aspirates (15 mL) from the iliac crest and lesion were captured in tubes containing EDTA. To prevent hemodilution of samples, native first pull aspirates (5 mL) were transferred to hematopathology for microscopic evaluation and confirmation of diagnosis as well as fluorescence-activated cell sorting (FACS) for immunophenotyping and assessment of MRD. The remaining 10 mL aspirate was subjected to CD138+ selection using magnetic beads according to manufacturer’s instructions. The CD138+ fraction was afterward analyzed with fluorescence in situ hybridization (FISH) and WES. DNA was extracted from CD138− fraction and was used for TCRseq (supplemental Figure 1). The workflow is summarized in supplemental Figure 1.
FACS
Immunophenotyping on fresh bone marrow aspirates from the iliac crest and corresponding lesions was performed as described previously,10,11 testing epitopes that are summarized in supplemental Table 1 to identify immune cell phenotypes. MRD (threshold 10−5) was assessed as described previously,12 using 3 tubes of 4 fluorochromes per tube, testing for the expression of CD38, CD138, CD45, CD56, CD19, CD117, CD28, and cytoplasmic κ and λ.
FISH
FISH analyses were performed on CD138-purified PCs, counting at least 100 nuclei per sample and using probes for: 1q, 1p, 5q, 9 satellite III, del13q, 15 α satellite, del17p, t(4;14), t(11;14), t(14;16), and breakapart probes for immunoglobulin H (IgH) as well as MYC.
WES
DNA was extracted from frozen PCs using kits according to manufacturer’s instructions (DNeasy kit; Qiagen, Hilden, Germany) for bulk WES. Oral swabs (oragene⋅DNA; DNA genotek, Ontario, Canada) were collected for germline controls. After quality check (Quibit Fluorometric Quantification DNA and RNA Assay kits; Thermo Fisher, MA), samples were subjected to WES. SureSelect XT Low Input Target Enrichment System (Agilent Inc, CA) was used for individual exome capturing of each DNA sample. DNA was sheared using a Covaris S220 (Covaris Inc., MA) followed by end repair, P5 adaptor ligation, and 10 cycles of polymerase chain reaction (PCR) to complete the P7 adapter. Unique dual-indexed libraries were purified with AMPureXP beads (Beckman Coulter, CA) and validated for appropriate size on a Tapestation 4200 DNA1000 screentape (Agilent Inc., CA). The purified library was then hybridized to the SureSelectXT Human All Exon V7 Capture library (Agilent Inc., CA). Afterward, the hybridized regions were bound to streptavidin magnetic beads and washed to remove any nonspecific bound products. Eluted library underwent a second 10-cycle PCR amplification to generate enough material for sequencing. Final libraries were purified, measured by Tapestation 4200 DNA1000 screentape, and quantitated using KAPA qPCR (KAPA Biosystems, Basel, Switzerland). Individual libraries were pooled in equimolar fashion at 2 nM final concentration. Each pool was denatured and diluted to 350 pM with 1% PhiX control library added. The resulting pool was then loaded into the appropriate NovaSeq Reagent cartridge and sequenced on a NovaSeq6000 following the manufacturer's recommended protocol (Illumina Inc., CA).
High quality paired-end reads passing Illumina RTA filter were initially processed against the National Center for Biotechnology Information human reference genome (GRCh37) using publicly available bioinformatic tools13,14 and Picard (http://picard.sourceforge.net/). Putative single-nucleotide variants and insertions and deletions were identified by running the variation detection module of Bambino.15 All putative single-nucleotide variants were further filtered based on a standard set of criteria to remove the following common types of false calls: (1) The alternative allele was present in the matching normal sample, and the contingency between the tumor and normal samples was not statistically significant. (2) The mutant alleles were only present in 1 strand, and the strand bias was statistically significant. (3) The putative mutation occurred at a site with systematically dropped base quality scores. (4) The reads harboring the mutant allele were associated with poor mapping quality. Ambiguous cases were manually inspected to ensure accuracy. Putative insertions and deletions were evaluated by a realignment process to filter out potential false calls introduced by unapparent germline events, mapping artifacts, and homopolymer. The identified somatic mutations were compared with the public human germline databases, including dbSNP,16 1000 Genomes Project,17 and the National Heart, Lung, and Blood Institute’s Exome Sequencing Project, to further exclude remaining germline polymorphisms. All mutations were annotated using ANNOVAR18 with the National Center for Biotechnology Information RefSeq database. Results were visualized with GenVisR in R.19 Overlap between mutations from bone marrow and lesion were compared by calculating Jaccard indices. To identify clinically relevant and druggable mutations, the Drug Gene Interaction Database was queried using the rDGIdb package in R.20
TCRseq
Sequencing libraries were generated using the immunoSeq kit from Adaptive Biotechnologies. Input DNA amounts (1800 ng) were determined using the recommendations in the kit based on the anticipated percentage of T cells for the sample type. The first round of PCR was accomplished using the proprietary PCR primer mix included with the kit. A positive control reaction, provided in the kit, and a negative control reaction were included with each sample batch. The first-round PCR was purified using the PCR Cleanup beads provided in the kit. A second round of PCR was performed to generate uniquely barcoded sequencing libraries utilizing the barcode primer plate included in the kit. Final library purification was done using the PCR Cleanup beads provided. Final libraries were validated for appropriate size on a 4200 TapeStation D1000 Screentape (Agilent Technologies, Inc.). Samples were pooled volumetrically, and the final pool was quantitated using KAPA Biosystems qPCR kit. The pool was denatured and diluted to 1.0 pM with 20% PhiX control library added. The resulting pool was then loaded into a NextSeq500 MidOutput 150 cycle Reagent cartridge and sequenced on a NextSeq500 following the manufacturer's recommended protocol (Illumina Inc.) with 156 Read1 cycles and 15 Index1 cycles utilizing the custom primers included in the immunoSeq kit.
Downstream analyses were performed with immunoSEQ Analyzer from Adaptive biotechnologies and the immunarch R package.21 The number of unique T-cell receptor (TCR) clones in repertoires (richness) was estimated by calculating chao1 indices. Simpson’s D was calculated to compare repertoire diversities. To analyze overlap between TCR repertoires from bone marrow and lesion, the Morisita-Horn Index was calculated. TCR data from 10 healthy individuals between ages 65 and 75 were downloaded from Adaptive biotechnologies public domain (https://clients.adaptivebiotech.com/immuneaccess).
Statistical analyses
Statistical analyses were performed in R. A two-tailed, nonparametric unpaired Wilcoxon rank-sum test was performed to compare independent samples. A paired Wilcoxon rank-sum test was used for comparison of bone marrow and lesion as well as pre- and posttherapy samples in individual patients. P values were considered significant if <.05.
Results
Spatial heterogeneity occurs in patients with extramedullary MM
In total, 18 patients underwent bone marrow aspirates and imaging-guided biopsies of paired MM lesions in a prospective clinical trial. Ten patients had NDMM, and 8 patients had RRMM. Extramedullary disease (EMD), defined by growth without connection to bone or outgrowth from cortical bone, was observed in 4 patients (1 NDMM, 3 RRMM). Table 1 and supplemental Table 2 summarizes patient characteristics of the analyzed cohort. All patients who underwent imaging-guided biopsies were evaluable for the study, and no dropouts occurred due to sample quality. No complications were observed during or after samples acquisition.
Baseline characteristics
| Patient . | Sex . | Disease status . | Subtype . | Location . | EMD . | Histology % PCs . | FACS . | Immunoparesis . | FISH . | |
|---|---|---|---|---|---|---|---|---|---|---|
| % PCs after sorting . | Viability . | |||||||||
| NDMM01 | Male | New | IgG κ | Bone marrow | N | 10 | 96 | 92 | No | Normal |
| L2 | 20 | 96 | 90 | |||||||
| NDMM02 | Male | New | IgG λ | Bone marrow | N | 70 | 97 | 98 | Yes | HD, del13, del15, IgH unknown |
| Iliac crest (L) | 60 | 99 | 94 | |||||||
| NDMM03 | Female | New | λ | Bone marrow | N | 10 | 91 | 97 | Yes | IgH unknown |
| Sacrum | 50 | 78 | 96 | |||||||
| NDMM04 | Female | New | IgG κ | Bone marrow | N | 25 | 95 | 89 | No | HD (both) |
| Iliac crest (R) | 25 | 72 | 92 | |||||||
| NDMM05 | Female | New | IgA κ | Bone marrow | N | 60 | 100 | 100 | Yes | del13, del16, del17, gain8, gain15, t(11;14) |
| T10 | 80 | 100 | 97 | |||||||
| L4 | 80 | 100 | 97 | |||||||
| NDMM06 | Female | New | IgG κ | Bone marrow | N | 30 | 96 | 89 | No | del13, del16, t(11;14) (both) |
| L4 | 30 | 92 | 90 | |||||||
| NDMM07 | Male | New | IgG κ | Bone marrow | N | 95 | 100 | 99 | Yes | del1p, HD |
| Iliac crest (L) | 95 | 100 | 99 | |||||||
| NDMM08 | Male | New | κ | Bone marrow | Y | 4 | 92 | 45 | Yes | Normal (both) |
| Clavicle | 100 | NA | NA | |||||||
| NDMM09 | Female | New | IgG κ | Bone marrow | N | 1 | 77 | 91 | No | Normal |
| L3 | 10 | 79 | 89 | |||||||
| NDMM10 | Female | New | κ | Bone marrow | N | MRD neg | 68 | 44 | No | Normal (both) |
| Iliac crest (R) | MRD pos | 70 | 88 | |||||||
| RRMM01 | Female | Relapse | IgA κ | Skin thigh (R) | Y | 100 | NA | NA | Yes | Normal |
| Scaral lesion | 100 | |||||||||
| RRMM02 | Male | Relapse | IgA κ/λ | Bone marrow | Y | 5 | 91 | 81 | No | gain1, t(MYC) |
| Clavicle (R) | 100 | NA | 92 | |||||||
| RRMM03 | Male | Relapse | λ | Bone marrow | N | 4 | 85 | 71 | No | del13, t(11;14) in OL negative in bone marrow |
| L3 | 50 | 95 | 40 | |||||||
| RRMM04 | Female | Relapse | IgG κ | Bone marrow | N | 79 | 99 | 98 | No | HD (gain1 3 and 4 copies), del4, del13, del 16, t(MYC) (both) |
| T12 | 83 | 98 | 91 | |||||||
| RRMM05 | Male | Relapse | IgG κ | Bone marrow | N | 15 | 97 | 87 | Yes | del16, del17 (both) |
| Iliac crest (R) | 15 | 99 | 90 | |||||||
| RRMM06 | Male | Relapse | κ | Bone marrow | Y | 35 | 99 | 99 | Yes | |
| Paravertebral | 100 | NA | NA | |||||||
| RRMM07 | Male | Relapse | IgD κ | Bone marrow (L) | N | 20 | 96 | 83 | Yes | TP53 del and 1q gain for both |
| Bone marrow (R) | 82 | 96 | 72 | |||||||
| RRMM08 | Male | Relapse | IgG λ | Bone marrow | N | 60 | 87 | 90 | Yes | gain5-17 3-4 copies, gain1 5-7 copies (both) |
| Iliac crest (R) | 60 | 94 | 85 | |||||||
| Patient . | Sex . | Disease status . | Subtype . | Location . | EMD . | Histology % PCs . | FACS . | Immunoparesis . | FISH . | |
|---|---|---|---|---|---|---|---|---|---|---|
| % PCs after sorting . | Viability . | |||||||||
| NDMM01 | Male | New | IgG κ | Bone marrow | N | 10 | 96 | 92 | No | Normal |
| L2 | 20 | 96 | 90 | |||||||
| NDMM02 | Male | New | IgG λ | Bone marrow | N | 70 | 97 | 98 | Yes | HD, del13, del15, IgH unknown |
| Iliac crest (L) | 60 | 99 | 94 | |||||||
| NDMM03 | Female | New | λ | Bone marrow | N | 10 | 91 | 97 | Yes | IgH unknown |
| Sacrum | 50 | 78 | 96 | |||||||
| NDMM04 | Female | New | IgG κ | Bone marrow | N | 25 | 95 | 89 | No | HD (both) |
| Iliac crest (R) | 25 | 72 | 92 | |||||||
| NDMM05 | Female | New | IgA κ | Bone marrow | N | 60 | 100 | 100 | Yes | del13, del16, del17, gain8, gain15, t(11;14) |
| T10 | 80 | 100 | 97 | |||||||
| L4 | 80 | 100 | 97 | |||||||
| NDMM06 | Female | New | IgG κ | Bone marrow | N | 30 | 96 | 89 | No | del13, del16, t(11;14) (both) |
| L4 | 30 | 92 | 90 | |||||||
| NDMM07 | Male | New | IgG κ | Bone marrow | N | 95 | 100 | 99 | Yes | del1p, HD |
| Iliac crest (L) | 95 | 100 | 99 | |||||||
| NDMM08 | Male | New | κ | Bone marrow | Y | 4 | 92 | 45 | Yes | Normal (both) |
| Clavicle | 100 | NA | NA | |||||||
| NDMM09 | Female | New | IgG κ | Bone marrow | N | 1 | 77 | 91 | No | Normal |
| L3 | 10 | 79 | 89 | |||||||
| NDMM10 | Female | New | κ | Bone marrow | N | MRD neg | 68 | 44 | No | Normal (both) |
| Iliac crest (R) | MRD pos | 70 | 88 | |||||||
| RRMM01 | Female | Relapse | IgA κ | Skin thigh (R) | Y | 100 | NA | NA | Yes | Normal |
| Scaral lesion | 100 | |||||||||
| RRMM02 | Male | Relapse | IgA κ/λ | Bone marrow | Y | 5 | 91 | 81 | No | gain1, t(MYC) |
| Clavicle (R) | 100 | NA | 92 | |||||||
| RRMM03 | Male | Relapse | λ | Bone marrow | N | 4 | 85 | 71 | No | del13, t(11;14) in OL negative in bone marrow |
| L3 | 50 | 95 | 40 | |||||||
| RRMM04 | Female | Relapse | IgG κ | Bone marrow | N | 79 | 99 | 98 | No | HD (gain1 3 and 4 copies), del4, del13, del 16, t(MYC) (both) |
| T12 | 83 | 98 | 91 | |||||||
| RRMM05 | Male | Relapse | IgG κ | Bone marrow | N | 15 | 97 | 87 | Yes | del16, del17 (both) |
| Iliac crest (R) | 15 | 99 | 90 | |||||||
| RRMM06 | Male | Relapse | κ | Bone marrow | Y | 35 | 99 | 99 | Yes | |
| Paravertebral | 100 | NA | NA | |||||||
| RRMM07 | Male | Relapse | IgD κ | Bone marrow (L) | N | 20 | 96 | 83 | Yes | TP53 del and 1q gain for both |
| Bone marrow (R) | 82 | 96 | 72 | |||||||
| RRMM08 | Male | Relapse | IgG λ | Bone marrow | N | 60 | 87 | 90 | Yes | gain5-17 3-4 copies, gain1 5-7 copies (both) |
| Iliac crest (R) | 60 | 94 | 85 | |||||||
FLC, free light chain; Hb, hemoglobin; HD, hyperdiploid; L, left, LDH, lactate dehydrogenase; N, no; NA, not available; R, right; Y, yes.
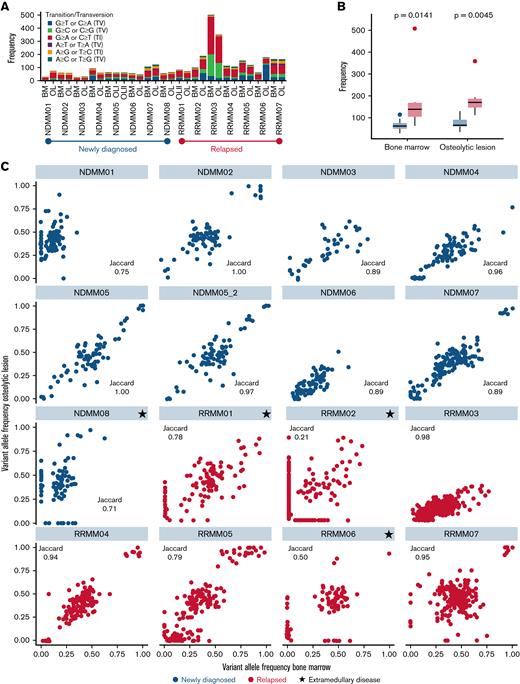
First, we investigated the malignant PC compartment by WES. No significant differences in total number of mutations and transitions/transversions were found between bone marrow and lesions for both NDMM and RRMM (Figure 1A). Significantly more mutations were detected in the bone marrow and lesions from patients with RRMM compared with NDMM (Figure 1B). Particularly in patients with NDMM (8 out of 10), we found a high concordance between mutations in PCs from the bone marrow and lesions with Jaccard indices of ∼0.9 and higher (Figure 1C). The least overlap among newly diagnosed patients was observed in patient NDMM08, who had a history of solitary plasmacytoma and progressed into systemic MM with a new EMD lesion originating from the left humerus and progressive serum free light chains (Figure 2C). Also, in 4 out of 7 patients with RRMM, ∼80% of mutations were shared between locations. Again, the least overlap was observed in patients with EMD (RRMM01, RRMM02, and RRMM06, Figure 1C). The entire list of mutations detected in bone marrow and lesions in NDMM and RRMM can be found in supplemental Material.
WES of the PC compartment from paired samples. (A) Number of total mutations as well as transitions and transversions from bone marrow and lesion (OL) for each individual patient. No significant differences between lesion and marrow were found for number of mutations and transitions/transversions in both NDMM and RRMM. (B) Significantly more mutations were found in patients with RRMM compared with NDMM in both locations. (C) Scatterplots of variant allele frequencies in the bone marrow (x-axis) and lesion (y-axis) in NDMM (blue) and RRMM (red). Jaccard indices were calculated to quantify overlap between paired samples. The largest numbers of unshared mutations were found in patients with EMD (indicated by black stars). BM, bone marrow.
WES of the PC compartment from paired samples. (A) Number of total mutations as well as transitions and transversions from bone marrow and lesion (OL) for each individual patient. No significant differences between lesion and marrow were found for number of mutations and transitions/transversions in both NDMM and RRMM. (B) Significantly more mutations were found in patients with RRMM compared with NDMM in both locations. (C) Scatterplots of variant allele frequencies in the bone marrow (x-axis) and lesion (y-axis) in NDMM (blue) and RRMM (red). Jaccard indices were calculated to quantify overlap between paired samples. The largest numbers of unshared mutations were found in patients with EMD (indicated by black stars). BM, bone marrow.
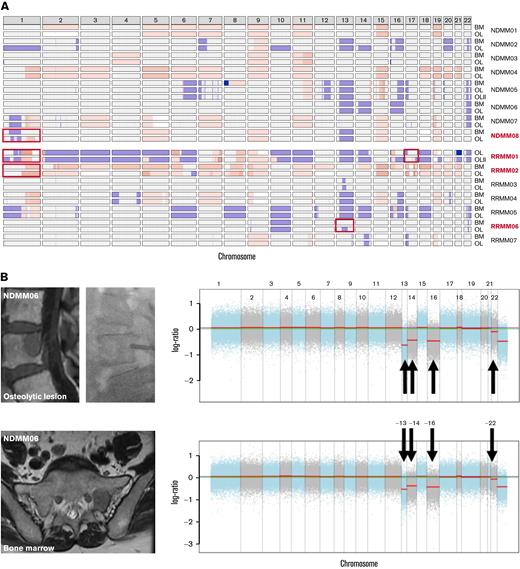
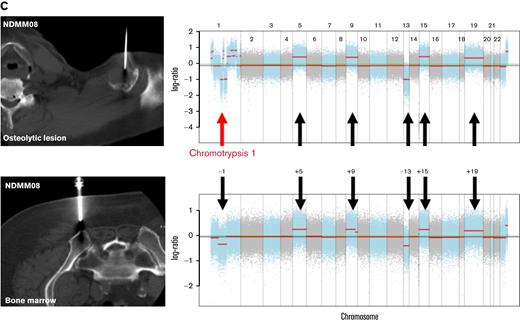
CNVs from WES. (A) Copy number plot using cnSpec from GenVisR19 showing gains (red) and losses (blue) for every chromosome (columns) and location per patient. Although high concordance between both locations was observed in most patients, significant differences (depicted by red boxes) were found in patients with EMD (in red). (B) Example of a newly diagnosed patient (NDMM06) with deletions of chromosomes 13, 14, 16, and 22 that were detected in the bone marrow (lower panel) and the biopsied vertebral lesion (upper panel). (C) Example of a patient with a hyperdiploid karyotype and a history of solitary plasmacytoma who showed progressive disease with a new lesion in the left humerus that extended into the extramedullary space. Although high concordance was observed between gains of chromosomes 5, 9, 15, and 19 as well as deletion of chromosome 13, differences were found for chromosome 1. Although a deletion was detected in the bone marrow (lower panel), oscillating copy numbers suggestive of chromotrypsis of chromosome 1 were assessed in the lesion (upper panel). BM, bone marrow; OL, lesion.
CNVs from WES. (A) Copy number plot using cnSpec from GenVisR19 showing gains (red) and losses (blue) for every chromosome (columns) and location per patient. Although high concordance between both locations was observed in most patients, significant differences (depicted by red boxes) were found in patients with EMD (in red). (B) Example of a newly diagnosed patient (NDMM06) with deletions of chromosomes 13, 14, 16, and 22 that were detected in the bone marrow (lower panel) and the biopsied vertebral lesion (upper panel). (C) Example of a patient with a hyperdiploid karyotype and a history of solitary plasmacytoma who showed progressive disease with a new lesion in the left humerus that extended into the extramedullary space. Although high concordance was observed between gains of chromosomes 5, 9, 15, and 19 as well as deletion of chromosome 13, differences were found for chromosome 1. Although a deletion was detected in the bone marrow (lower panel), oscillating copy numbers suggestive of chromotrypsis of chromosome 1 were assessed in the lesion (upper panel). BM, bone marrow; OL, lesion.
Because chromosomal aberrations are major drivers of disease progression and important prognostic factors in MM, 22 we inferred copy number variations (CNVs) from WES. Although patients with strictly intramedullary growth showed a high concordance between CNVs from bone marrow and lesion (Figure 2A-B), significant differences between both locations were assessed in patients with EMD (Figure 2A,C). Importantly, these changes included chromosomes with prognostic implications like site-specific losses and amplifications of chromosome 1 (NDMM08, RRMM01, and RRMM02) as well as deletion of chromosome 17p (RRMM01).
Immune repertoire in newly diagnosed and relapsed disease
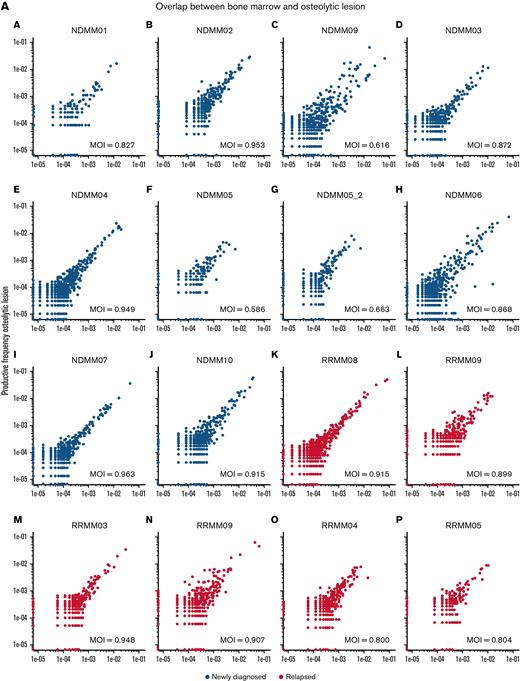
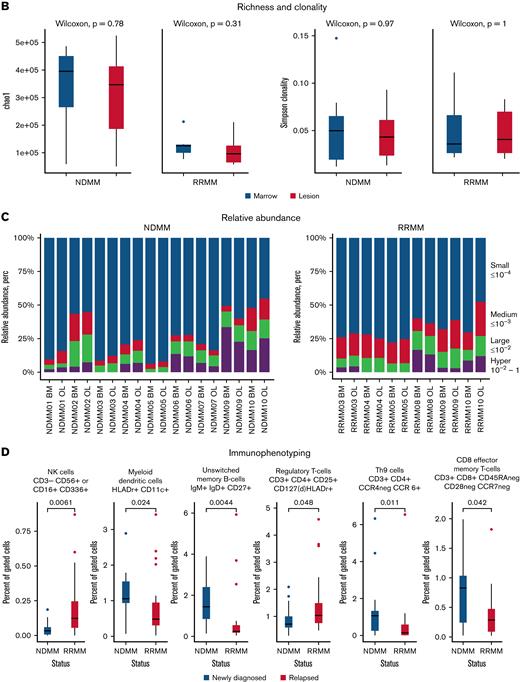
Next, we investigated the immune microenvironment of the bone marrow and MM lesions in newly diagnosed and relapsed disease with TCRseq and NGF. In accordance with results from WES, we found a strong overlap between TCR repertoires from both locations in NDMM and RRMM as indicated by Morisita indices (Figure 3A). Analysis of estimated TCR richness (chao1 estimator) and clonality also showed no significant differences between bone marrow and lesion, regardless of disease status (Figure 3B). Significantly richer TCR repertoires were observed in NDMM compared with RRMM (P = .0031). The majority of TCR clones in NDMM and RRMM accounted for a small proportion of the total number of T cells (≤10−4), and no significant differences in relative abundancies were detected between bone marrow and lesion (Figure 3C).
NGF and TCRseq from paired samples in newly diagnosed and relapsed patients. (A) Scatterplots of productive TCR frequencies in bone marrow (x-axis) and lesions (y-axis) for patients with NDMM (blue) and RRMM (red). Repertoire overlaps were compared by calculating Morisita indices (MOI). In NDMM and RRMM, TCR repertoires showed significant overlap in accordance with findings from WES. (B) Comparison of the actual number of detected TCR sequences (richness estimated by chao1 indices) and clonality (Simpson clonality) showed no significant differences between both locations in NDMM and RRMM. (C) There were also no differences between both locations regarding relative abundance because distributions between lesion and marrow of small (blue), medium (red), large (green), and hyperexpanded (purple) TCR clones were comparable in NDMM and RRMM. (D) Although NGF showed no significant differences between lesion and marrow, patients with RRMM harbored fewer myeloid dendritic cells, unswitched memory B cells, Th9 cells, and CD8 effector memory T cells and more NK cells and regulatory T cells compared with NDMM. This underlines the immunosuppressive capabilities of RRMM.
NGF and TCRseq from paired samples in newly diagnosed and relapsed patients. (A) Scatterplots of productive TCR frequencies in bone marrow (x-axis) and lesions (y-axis) for patients with NDMM (blue) and RRMM (red). Repertoire overlaps were compared by calculating Morisita indices (MOI). In NDMM and RRMM, TCR repertoires showed significant overlap in accordance with findings from WES. (B) Comparison of the actual number of detected TCR sequences (richness estimated by chao1 indices) and clonality (Simpson clonality) showed no significant differences between both locations in NDMM and RRMM. (C) There were also no differences between both locations regarding relative abundance because distributions between lesion and marrow of small (blue), medium (red), large (green), and hyperexpanded (purple) TCR clones were comparable in NDMM and RRMM. (D) Although NGF showed no significant differences between lesion and marrow, patients with RRMM harbored fewer myeloid dendritic cells, unswitched memory B cells, Th9 cells, and CD8 effector memory T cells and more NK cells and regulatory T cells compared with NDMM. This underlines the immunosuppressive capabilities of RRMM.
Immune repertoires were furthermore characterized with NGF. In agreement with WES and TCRseq, no significant differences in the cellular composition of the non-PC compartment in the bone marrow and lesion were found. Compared with NDMM, the bone marrow of RRMM harbored less frequently myeloid dendritic cell, unswitched memory B cells, Th9 cells, and CD8 effector memory T cells but more natural killer (NK) cells and regulatory T cells (Tregs; Figure 3D).
Effects of induction therapy on immune repertoires in NDMM
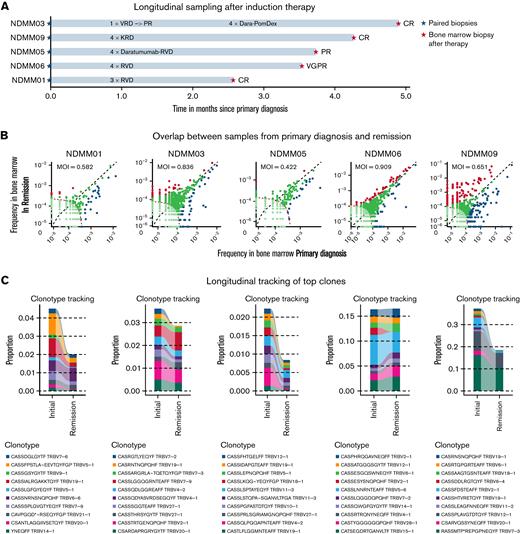
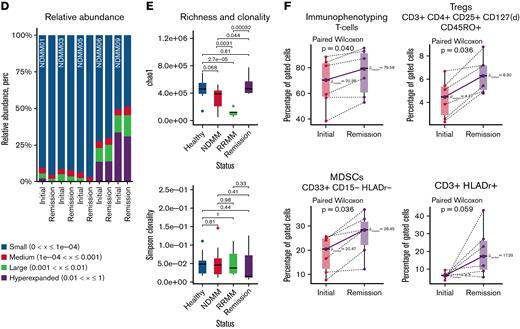
To study the effects of induction therapy on the immune repertoire, longitudinal samples were acquired before and after treatment with an IMiD, PI, and steroids in 5 patients (Figure 4A). Significant expansion of TCR clones after therapy were observed in all 5 patients, regardless of depth of remission after induction therapy (Figure 4B). Less overlap of TCR repertoires between primary diagnosis and remission was observed in 4 out of 5 patients. Longitudinal tracking showed no expansion but reduction of initially present top 10 clones (Figure 4C). No differences in relative TCR abundance were found between both timepoints (Figure 4D). When analyzing TCR repertoire richness and clonality, we found significant differences in richness between healthy individuals and NDMM as well as NDMM and RRMM, whereas no differences in clonality were observed (Figure 4E). After induction therapy, TCR repertoire richness reached levels of healthy individuals (Figure 4E). NGF demonstrated significantly more Tregs and MDSCs after treatment in all patients, regardless of remission (Figure 4F).
Longitudinal analyses after induction therapy. (A) In 5 patients, bone marrow samples were collected after induction therapy. All patients received combination treatment with an IMiD, PI, and dexamethasone. Patient NDMM03 was changed from bortezomib, lenalidomide, dexamethasone (VRD) to daratumumab, pomalidomide, dexamethasone (Dara-PomDex) due to suboptimal response prior to intended stem cell collection. NDMM05 received daratumumab–lenalidomide, bortezomib, dexamethasone (RVD) because of cytogenetically defined high-risk disease (deletion 17p). (B) Scatterplots showing TCR frequencies and repertoire overlap between bone marrow samples at primary diagnosis (x-axis) and after therapy (y-axis). Significant differences of expanded clones were detected at primary diagnosis (blue) and after therapy (red). The patient with the least reduction of M-protein (NDMM05) showed least expanded clones. (C) Longitudinal tracking of top 10 TCR clones at primary diagnosis showed a reduction of clone size in 4 out of 5 patients. (D) No significant changes in relative abundance were observed. (E) Although no differences in TCR repertoire clonality was observed when comparing healthy individuals with NDMM, RRMM, and patients in remission after therapy, repertoire richness as estimated by chao1 indices was significantly lower in RRMM compared with NDMM and healthy individuals. Upon induction therapy, richness reached levels of healthy individuals. (F) Longitudinal immunophenotyping showed more CD3+ cells after induction therapy. Further analysis revealed significantly more CD3+HLADr+ cells and Tregs. Additionally, more myeloid derived suppressor cells (MDSCs) were detected, regardless of depth of response. Because Tregs and MDSCs are associated with immunosuppression in myeloma and were expanded after therapy, it raises the question of whether early changes upon induction therapy are caused by direct antimyeloma effects and not immunomodulation. KRD, carfilzomib/lenalidomide/dexamethasone.
Longitudinal analyses after induction therapy. (A) In 5 patients, bone marrow samples were collected after induction therapy. All patients received combination treatment with an IMiD, PI, and dexamethasone. Patient NDMM03 was changed from bortezomib, lenalidomide, dexamethasone (VRD) to daratumumab, pomalidomide, dexamethasone (Dara-PomDex) due to suboptimal response prior to intended stem cell collection. NDMM05 received daratumumab–lenalidomide, bortezomib, dexamethasone (RVD) because of cytogenetically defined high-risk disease (deletion 17p). (B) Scatterplots showing TCR frequencies and repertoire overlap between bone marrow samples at primary diagnosis (x-axis) and after therapy (y-axis). Significant differences of expanded clones were detected at primary diagnosis (blue) and after therapy (red). The patient with the least reduction of M-protein (NDMM05) showed least expanded clones. (C) Longitudinal tracking of top 10 TCR clones at primary diagnosis showed a reduction of clone size in 4 out of 5 patients. (D) No significant changes in relative abundance were observed. (E) Although no differences in TCR repertoire clonality was observed when comparing healthy individuals with NDMM, RRMM, and patients in remission after therapy, repertoire richness as estimated by chao1 indices was significantly lower in RRMM compared with NDMM and healthy individuals. Upon induction therapy, richness reached levels of healthy individuals. (F) Longitudinal immunophenotyping showed more CD3+ cells after induction therapy. Further analysis revealed significantly more CD3+HLADr+ cells and Tregs. Additionally, more myeloid derived suppressor cells (MDSCs) were detected, regardless of depth of response. Because Tregs and MDSCs are associated with immunosuppression in myeloma and were expanded after therapy, it raises the question of whether early changes upon induction therapy are caused by direct antimyeloma effects and not immunomodulation. KRD, carfilzomib/lenalidomide/dexamethasone.
Clinical significance of imaging-guided biopsies
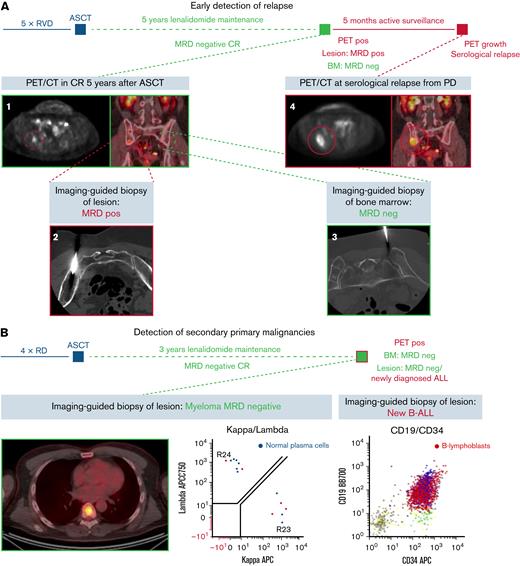
Beyond enabling the evaluation of spatiotemporal immunogenomic heterogeneity, imaging-guided biopsies had direct clinical consequences for the patients enrolled in our study (Figure 5). It still needs to be clarified whether imaging and methods from bone marrow like NGF and next generation sequencing (NGS) provide complimentary or additive information on MRD status. There is evidence that double negativity from both methods is superior to being MRD− only by bone marrow assessment.23,24 Here, we showed differing MRD findings in a patient with a new lesion from PET/CT (Figure 5A). After 5 cycles of RVD and high-dose chemotherapy followed by autologous stem cell transplantation, the patient went into MRD− CR, which was maintained during 5 years of lenalidomide maintenance therapy. Routine follow-up with PET/CT showed a new lesion in the caudal right ilium (Figure 5A1). Although NGF of a bone marrow aspirate from the right iliac crest showed persistent MRD negativity (Figure 5A3), analysis of an imaging-guided biopsy of the PET+ lesion revealed monoclonal PCs (Figure 5A2). Because no other signs for disease activity were found at that timepoint, the patient was actively surveilled. Repeated PET/CT after 5 months showed progression of the respective lesion (Figure 5A4). Serological evaluation confirmed relapse from MRD− CR by reappearance of monoclonal protein, and systemic treatment was reinitiated.
Clinical significance of imaging-guided biopsies. (A) Early, site-specific detection of relapse in a patient without MRD (MRD−) complete response (CR) after 5 cycles of RVD (lenalidomide, bortezomib, dexamethasone), high-dose chemotherapy, and autologous stem cell transplantation (ASCT) followed by 5 years of lenalidomide maintenance. Routine PET/CT showed a small PET+ lesion in the caudal right pelvis (A1). Although flow cytometry showed no signs for MRD in the iliac crest (A3), MRD positivity was confirmed by imaging-guided biopsy of the lesion (A2). Because the patient had no other signs for disease activity, active surveillance was continued. Repeated PET/CT 5 months later showed progression of the biopsied lesion. At that time point, serological relapse was also confirmed, and systemic therapy was initiated. (B) The necessity to confirm findings from PET/CT with biopsies is demonstrated by a case of secondary B-cell acute lymphoblastic leukemia (B-ALL) in a patient in MRD− CR after 4 cycles of lenalidomide and dexamethasone, ASCT, and 3 years of lenalidomide maintenance. Although imaging-guided biopsy of a newly emerged lesion showed no signs for monoclonal PCs, B lymphoblasts were detected. The patient was treated with induction therapy and allogeneic transplantation, and both conditions remain in CR at the moment. (C) To investigate whether mutations from WES are accessible for therapeutic intervention, the Drug Gene Interaction database was queried. More interactions were found in PCs from lesion (OL), which emphasizes that personalized treatment approaches need to be monitored with whole-body imaging. (D) Also, prognostic factors were detected by imaging-guided biopsies (eg, TP53 mutation in a vertebral body of patient RRMM03). Lollipot plot shows all TP53 mutations in the analyzed cohort. Also, mutations associated with drug resistance (eg, PSMC3 associated with resistance to proteasome inhibitors in RRMM05) were detected by imaging-guided biopsies. BM, bone marrow.
Clinical significance of imaging-guided biopsies. (A) Early, site-specific detection of relapse in a patient without MRD (MRD−) complete response (CR) after 5 cycles of RVD (lenalidomide, bortezomib, dexamethasone), high-dose chemotherapy, and autologous stem cell transplantation (ASCT) followed by 5 years of lenalidomide maintenance. Routine PET/CT showed a small PET+ lesion in the caudal right pelvis (A1). Although flow cytometry showed no signs for MRD in the iliac crest (A3), MRD positivity was confirmed by imaging-guided biopsy of the lesion (A2). Because the patient had no other signs for disease activity, active surveillance was continued. Repeated PET/CT 5 months later showed progression of the biopsied lesion. At that time point, serological relapse was also confirmed, and systemic therapy was initiated. (B) The necessity to confirm findings from PET/CT with biopsies is demonstrated by a case of secondary B-cell acute lymphoblastic leukemia (B-ALL) in a patient in MRD− CR after 4 cycles of lenalidomide and dexamethasone, ASCT, and 3 years of lenalidomide maintenance. Although imaging-guided biopsy of a newly emerged lesion showed no signs for monoclonal PCs, B lymphoblasts were detected. The patient was treated with induction therapy and allogeneic transplantation, and both conditions remain in CR at the moment. (C) To investigate whether mutations from WES are accessible for therapeutic intervention, the Drug Gene Interaction database was queried. More interactions were found in PCs from lesion (OL), which emphasizes that personalized treatment approaches need to be monitored with whole-body imaging. (D) Also, prognostic factors were detected by imaging-guided biopsies (eg, TP53 mutation in a vertebral body of patient RRMM03). Lollipot plot shows all TP53 mutations in the analyzed cohort. Also, mutations associated with drug resistance (eg, PSMC3 associated with resistance to proteasome inhibitors in RRMM05) were detected by imaging-guided biopsies. BM, bone marrow.
Imaging-guided biopsies did not only reveal relapse from MRD negativity but also detected secondary primary malignancies (SPM). In a patient treated with 4 cycles of lenalidomide and dexamethasone followed by high-dose chemotherapy and autologous transplant, MRD− CR was maintained with lenalidomide for 3 years until multiple new PET+ lesions occurred. (Figure 5B). NGF showed no signs for monoclonal PCs in bone marrow aspirates from the iliac crest and a biopsied lesion. Although no malignant cells were found in the peripheral blood or the random bone marrow sample, NGF detected B lymphoblasts in aspirates from a PET+ lesion. The patient was treated with induction chemotherapy and allogeneic stem cell transplantation and currently shows no signs for MM or B-cell acute lymphocytic leukemia.
In addition to the detection of relapse from MRD negativity and SPMs, imaging-guided biopsies might have prognostic and predictive value. To investigate whether unshared mutations might be accessible for therapeutic interventions, the Drug Gene Interaction Database was queried for clinically actionable mutations (Figure 5C).20 More interactions were found in lesions compared with the bone marrow, underlining that response to targeted therapies needs to be assessed with whole-body imaging. Furthermore, we showed spatially divergent results for relevant prognostic markers (eg, chromosome 1 aberrations [Figure 2A] and TP53 mutations [RRMM03, Figure 5D]) as well as mutations associated with drug resistance (eg, PSMC3 in RRMM05,25Figure 5D).
Discussion
In the current study, we demonstrated spatial heterogeneity based on WES, especially in MM patients with EMD, as well as increased mutational burden accompanied by immune dysregulation in RRMM compared with NDMM. After IMiD- and PI-based induction therapies, we identified a richer TCR repertoire compared with primary diagnosis and identified subsets of immune cells that are expanded by treatment. Lastly, we showed that imaging-guided biopsies of MM lesions are of diagnostic, prognostic, and predictive relevance because site-specific detection of locoregional MRD and SPM, as well as mutations associated with drug susceptibility and resistance, had direct clinical consequences.
MM is a spatially divergent disease, and findings from whole-body imaging showing focal lesions serve as surrogate for clonal heterogeneity.8 In agreement with a recent study in heavily pretreated patients with MM demonstrating a high mutational load and increased genomic instability,26 we found significantly more mutations in RRMM compared with NDMM. It could have been expected that spatial genomic heterogeneity is more prevalent in patients with higher mutational load. However, based on our previous work, we included additional patients with EMD in the current analysis. Therefore, we were able to show significant spatial heterogeneity in patients with EMD, regardless of mutational load or disease status, and limited spatial heterogeneity in patients with strictly intramedullary MM.
In accordance with the first study that demonstrated spatial heterogeneity especially in larger lesions,8 we found most unshared mutations between the bone marrow and lesions in patients with EMD. MM cells usually reside within the bone marrow and are highly dependent on survival signals from the surrounding tumor microenvironment.27 Once MM cells gain access to the vasculature, extramedullary spread becomes possible and most likely makes them independent from survival signals from bone marrow cells.28 Besides mutations in genes that have been associated with advanced disease like BRAF, NRAS, KRAS, and MYC, we also found additional chromosomal aberrations in extramedullary lesions like amplifications of chromosome 1. In the past, several studies showed an association between high-risk cytogenetics and EMD. However, these analyses were restricted to either bone marrow samples, cells derived from EMD,29 or single case reports.30,31 These studies left the question unanswered of which occurs first: high-risk cytogenetics or EMD. Because we were able to perform biopsies on the same day and showed high-risk features in EMD compared with bone marrow, we provide first evidence that EMD might represent an evolving clone from intramedullary disease. Beyond their biological implications, these findings are of prognostic relevance because the respective changes have been associated with adverse outcome in MM32 and would have been missed by sampling solely from the iliac crest.
Our findings from NGF and TCRseq to study the non-PC compartment are in agreement with data from WES because we found a strong overlap in TCR repertoires and no significant differences between bone marrow and lesion environment. This also supports that intramedullary MM is highly dependent on the interplay with the surrounding host cells, whereas EMD does not rely on supporting signals from the bone marrow microenvironment. This is supported by findings from histology showing subtotal PC infiltration in all patients with EMD in our current study. However, our results on the immune repertoire from TCRseq and NGF from different locations need to be interpreted with caution: Although we aimed at using first-pull aspirates(maximum, 5 mL) for NGF and the CD138− fraction isolated from additional 10 mL aspirate for TCRseq analyses, we cannot rule out contamination of samples by peripheral blood. This could also provide an explanation for the overlapping immune repertoire from both locations and the small proportion of productive TCR clones because TCR sequences could be derived from naïve T cells from contaminating peripheral blood. Therefore, novel techniques like single-cell TCRseq33 and spatial transcriptomics will be implemented to investigate the interface between lesions and the immune system in the future.
Because there is a strong connection between nonsynonymous mutations and expanded TCR clones in solid tumors, like non–small-cell lung cancer,7 we compared TCR repertoires from NDMM and RRMM. In contrast to previous studies in solid tumors, we found no correlation between mutational burden and relative TCR abundance. Compared with RRMM, patients with NDMM harbored fewer mutations and exhibited a richer TCR repertoire. NGF showed significant differences between RRMM and NDMM: fewer effector T cells and unswitched memory B cells and myeloid dendritic cell, but more NK cells and Tregs were observed in RRMM. Immune dysregulation creating an immunosuppressive environment is an established concept in progressive MM,27,34 and increased numbers of Tregs and NK cells have been described during the natural history of the disease with single-cell RNA sequencing.3 This might have clinical implications. Although in some metastatic solid tumors mutational burden can be correlated to response and superior survival after checkpoint inhibition,5,6 studies of PD-1/PD-L1 inhibitors have been underwhelming in MM.35,36 It can be hypothesized that progressive immunoparesis and the missing connection between mutational burden and antitumor immunity might be a reason for the detrimental effects of checkpoint inhibition in MM.
In longitudinal samples after induction therapy with an IMiD, PI, and dexamethasone, we observed a richer TCR repertoire but no significant expansion of distinct TCR clones. In parallel to the spatial analysis of the TCR repertoire, bulk TCR sequencing of naïve T cells from contaminating peripheral blood might have introduced bias into this analysis. However, regardless of depth of response, we observed an expansion of Tregs and MDSCs that have been reported to promote immune evasion.27,34 This raises the question of whether early effects of combination induction therapy are mainly driven by direct antimyeloma effects and not immunomodulation. Future longitudinal analyses after autologous transplantation and maintenance therapy will follow to elucidate this controversial finding. However, this hypothesis is supported by a recent study that showed no beneficial effect of adding elotuzumab to RVD induction therapy.37 Elotuzumab promotes, among others, long-term effects on NK activity in MM.38 To prove our hypotheses on immune dysregulation in RRMM and longitudinal changes after therapy, future in vitro studies exploring the roles of Tregs, MDSCs, and NK cells in the respective patients are warranted.
There is still a controversial discussion of whether findings from whole-body imaging are complimentary to MRD assessment from the bone marrow. Two large prospective trials demonstrated beneficial effects of normalized PET studies in MRD− patients. However, both studies showed discordant PET findings in MRD− patients in up to 25%.23,24 We were able to demonstrate that imaging-guided biopsies are of diagnostic, prognostic, and predictive relevance because we were able to detect MRD as well as SPM in MRD− patients based on PET findings. Our results emphasize that repeated PET/CTs in MRD− patients should be performed (eg, every 12 months) to rule out progression or SPMs.
Lastly, we show that spatial heterogeneity might be the reason for lack of response to targeted approaches. Imaging-guided biopsies revealed PC clones with different drug susceptibilities, like spatially divergent BRAF and RAS mutations. Spatially divergent clones have been described under BRAF inhibition in MM.39 However, in our study, participants were treatment naïve to targeted approaches. This underlines the importance of whole-body imaging and guided biopsies of the respective lesions in patients with residual disease after personalized treatments and MRD− patients.
Taken together, we show spatiotemporal differences of the PC and non-PC compartment, especially in progressive and extramedullary myeloma, with clinical consequences resulting from imaging-guided biopsies of MM lesions.
Acknowledgments
The authors would like to thank their health care assistants Amy McCracken and Mahfoudh Saleh for their support during acquisition of the samples and preparing and caring for our patients. Special thanks to L. Shawn Matott for his continuous support regarding any problems with the High-Performance Computing environment at Roswell Park and the University of Buffalo.
This work is supported by a grant from the German Cancer Aid, covering the salary of M.M., and an Institutional Educational Research Grant from Celgene Corporation. The Genomics, Biostatistics & Bioinformatics, and Flow & Image Cytometry Shared Resources are supported by the National Institutes of Health (NIH) National Cancer Institute (NCI) Cancer Center Support Grant P30-CA16056. H.M. is funded by F32-CA239356. Institutional research support was provided by Celgene.
Authorship
Contribution: M.M., A.M.A.M., P.L.M., and J.H. contributed to conception and design of the study; M.M., A.M.A.M., A.B., R.A., A.W.B., C.R., K.C., H.M., P.K.W., J.T., J.L., S.T.G., P.S., P.L.M., and J.H. contributed to acquisition of data (acquired and managed patients, provided facilities, FISH, flow cytometry, etc); M.M., A.M.A.M., J.W., L.W., Q.H., N.H., M.S., N.M., S.L., P.L.M., and J.H. contributed to analysis and interpretation of data (eg, statistical analysis, biostatistics, and computational analysis); and all authors contributed to writing, review, and/or revision of the manuscript.
Conflict-of-interest disclosure: P.L.M. reports advisory board membership and consulting fees from BlueBird Biotech, Bristol Myers Squibb, Celgene, Fate Therapeutics, Janssen, Juno, Karyopharm, Magenta Therapeutics, Sanofi, and Takeda; and honoraria from BlueBird Biotech, Bristol Myers Squibb, Celgene, Fate Therapeutics, Janssen, Juno, Karyopharm, Magenta Therapeutics, Sanofi, and Takeda. J.H. reports advisory board membership and honoraria from Adaptive, Amgen, Bristol Myers Squibb, Celgene, GlaxoSmithKline, Janssen, Oncotracker, Oncopeptide, Skyline, and Takeda. M.M. reports advisory board membership and honoraria from Amgen, BMS, Celgene, and Takeda. The remaining authors declare no competing financial interests.
Correspondence: Maximilian Merz, Myeloma Service, Department of Medicine, Roswell Park Comprehensive Cancer Center, Elm and Carlton St, Buffalo, NY 14263; e-mail: maximilian.merz@medizin.uni-leipzig.de; and Jens Hillengass, Myeloma Service, Department of Medicine, Roswell Park Comprehensive Cancer Center, Elm and Carlton St, Buffalo, NY 14263; e-mail: jens.hillengass@roswellpark.org.
References
Author notes
Parts of this work were awarded with a Conquer Cancer Award at the 2020 American Society of Clinical Oncology (ASCO) meeting on 29-31 May 2020 and presented at the American Society of Hematology (ASH) meetings on 5-8 December 2020 and 11-14 December 2021.
WES data from 10 patients were part of a previous analysis, including single-cell RNA sequencing.9 Access to WES and TCR raw data as well as R code created for this analysis can be requested from the corresponding author, Maximilian Merz (maximilian.merz@medizin.uni-leipzig.de) and Jens Hillengass (jens.hillengass@roswellpark.org).
The full-text version of this article contains a data supplement.
S.L., P.L.M., L.W., and J.H. share senior authorship.