Key Points
Germ line Runx1 mutations deregulate inflammatory cytokines in bone marrow and predispose to hematologic malignancies.
Runx1R188Q/+ LT-HSCs have competitive advantage in Runx1R188Q/+ recipients, raising concerns on the use of gene-editing corrective therapies.
Abstract
Germ line mutations in the RUNX1 gene cause familial platelet disorder (FPD), an inherited disease associated with lifetime risk to hematopoietic malignancies (HM). Patients with FPD frequently show clonal expansion of premalignant cells preceding HM onset. Despite the extensive studies on the role of RUNX1 in hematopoiesis, its function in the premalignant bone marrow (BM) is not well-understood. Here, we characterized the hematopoietic progenitor compartments using a mouse strain carrying an FPD-associated mutation, Runx1R188Q. Immunophenotypic analysis showed an increase in the number of hematopoietic stem and progenitor cells (HSPCs) in the Runx1R188Q/+ mice. However, the comparison of Sca-1 and CD86 markers suggested that Sca-1 expression may result from systemic inflammation. Cytokine profiling confirmed the dysregulation of interferon-response cytokines in the BM. Furthermore, the expression of CD48, another inflammation-response protein, was also increased in Runx1R188Q/+ HSPCs. The DNA-damage response activity of Runx1R188Q/+ hematopoietic progenitor cells was defective in vitro, suggesting that Runx1R188Q may promote genomic instability. The differentiation of long-term repopulating HSCs was reduced in Runx1R188Q/+ recipient mice. Furthermore, we found that Runx1R188Q/+ HSPCs outcompete their wild-type counterparts in bidirectional repopulation assays, and that the genetic makeup of recipient mice did not significantly affect the clonal dynamics under this setting. Finally, we demonstrate that Runx1R188Q predisposes to HM in cooperation with somatic mutations found in FPDHM, using 3 mouse models. These studies establish a novel murine FPDHM model and demonstrate that germ line Runx1 mutations induce a premalignant phenotype marked by BM inflammation, selective expansion capacity, defective DNA-damage response, and predisposition to HM.
Introduction
Familial platelet disorder with associated hematopoietic malignancy (FPDHM; also called FPDMM, OMIM601399) is a rare, autosomal dominant disorder characterized by life-long thrombocytopenia and autoimmune complications with variable expressivity.1,2 Patients with FPD have high lifetime risk (35%-50%) to HMs with an average age at onset of 33 years (range, 4-741,3). These HMs are predominantly myelodysplastic syndrome (MDS) and acute myeloid leukemia, although cases of lymphoid leukemia and multiple myeloma have also been reported.4-7 FPDHM is caused by germ line mutations in the RUNX1 gene, which encodes the DNA binding subunit of the heterodimeric RUNX1/CBFβ transcription factor.2 This transcription factor has tumor suppressor function and is a frequent target of mutations in a variety of hematologic malignancies.8-10
Runx1 and Cbfβ are essential for the development of embryonic definitive hematopoiesis,11-13 and Runx1 regulates adult hematopoietic differentiation in multiple compartments, including the myeloid, megakaryocytic, and lymphoid lineages.14,15 Hematopoietic Runx1-loss reduces lymphoid differentiation, megakaryocyte maturation, and platelet counts and increases the myeloid progenitor cells.14,16,17 However, it does not affect the frequency of long-term hematopoietic stem cells (LT-HSCs) or induce leukemia in mice.
During the premalignant phase of the disease, patients with FPDHM have higher rate of clonal hematopoiesis than in the general population and a cumulative risk of 80% in having detectable clones with mutations by the age of 50 years.18 These clones may remain stable for years before disease onset, and accumulate somatic mutations, including RUNX1, BCOR, TET2, or in components of signal transduction pathways.4 However, the defects in hematopoietic function during the premalignant period that may predispose to FPDHM are poorly understood. In this study, we combine functional and molecular assays in mice carrying Runx1R188Q germ line mutation, corresponding to the FPDHM-associated pathogenic mutation RUNX1-R201Q, to determine critical alterations in premalignant hematopoiesis.19 We used bidirectional repopulation assays to determine the relative expansion capacity of wild-type (WT) and Runx1R188Q hematopoietic stem and progenitor cells (HSPCs), and the role of the genetic background of the recipient mice in their expansion. Finally, we used 3 mouse models to determine the predisposition to HM in Runx1R188Q/+ mice. These studies demonstrate that Runx1R188Q/+ germ line mutation triggers inflammation in the bone marrow (BM), reduces DNA-damage response (DDR) activity, and predisposes to HM in cooperation with somatic mutations.
Material and methods
Mouse strains
The mice were maintained at the University of Massachusetts Chan Medical School animal facility, which is accredited by the American Association for Laboratory Animal Care. The experiments have been approved by the University of Massachusetts Chan Medical School Institutional Animal Care and Use Committee (protocol #202100194, L.H.C.). Animal procedures adhere to the US Public Health Service Policy on Humanae Care and Use of Laboratory Animals.
Generation of Runx1R188Q/+ (C57BL/6J-Runx1<tm1Lhc>R188Q) mice
To target the Runx1R188Q allele, a mix of ribonucleoproteins (Cas9/R188Q–single-guide RNA) and R188Q-HR oligomer (supplemental Table 1) was microinjected into the pronuclei of fertilized C57BL/6J × C57BL/6N embryos and then surgically transferred into recipient females. Tail DNA of founders was used for the identification of mice with the expected edited allele, using polymerase chain reaction (PCR) amplification or Sanger sequencing. Selected founders were crossed to C57BL/6N mice, and Runx1R188Q/+ F1 progeny were validated by Sanger sequencing and by PCR/Apa1 digestion, as described below and illustrated in supplemental Figure 1A. F1 mice were backcrossed over 10 generations and kept in C57BL/6N strain (Taconic Farms).
Statistical analysis
Standard deviation in scatter plots and in vivo experiments were calculated using GraphPad. Statistical significance was calculated by using unpaired, 2-tailed t test, ∗P < .05 or ∗∗P < .005. Cytokine levels in Runx1R188Q/+ BM were compared with that in the WT by calculating fold change for each replicate over average expression in WT samples. Statistical significance for cytokine profiling and cytokines in peripheral blood was calculated using unpaired, 2-tailed t test. The estimation of median latency of HM and P values were estimated using log-rank test.
Additional material and methods can be found in the supplemental Information.
Results
Runx1R188Q/+ mice expresses normal levels of Runx1R188Q protein in BM progenitor cells
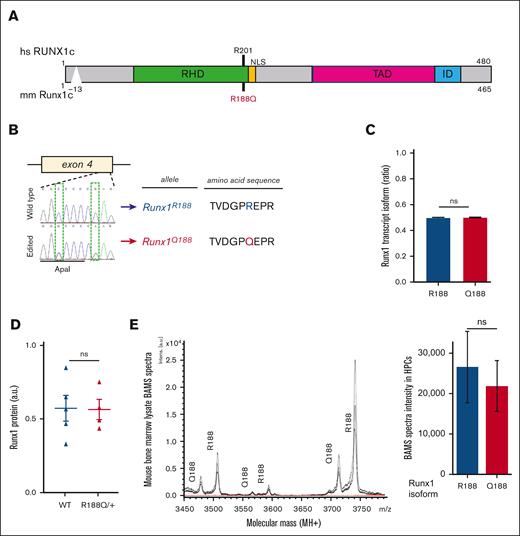
To better understand the role of germ line RUNX1 mutations in premalignant hematopoiesis, we generated a mouse strain with the Runx1R188Q allele. We created a G>A substitution in Runx1 exon 4 that changes the amino acid arginine (R) to glutamine (Q) at position 188, using a CRISPR/Cas9 gene-editing strategy (Figure 1A-B). The murine R188 amino acid, which corresponds to amino acid R201 in human RUNX1c, makes direct contact with a guanine nucleotide in the RUNX1 consensus binding site TGYGGT, and the R188Q mutation abrogates DNA binding activity.20,21 The R201Q missense mutation has been reported as a germ line mutation in FPDHM,2,22 and as a somatic mutation in leukemia.23,24 In addition, we introduced a C>G silent modification at the third position of the codon that encodes for glycine-186 to introduce an Apa1 restriction site and destroy the PAM sequence (Figure 1B). Potential off-target mutations were estimated in F1 tail-snip DNA using CRISPRseek package.25 We found no off-target effects at the 16 predicted loci (considering 1-3 mismatches) when tested by PCR-sequencing (supplemental Table 2). The Runx1R188Q/+ mice were born in mendelian ratios and were healthy, whereas Runx1R188Q/R188Q homozygous were embryonic lethal, as reported in Runx1−/− genotype (supplemental Table 311,12).
Runx1R188Q/+ mice express similar levels of Runx1R188 and Runx1Q188 in hematopoietic cells. (A) Schematic representation of RUNX1 protein indicating human (hs) and mouse (mm) amino acids and the R188Q mutation. The RUNT homology domain (RHD), nuclear localization signal (NLS), transactivation domain (TAD), and the inhibitory domain (ID) are shown. (B.) Sanger sequencing of region surrounding the edited site in exon 4 from WT and Runx1R188Q/+ tail DNA, and respective amino acid sequences. (C) Quantification of Runx1R188 (blue) and Runx1Q188 (red) transcript isoform levels (relative ratio) from Runx1R188Q/+ BM cells as estimated by Illumina sequencing. (D) Quantification of Runx1 protein levels in lysates from Runx1R188Q/+ BM cells, as estimated by western blot densitometric analysis. (E) Spectra analysis (left) and quantification (right) of Runx1 protein isoform levels from Runx1R188Q/+ BM cells, as estimated by Bead Assisted Mass Spectrometry (BAMS). ns, not significant.
Runx1R188Q/+ mice express similar levels of Runx1R188 and Runx1Q188 in hematopoietic cells. (A) Schematic representation of RUNX1 protein indicating human (hs) and mouse (mm) amino acids and the R188Q mutation. The RUNT homology domain (RHD), nuclear localization signal (NLS), transactivation domain (TAD), and the inhibitory domain (ID) are shown. (B.) Sanger sequencing of region surrounding the edited site in exon 4 from WT and Runx1R188Q/+ tail DNA, and respective amino acid sequences. (C) Quantification of Runx1R188 (blue) and Runx1Q188 (red) transcript isoform levels (relative ratio) from Runx1R188Q/+ BM cells as estimated by Illumina sequencing. (D) Quantification of Runx1 protein levels in lysates from Runx1R188Q/+ BM cells, as estimated by western blot densitometric analysis. (E) Spectra analysis (left) and quantification (right) of Runx1 protein isoform levels from Runx1R188Q/+ BM cells, as estimated by Bead Assisted Mass Spectrometry (BAMS). ns, not significant.
Analysis of transcript levels in Runx1R188Q/+ BM cells revealed that Runx1R188 and Runx1Q188 alleles were expressed at similar levels (Figure 1C; supplemental Table 4). The levels of Runx1 protein in WT and Runx1R188Q/+ BM cells were also found to be similar using immunoblotting (Figure 1D; supplemental Figure 1B; supplemental Table 4). We used bead-assisted mass-spectrometry, a bioanalytical method that combines affinity capture with matrix-assisted laser desorption/ionization mass spectrometry, to accurately quantify the expression levels of Runx1R188 and Runx1Q188 isoforms in the hematopoietic cells.26 This analysis confirmed that the expression of Runx1R188 and Runx1Q188 protein isoforms were similar and comparable to the synthetic peptide controls (Figure 1E; supplemental Figure 1C; supplemental Table 5). These results confirm that the R188Q mutation does not alter RUNX1 transcript and protein stability in BM cells, as previously reported in vitro.20
Runx1R188Q/+ BM has increased HSPCs and inflammatory cytokines
Individuals with FPDHM may develop clonal hematopoiesis years before they succumb to HM.3,4 To better understand the hematopoietic alterations caused by germ line RUNX1 mutations, we studied the composition of the hematopoietic progenitor cells in the BM of 12-weeks-old mice. The Runx1R188Q/+ BM showed a significant increase in cellularity (P = .004; Figure 2A), and in the immunophenotypic HSPCs (LKS+; Lineage−, c-kit+, and Sca1+; Figure 2B). This expansion included the LT-HSCs (LKS+CD34−FLT3−) and short-term (ST-HSCs; LKS+CD34+FLT3−) HSCs, as well as in the multipotential progenitor cells (MPPs; LKS+CD34+FLT3+) of Runx1R188Q/+ mice (3.85 × 104 R188Q/+ vs 1.82 × 104 WT cells; P = .001) with a proportional increase in the herein subcompartments (Figure 2C; supplemental Figure 2A). Furthermore, the median fluorescence intensity of Sca1 was significantly increased in Runx1R188Q/+ LKS+ cells, indicating an increase in cell-surface Sca1 protein of HSPCs (Figure 2D). Surprisingly, the expression levels of Ly6a transcript (encoding Sca1 protein) were not changed (Figure 2E), suggesting that this increase may be caused by a posttranscriptional regulatory mechanism.
Analysis of Runx1R188Q/+ BM HSPCs using Sca-1 and CD86 markers and cytokine profiling. (A) BM cellularity of WT and Runx1R188Q/+ mice. (B-C) Immunophenotypic analysis of BM HSPCs using Sca1 and CD86, including LKS+ and LK86+ (B), and LT-HSCs, ST-HSCs, and MPPs (C); (D-E) Quantification of Sca1 protein levels in cell membrane (median fluorescence intensity [MFI], D), and transcript (Ly6a) levels € in LKS+ cells. (F) Representative flow cytometry plots for the analysis of LKS and LK86 marked HSPCs; blue shade represents LK86+ (left panels) and LKS+ (right panels). (G) Relative expression levels of 25 cytokines in BM fluid (Runx1R188Q/+/WT; n = 6 per group). (H-I) Quantification of number of myeloid progenitor cells using Sca-1 (LKS-) and CD86 (LK86-) markers (H), and in common myeloid progenitor (CMPs), GMPs, and MEPs (I). ∗P < .05, ∗∗P < .005; ns, not significant.
Analysis of Runx1R188Q/+ BM HSPCs using Sca-1 and CD86 markers and cytokine profiling. (A) BM cellularity of WT and Runx1R188Q/+ mice. (B-C) Immunophenotypic analysis of BM HSPCs using Sca1 and CD86, including LKS+ and LK86+ (B), and LT-HSCs, ST-HSCs, and MPPs (C); (D-E) Quantification of Sca1 protein levels in cell membrane (median fluorescence intensity [MFI], D), and transcript (Ly6a) levels € in LKS+ cells. (F) Representative flow cytometry plots for the analysis of LKS and LK86 marked HSPCs; blue shade represents LK86+ (left panels) and LKS+ (right panels). (G) Relative expression levels of 25 cytokines in BM fluid (Runx1R188Q/+/WT; n = 6 per group). (H-I) Quantification of number of myeloid progenitor cells using Sca-1 (LKS-) and CD86 (LK86-) markers (H), and in common myeloid progenitor (CMPs), GMPs, and MEPs (I). ∗P < .05, ∗∗P < .005; ns, not significant.
Inflammation has been reported to induce Sca1 expression in LSK− (Lineage−, c-kit+, Sca1−) hematopoietic progenitor cells or increase its expression in LSK+ cells.27,28 To determine whether the observed increase in LKS+ cells results from an inflammation-mediated increase in Sca1, we reanalyzed this compartment by replacing Sca1 with CD86 (Figure 2F), a marker expressed in the HSPCs and that is not affected by inflammation.29 The fraction of LK86+Runx1R188Q/+ HSPCs was significantly increased when compared with that of WT group (Figure 2B-C), although at significantly lower levels than when compared with Sca1, indicating that the Sca-1 increase in HSPCs may be driven by inflammation in this context and suggesting that the Runx1R188Q/+ mutation may increases immunophenotypic HSPCs through inflammation-dependent and -independent mechanisms. Furthermore, the median fluorescence intensity of CD48, an inflammation-response cell-surface marker regulated in Runx1-knockout mice,16,30 was increased in Runx1R188Q/+ ST-HSCs and MPPs (supplemental Figure 2B), supporting the hypothesis that Runx1R188Q/+ HSPCs are modulated by deregulated inflammation in the BM.
To test whether the Runx1R188Q/+ BM has an inflammatory microenvironment, we quantified the levels of 36 inflammatory cytokines in the BM serum of WT and Runx1R188Q/+ mice (n = 6 per group). The expression of most cytokines detected was deregulated two- to sixfold (Figure 2G; supplemental Table 6), predominantly in cytokines regulated by interferon and tumor necrosis factor α pathways. These include significant increase in chemokines Cxcl10/IP-10 and Ccl5/Rantes (XCxcl10: 2.4 and XCcl5: 1.9-fold-increase, respectively, P < .05), known to influence HSC differentiation, promote hematopoietic regeneration, and cause myeloid bias in mice.31,32 Our results indicate that the germ line Runx1-R188Q mutation induce low-dose inflammation in the murine BM.
The immunophenotypic analysis of LKS− (lineage−, c-kit+, Sca-1−) cells revealed a significant increase in common myeloid and granulocytic-monocytic (GMP) progenitor cells in Runx1R188Q/+ mice (Figure 2H-I), indicating that the Runx1-R188Q–mediated expansion of premyeloerythroid progenitor cells in the BM is independent of changes in Sca-1 levels.
Runx1R188Q/+ hematopoietic progenitor cells have enhanced myeloid progenitors and defective DDR
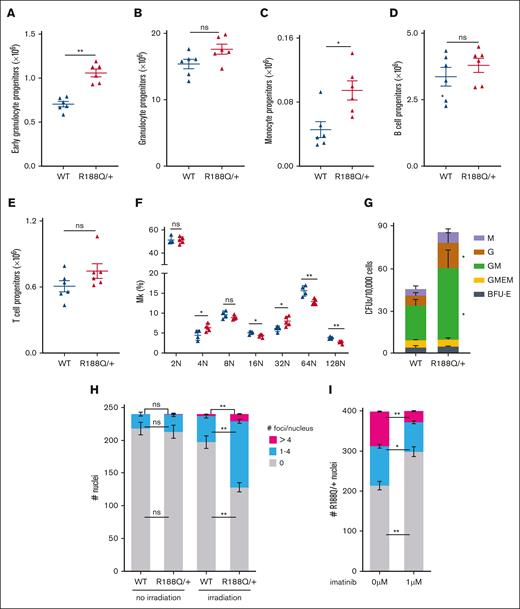
The increase in LKS− cells, prompted us to analyze the frequency of GMP-derived progenitors in the BM. Immunophenotypically, the early granulocyte progenitor (Mac1+, Gr-1low) and monocyte progenitor (Gr1−Mac1+) cells were significantly increased, albeit no changes were observed in the granulocyte progenitor (Gr1+Mac1+) cells (Figure 3A-C; supplemental Figure 3A). Conversely, the frequency of lymphoid B–cell and T-cell progenitor cells was similar in both genetic groups (Figure 3D-E). The analysis of megakaryocyte progenitors using Cd41 and Hoechst staining revealed that a heterozygous Runx1R188Q mutation causes a modest but significant reduction in megakaryocytic maturation, with increase in 4N and 32N in detriment of mature forms (P < .01; Figure 3F; supplemental Figure 3B). Functionally, the colony-forming unit capacity of Runx1R188Q/+ BM cells increased 100% (Avewt = 45/1 × 104 BM cells; AveR188Q/+ = 85/1 × 104 BM cells; P = .019), caused by an increase in granulocyte, monocyte, and granulocyte/monocyte colonies (Figure 3G).
Analysis of hematopoietic progenitor cells in Runx1R188Q/+ BM. (A-C) Flow cytometry analysis of WT and Runx1R188Q/+ BM cells for early (Mac1+Gr1low, A) and late (Mac1+Gr1+, B) granulocytic progenitor cells, and monocytic progenitor cells (C). (D) Colony-forming unit (CFU) assay using BM cells from WT and Runx1R188Q/+ mice. Graph depicts monocyte (M), granulocyte (G), granulocyte/monocyte (GM), granulocyte/monocyte/erythroid/megakaryocytic, and burst forming colony (BFU) -erythroid colonies. (E-F) Flow cytometry analysis for B and T cells in WT and Runx1R188Q/+ BM cells. (G) Ploidy analysis in megakaryocytes of WT and Runx1R188Q/+ BM. (H-I) Quantification of nuclei with 53bp1-positive foci in WT and Runx1R188Q/+ Lindep BM cells, 2 hours after 2 Gy ionizing radiation (H), and pretreated for 1 hour with imatinib (I). ∗P < .05, ∗∗P < .005; ns, not significant.
Analysis of hematopoietic progenitor cells in Runx1R188Q/+ BM. (A-C) Flow cytometry analysis of WT and Runx1R188Q/+ BM cells for early (Mac1+Gr1low, A) and late (Mac1+Gr1+, B) granulocytic progenitor cells, and monocytic progenitor cells (C). (D) Colony-forming unit (CFU) assay using BM cells from WT and Runx1R188Q/+ mice. Graph depicts monocyte (M), granulocyte (G), granulocyte/monocyte (GM), granulocyte/monocyte/erythroid/megakaryocytic, and burst forming colony (BFU) -erythroid colonies. (E-F) Flow cytometry analysis for B and T cells in WT and Runx1R188Q/+ BM cells. (G) Ploidy analysis in megakaryocytes of WT and Runx1R188Q/+ BM. (H-I) Quantification of nuclei with 53bp1-positive foci in WT and Runx1R188Q/+ Lindep BM cells, 2 hours after 2 Gy ionizing radiation (H), and pretreated for 1 hour with imatinib (I). ∗P < .05, ∗∗P < .005; ns, not significant.
The hematopoietic progenitor cells derived from induced-pluripotent stem cells of a patient with FPDHM with the RUNX1R201Q mutation showed reduced DDR in vitro.33 Similarly, we found that the Runx1R188Q/+ BM hematopoietic progenitor cells showed a reduced DDR after irradiation, with a significant accumulation of 53bp1-positive foci in Runx1R188Q/+ nuclei, suggesting that the activity of DNA-damage repair complexes is sensitive to Runx1 dosage (Figure 3H; supplemental Figure 3C). Furthermore, because Runx1 function depends on the balance between active and inactive Runx1 proteins, and inhibition of Abelson nonreceptor tyrosine kinase (ABL) can activate RUNX1 by dephosphorylation of tyrosine residues at the C-terminal inhibitory domain,34,35 we estimated the number of foci-positive nuclei in Runx1R188Q/+ cells pretreated with the ABL inhibitor, imatinib. The number of nuclei with unresolved foci was restored to levels similar to that of control group (Figure 3I), indicating that RUNX1-mediated regulation of DDR is regulated by the tyrosine kinase ABL.
Runx1R188Q/+ mice have mild leukopenia and platelet dysfunction
The alterations observed in the BM cells prompted us to evaluate the composition of peripheral blood leukocytes in Runx1R188Q/+ mice. The total count of white blood cells in circulation was significantly reduced (Figure 4A), primarily caused by a significant reduction in B cells and T cells (Figure 4B-C). In addition, a trend to reduced neutrophils and monocytes was also evident, although it was statistically not significant (Figure 4D-F).
Runx1R188Q/+ peripheral blood cells have reduced lymphocytes and platelets activity. (A-F) Quantification of WBCs (A), B cells (B), and T cells (C), c-kit (D), neutrophils (Gr1+Mac+, E), monocytes (Mac1+Gr1−, F) by flow cytometry. (G-J) Quantification of platelet activity after thrombin activation, including levels of fibrinogen receptor (CD41/61) in membrane (G), translocation of P-selectin from alpha granules (H), mepacrine uptake by dense granules (I), and serotonin release from dense granules (J). ∗P < .05, ∗∗P < .005; ns, not significant.
Runx1R188Q/+ peripheral blood cells have reduced lymphocytes and platelets activity. (A-F) Quantification of WBCs (A), B cells (B), and T cells (C), c-kit (D), neutrophils (Gr1+Mac+, E), monocytes (Mac1+Gr1−, F) by flow cytometry. (G-J) Quantification of platelet activity after thrombin activation, including levels of fibrinogen receptor (CD41/61) in membrane (G), translocation of P-selectin from alpha granules (H), mepacrine uptake by dense granules (I), and serotonin release from dense granules (J). ∗P < .05, ∗∗P < .005; ns, not significant.
Patients with FPDHM frequently have mild to moderate thrombocytopenia and prolonged bleeding, caused by reduced platelet function.36 These platelets are typically of normal size but with reduced granules and defective aggregation capacity. We found that the number and size of platelets in Runx1R188Q/+ mice were not significantly changed, albeit a trend to reduced numbers was observed (supplemental Figure 4A-B). The platelet function was significantly reduced, as measured by fibrinogen receptor activation (CD41/CD61) in membrane after thrombin treatment (Figure 4G). Similarly, the platelets were dysfunctional, as evidenced by a significantly reduced translocation of P-selectin by the α-granules, and mepacrine retention or serotonin secretion by the dense granules (Figure 4H-J). These phenotypes parallel the defects found in platelets from patients with FPDHM and show that Runx1R188Q/+ mice have reduced megakaryocyte maturation and platelet function.37
The Runx1R188Q/+ myeloid and lymphoid progenitors have higher engraftment capacity
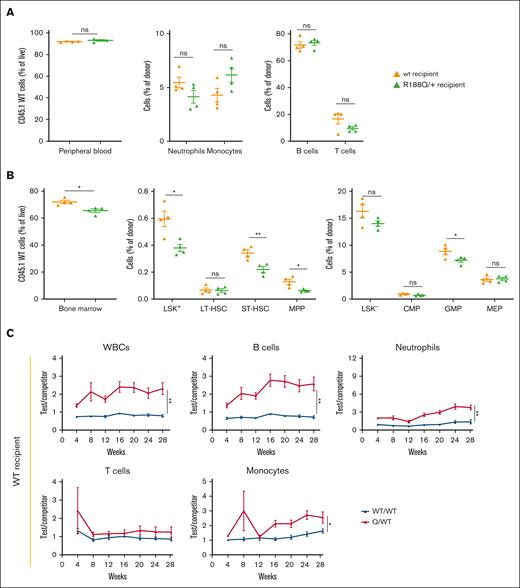
To understand whether Runx1-R188Q expressing nonhematopoietic cells regulate HSPC differentiation, we tested the long-term repopulation capacity (LT-RC) of WT BM cells transplanted into WT or Runx1R188Q/+ recipient mice, using a bidirectional noncompetitive repopulation assay (supplemental Figure 5A). Time-course analysis (4-24 weeks) of peripheral blood leukocytes revealed that WT donor cells had similar contribution in both recipient genotypes (Figure 5A; supplemental Figure 5B). The HSPC analysis at week 24 revealed no changes in LT-HSCs, indicating that the engraftment and LT-RC of LT-HSCs is not affected by the genotype of the recipient. However, the ability of LT-HSCs to differentiate to ST-HSCs, MPPs, and GMPs was significantly reduced in the Runx1R188Q/+ recipient mice (Figure 5B), suggesting that WT LT-HSCs have defective differentiation capacity when transplanted in Runx1R188Q/+ recipient mice.
Bidirectional repopulation assays of RunxR188Q/+ hematopoietic cells. (A-B) Analysis of contribution of WT donor cells (CD45.1) to peripheral blood (A) and BM (B) after 24 weeks, into WT (orange) or RunxR188Q/+ (green) recipient mice (CD45.2), using bidirectional noncompetitive repopulation assays. (C-D) Time-course analysis of donor cell contribution (test/competitor: WT/WT in blue, Runx1R188Q/+ /WT in red) to peripheral blood in wt (C) and Runx1R188Q/+ (D) recipient mice, in bidirectional competitive repopulation assay (BCRA). ∗P < .05, ∗∗P < .005; ns, not significant.
Bidirectional repopulation assays of RunxR188Q/+ hematopoietic cells. (A-B) Analysis of contribution of WT donor cells (CD45.1) to peripheral blood (A) and BM (B) after 24 weeks, into WT (orange) or RunxR188Q/+ (green) recipient mice (CD45.2), using bidirectional noncompetitive repopulation assays. (C-D) Time-course analysis of donor cell contribution (test/competitor: WT/WT in blue, Runx1R188Q/+ /WT in red) to peripheral blood in wt (C) and Runx1R188Q/+ (D) recipient mice, in bidirectional competitive repopulation assay (BCRA). ∗P < .05, ∗∗P < .005; ns, not significant.
We functionally studied the LT-RC of Runx1R188Q/+ HSPCs, using a bidirectional competitive repopulation assay (supplemental Figure 5C). The white blood cell count in Runx1R188Q/+ test group displayed a statistically significant growth advantage from the early time point after engraftment in both recipient mice, which was sustained for 28 weeks (Figure 5C-D; supplemental Figure 5D). The observed increase was primarily caused by the expansion of Runx1R188Q/+ B cells, neutrophils, and monocytes, whereas the contribution of T cells was unchanged.
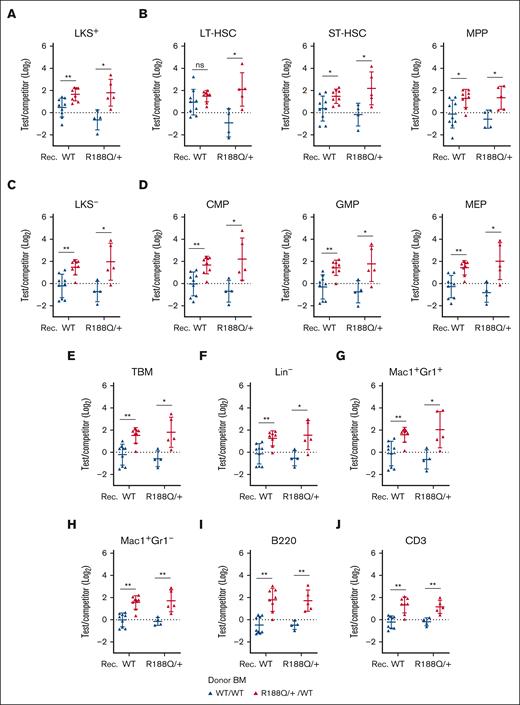
Analysis of LT-RC at week 28 in the BM revealed that the Runx1R188Q/+ LKS+ cells outcompeted WT cells in both recipient groups (Figure 6A). Whitin this compartment, the Runx1R188Q/+ LT-HSCs were increased in the Runx1R188Q/+ but not in the WT recipients, suggesting a differential engraftment capacity or that Runx1R188Q/+ recipient–derived cell signals may drive HSC function. Notably, this increase correlated with the initiation of the differentiation program (ST-HSCs and MPPs; Figure 6B). Analysis of the LKS− compartment confirmed myeloexpansion of Runx1R188Q/+ progenitor donor cells in both recipient genotypes (Figure 6C-D). Furthermore, the Runx1R188Q/+ BM late progenitor cells were also expanded in the lymphoid and myeloid compartments (Figure 6E-J).
Runx1R188Q/+ BM hematopoietic progenitor cells outcompete WT cells in BCRAs. (A-D) Contribution of donor (test/competitor: WT/WT in blue, Runx1R188Q/+ /WT in red) HSPCs, including LKS+ (A), LT-, ST-HSCs and MPPs (B), LKS- (C), and committed progenitor cells (CMPs, GMPs, and megakaryocytic-erythroid progenitors (MEPs), D). (E) estimation of total BM cells. (F-J) Contribution of donor (test/competitor) cells to committed progenitor cells in BM, including lineage-negative (Lin−) cells (F), neutrophil- (G) and monocyte-progenitors (H), B cells (I) and T cells (J). ∗P < .05, ∗∗P < .005.
Runx1R188Q/+ BM hematopoietic progenitor cells outcompete WT cells in BCRAs. (A-D) Contribution of donor (test/competitor: WT/WT in blue, Runx1R188Q/+ /WT in red) HSPCs, including LKS+ (A), LT-, ST-HSCs and MPPs (B), LKS- (C), and committed progenitor cells (CMPs, GMPs, and megakaryocytic-erythroid progenitors (MEPs), D). (E) estimation of total BM cells. (F-J) Contribution of donor (test/competitor) cells to committed progenitor cells in BM, including lineage-negative (Lin−) cells (F), neutrophil- (G) and monocyte-progenitors (H), B cells (I) and T cells (J). ∗P < .05, ∗∗P < .005.
Runx1R188Q predisposes to hematologic malignancies
To gain insights in the predisposition of Runx1R188Q/+ in HMs, we studied 3 experimental in vivo models. Considering that loss of the second RUNX1 allele is a somatic mutation found in a fraction of FPDHM,38 we first determined the HM latency in mice carrying the R188Q germ line mutation and the hematopoietic loss (Δ) of the second allele (Runx1R188Q/Δ). The Runx1R188Q/Δ mice succumbed to a variety of HMs with full penetrance (median latency, 37 weeks; Figure 7A). The pathology of disease was of predominantly MDS and myeloproliferative neoplasm (MDS/MPN), mixed with leukemic cells. Common features of MDS/MPN in the BM included hypercellular marrow, composed of myeloid dominant hematopoiesis, and some cases with reduced megakaryocytes with dysplastic forms. In addition, the BM presented scattered hemo or erythrophagocytotic macrophages, evidencing myeloid and erythroid progenitor cells with increased stress (Figure 7B). Frequently (4/6 cases), MDS/MPN cells were mixed with myeloid leukemia (ML) cells with different levels of leukemic blasts, as evidenced by histology analysis and functionally by the leukemia latency in secondary transplantation assays (supplemental Figure 7A). These mice showed splenomegaly with reduced red pulp and predominant infiltration by immature hematopoietic cells. Finally, 1 of 6 mice succumbed to T-cell leukemia/lymphoma, with thymoma with a predominant population of blasts with scant cytoplasm. Conversely, practically all control mice, either lacking 1 copy of Runx1 in the hematopoietic cells (Runx1+/Δ) or with the R188Q germ line mutation (Runx1R188Q/+) remained healthy for 78 weeks (experimental end point; Figure 7A), with the exception of 1 Runx1R188Q/+ mouse that developed lymphoma at 76 weeks. The Runx1R188Q/+mice remained healthy, and analysis at experimental end point revealed increased myeloid progenitors and hypolobulated megakaryocytes in spleen and BM, suggesting a progressive myeloproliferative phenotype.
Runx1R188Q/+ predisposes to a variety of hematologic malignancies in cooperation with other somatic mutations. (A) Kaplan-Meier survival curve of WT, Runx1R188Q/+, Runx1Δ/+, and Runx1R188Q/Δ mice. (B) Histology analysis of BM and peripheral blood of Runx1R188Q/Δ mice with HM. (C) Kaplan-Meier survival curve of mice with Runx1 mutations and NrasG12D/+ allele. (D) Histology analysis of BM, and thymus of Runx1R188Q/ΔNrasG12D/+ mice with HM. (E) Kaplan-Meier survival curve of mice treated with ethyl-nitrosourea (ENU). (F) Histology analysis of BM of ENU-treated Runx1R188Q/Δ mice.
Runx1R188Q/+ predisposes to a variety of hematologic malignancies in cooperation with other somatic mutations. (A) Kaplan-Meier survival curve of WT, Runx1R188Q/+, Runx1Δ/+, and Runx1R188Q/Δ mice. (B) Histology analysis of BM and peripheral blood of Runx1R188Q/Δ mice with HM. (C) Kaplan-Meier survival curve of mice with Runx1 mutations and NrasG12D/+ allele. (D) Histology analysis of BM, and thymus of Runx1R188Q/ΔNrasG12D/+ mice with HM. (E) Kaplan-Meier survival curve of mice treated with ethyl-nitrosourea (ENU). (F) Histology analysis of BM of ENU-treated Runx1R188Q/Δ mice.
Considering that somatic mutations in components of signaling transduction pathways are frequently found in FPDHM,4 we combined the Runx1 alleles with the NrasLSLG12D conditional knock in allele as a second approach. The 5 NrasLSLG12D/+Runx1R188Q/Δ mice succumbed to HM with a short median latency (14.7 weeks; Figure 7C). Four mice presented MDS/MPN pathology mixed with ML, as described above. One case showed T-cell leukemia, marked by anemia, and enlarged thymus and lymph nodes, with a monotonous population of blasts with scant cytoplasm, multiple mitotic forms and apoptotic bodies (Figure 7D). The NrasLSLG12D/+Runx1R188Q/+ group succumbed to HM (median latency, 54 weeks), 50% of which showed T-cell leukemia and 50% with MDS/MPN phenotype.
Lastly, we evaluated HM latency by inducing mutations with the chemical mutagen ethyl-nitrosourea as previously described.39 The Runx1R188Q/Δ mice (n = 4) succumbed to HM with a median latency of 16.1 weeks and complete penetrance (Figure 7E). Their pathology included anemia, splenomegaly with increased c-kit+/Mac1+Gr1+ immature cells, and a predominant dysplastic hypolobulated morphology in the BM (Figure 7F). The Runx1R188Q/+ mice (n = 11) succumbed to MDS/MPN or MDS/MPN with overlapping acute ML with a pathology similar to that of Runx1R188Q/Δ group, and a median latency of 48.1 weeks and complete penetrance.
The WT group succumbed to disease with a median latency of 61.7 weeks, marking the background disease pathology caused by ethyl-nitrosourea. This included lymphoid disease in 60% (3/5) of the mice, with enlarged thymus and increased T-cell progenitors in the BM. The remaining 40% (2/6) mice showed solid tumors in the liver, lymphopenia, and splenomegaly.
In sum, these studies demonstrate that the Runx1-R188Q mutation predisposes mice to HM, and that the addition of “cooperating” mutations, such as the loss of the second Runx1 allele and/or Nras-G12D can accelerate HM transformation in mice. In addition, these studies validate that Runx1 germ line mutations can predispose to a variety of HMs, as found in patients with FPD.
Discussion
Individuals with FPDHM frequently show clonal expansion of premalignant cells preceding HM onset. Despite the extensive studies of RUNX1 function in hematopoiesis, the alterations caused by germ line RUNX1-mutations in the premalignant BM are not well-understood. In this study, we investigated the role of the FPDHM-associated Runx1R188Q mutation in hematopoietic function using immunophenotypic and functional assays. We discovered that Runx1R188Q/+ remains healthy but with deregulated inflammatory cytokines and reduced DNA-damage repair capacity in the BM. Runx1R188Q/+ mice have increased LT-RC and predispose to HM in cooperation with somatic mutations.
The proinflammatory phenotype in Runx1R188Q/+ mice was marked by a low-dose deregulation of inflammation-associated cytokines in the extracellular fluid and the increase of cell-surface proteins Sca-1 and CD48 of HSPCs, in line with these results, the immunophenotypic increase in LKS+ cells has been reported in Runx1+/− and in hematopoietic Runx1-null mice (40,41). It is possible that the apparent increase in HSPCs may results in the mutant Runx1–mediated increase in Sca-1 expression of Sca1-low/negative hematopoietic progenitor cells. Of note, Sca1 can mediate inflammation-induced HSC proliferation and differentiation,42,43 suggesting that its upregulation may have functional effects in the Runx1R188Q/+ HSPCs. In addition, Runx1-null GMP/granulocytic progenitor cells have a hypersensitive inflammatory response to acute stress (eg, lipopolysaccharide treatment) via tumor necrosis factorα/nuclear factor κB pathway,44 and Runx1 regulates dendritic cell differentiation.45 However, the role of RUNX1 mutations in the expression and secretion of cytokines and chemokines in other cell types is poorly understood.
Healthy patients with FPDHM show variable platelet counts but frequently within the normal range.4 The platelet counts in mice depends on the levels of Runx1 expression, because they are within the normal range, Runx1 heterozygous mice and significantly reduced in Runx1-null mice.14,17 The Runx1R188Q/+ mice have a modest reduction in megakaryocyte maturation, and relatively normal platelet counts. Functionally, however, Runx1R188Q/+ platelets have a defective activation response to thrombin treatment. This defect was evidenced by the reduced levels of activated fibrinogen receptor in membrane, of serotonin release by the dense granules and of P-selectin translocation from the alpha granules to the membrane. These defects correlate with the deficiencies reported in platelets of patients with FPDHM37,46-48 and highlight that platelet function is highly sensitive to RUNX1 expression levels.
The functional analysis in transplantation assays demonstrated that Runx1R188Q/+ HSPCs have higher LT-RC than WT counterparts. Accordingly, a recent study reported the engraftment of RUNX1-edited HSPCs in rhesus macaques,49 underscoring the role of Runx1 mutations in preleukemic expansion and highlighting the concerns on the use of potential gene-editing therapies in premalignant FPDHM HSCs. In addition, this analysis reveals that Runx1R188Q/+ HSPCs have selective expansion in both genetic backgrounds, albeit the differences seen in HSPC compartment, indicating that nonhematopoietic cells have negligible impact on the long-term expansion of Runx1-mutant HSPCs. Interestingly, Runx1-loss in BM mesenchymal stem cells, which secrete cytokines and chemokines in the BM, did not affect HSC function,50 arguing that the source of BM inflammation may reside in the Runx1R188Q/+ HSC–derived hematopoietic progenitors and immune cells.
Runx1 attenuates DDR response in HSPCs through mechanisms poorly understood. For instance, hematopoietic cells expressing a C-terminus truncated RUNX1 protein have an increase in γH2AX foci and repression of Gadd45a expression, a protein that mediates DDR.51 In addition, hematopoietic cells derived from FPDHM–derived induced pluripotent stem cells carrying the Runx1R201Q mutation have reduced DDR response.33 We found that Runx1R188Q/+ BM hematopoietic progenitor cells have reduced DDR response, as marked by the quantification of 53bp1 positive nuclear foci. The results suggest that reduced RUNX1 expression or activity hampers the repair of DNA breaks, and overtime, promote the acquisition of somatic mutations in patients with premalignant FPDHM. Notably, treatment with the receptor tyrosine kinase inhibitor imatinib restores DDR response in Runx1R188Q/+ cells, suggesting that treatment of patients with premalignant FPDHM with tyrosine kinase inhibitors could delay HM onset. The mechanism by which imatinib restores DDR in Runx1R188Q/+ cells is unknown. The tyrosine kinase inhibitors could be restoring DDR response by increasing the pool of “active” RUNX1 proteins. Indeed, the non–receptor tyrosine kinase c-Abl, a target of imatinib and dasatinib, can inhibit RUNX1 function by tyrosine phosphorylation at its inhibitory domain.34,35 Alternatively, and considering the variety of Abl targets that interfere with DDR,52 it is possible that imatinib may regulate RUNX1-independent tyrosine kinase pathways.
The majority of patients with FPDHM have thrombocytopenia and other complication but do not develop HM in their lifetime. Similarly, the Runx1R188Q/+ mice have defective platelet function and alterations in hematopoietic cells but remain healthy for over 18 months, confirming that the Runx1R188Q mutation is not sufficient to trigger HM mice. We demonstrate that Runx1R188Q/+ mice predispose to HM in cooperation with somatic mutations using 3 models. The pathology of these mice is primarily MDS/MPN and ML, and a minority of lymphoid neoplasms.
In conclusion, we propose that the Runx1R188Q mutation confers an inflammatory BM environment that favors accumulation of somatic mutations overtime and predisposes to HM. Finally, the Runx1R188Q/+ strain is a valuable new model for mechanistic, functional, and therapeutic studies in FPDHM development, prevention, and treatment.
Acknowledgments
The authors thank Jennifer Miller and Snuhi Dasgupta for their technical contribution in the study. This study was supported by grants of the RUNX1 Research Program and the Alex’s Lemonade Stand Foundation for Childhood Cancer Research (18-07741, L.H.C.), the Alex’s Lemonade Stand Foundation for Childhood Cancer Research (IA-19-16573, L.H.C.), and the National Institutes of Health-National Cancer Institute (R01CA204979, L.H.C.).
Authorship
Contribution: M.H.A., M.H., A.C., and N.I. participated in experimental design, and conducted experiments; M.M. and A.A. evaluated cytokine levels in bone marrow; L.G. conducted the embryo injections and generated the Runx1 mutant founder mice; W.J.W. and G.P. conducted the pathology analysis of samples; L.J.Z. performed off-target effect and statistical analyses of the data; J.C.S. generated bead-assisted mass-spectrometry affinity reagents and conducted the mass spectrometry analysis; B.E. and P.P.L. participated in the interpretation of the data; S.A.W. participated in the experimental design of gene-editing experiments and provided reagents; L.H.C. conceived the experiments and led the experimental interpretation and manuscript preparation; all authors participated in the preparation of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lucio H. Castilla, Department of Molecular, Cell and Cancer Biology, University of Massachusetts Chan Medical School, Worcester, MA 01605; e-mail: lucio.castilla@umassmed.edu.
References
Author notes
Data are available upon reasonable request from the corresponding author, Lucio H. Castilla (lucio.castilla@umassmed.edu).
The full-text version of this article contains a data supplement.


![Analysis of Runx1R188Q/+ BM HSPCs using Sca-1 and CD86 markers and cytokine profiling. (A) BM cellularity of WT and Runx1R188Q/+ mice. (B-C) Immunophenotypic analysis of BM HSPCs using Sca1 and CD86, including LKS+ and LK86+ (B), and LT-HSCs, ST-HSCs, and MPPs (C); (D-E) Quantification of Sca1 protein levels in cell membrane (median fluorescence intensity [MFI], D), and transcript (Ly6a) levels € in LKS+ cells. (F) Representative flow cytometry plots for the analysis of LKS and LK86 marked HSPCs; blue shade represents LK86+ (left panels) and LKS+ (right panels). (G) Relative expression levels of 25 cytokines in BM fluid (Runx1R188Q/+/WT; n = 6 per group). (H-I) Quantification of number of myeloid progenitor cells using Sca-1 (LKS-) and CD86 (LK86-) markers (H), and in common myeloid progenitor (CMPs), GMPs, and MEPs (I). ∗P < .05, ∗∗P < .005; ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/23/10.1182_bloodadvances.2023010398/2/m_blooda_adv-2023-010398-gr2.jpeg?Expires=1769107543&Signature=FWdYTUlxPahjkmG94TW0kUeY9yRwP-DgzYRxL7rBqktW58ObDfasNs49wAQpvQ4KIBJK3BD2IjzTH~jiXVCdAQxOzBqDTeW7bulFjI6oM-yY18QfI-xIGSvzTbmItG0xzLTyciTrxUJD3jbc4~S~ksahzmP5zzhhK7tEqexuRK0tRnbqBLgOQnJf6cmwOCN8oWe8uLYCnlRkBOcaRZtkZBudm6qwphmOyH9PfMG44vWFIR09AzPTGuKJzDmKPJ~NNNSaHeX~DcgiI5BxRMd8BDC8eRGfaCFwigSbj-5kVk~hrGP94ZVx1lxFZsXmNA3yyeYSzEEMpsPlEQRw0s7mQg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)