Key Points
An in vitro stromal cell–free model that allows concurrent study of multiple human hematopoietic lineages was identified.
AHR signaling drives human HSPCs to differentiate toward granulocytes and monocytes and suppresses lymphoid and megakaryocyte lineages.
Abstract
In vitro models to study simultaneous development of different human immune cells and hematopoietic lineages are lacking. We identified and characterized, using single-cell methods, an in vitro stromal cell–free culture system of human hematopoietic stem and progenitor cell (HSPC) differentiation that allows concurrent development of multiple immune cell lineages. The aryl hydrocarbon receptor (AHR) is a ligand–activated transcription factor influencing many biological processes in diverse cell types. Using this in vitro model, we found that AHR activation by the highly specific AHR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin, drives differentiation of human umbilical cord blood–derived CD34+ HSPCs toward monocytes and granulocytes with a significant decrease in lymphoid and megakaryocyte lineage specification that may lead to reduced immune competence. To our knowledge, we also discovered for the first time, using single-cell modalities, that AHR activation decreased the expression of BCL11A and IRF8 in progenitor cells, which are critical genes involved in hematopoietic lineage specification processes at both transcriptomic and protein levels. Our in vitro model of hematopoiesis, coupled with single-cell tools, therefore allows for a better understanding of the role played by AHR in modulating hematopoietic differentiation.
Introduction
Hematopoietic stem and progenitor cells (HSPCs) differentiate to give rise to the host’s immune repertoire. Xenotransplantation may be a feasible process to study the differentiation of human HSPCs in vivo. However, direct study of human HSPCs and their modulation by individual components or perturbing agents without interference from other cell types is not currently feasible, and in vitro studies are used for these investigations. Different in vitro and ex vivo model systems using miscellaneous culture conditions have been developed over the past few decades to propagate proliferation of HSPCs and development of specific hematopoietic lineages.1-3 However, relatively few studies exist that have investigated development of both human lymphoid and myeloid lineages concurrently under the same conditions. In this study, we report the characterization and use of an in vitro stromal cell–free model system that allows temporal study of early human HSPC differentiation into multiple lineage–specified immune progenitor cells. We have previously used this model to investigate the role of the transcription factor, the aryl hydrocarbon receptor (AHR), in B cell development from human HSPCs.4 Using single-cell transcriptomics to guide us, we demonstrated that this model is also very useful in investigating the role of AHR in overall human hematopoiesis.
AHR is a ligand–activated transcription factor that plays highly context-dependent roles in regulating different physiological systems.5,6 AHR has been historically studied in the context of mediating toxicity of diverse xenobiotic compounds, such as polycyclic aromatic hydrocarbons, polychlorinated biphenyls, and polychlorinated dibenzo-p-dioxins. With time, its involvement in normal physiological processes, including within the immune system, has been established.7,8 Recent studies in mice and humans have demonstrated that HSPCs are susceptible to modulation by AHR signaling with antagonism of AHR signaling leading to increased proliferation and decreased differentiation of CD34+ HSPCs.9,10 Dysregulation of HSPC function can have profound consequences on immunocompetence and other hematopoietic functions of an organism. Previous in vivo studies in mice and in vitro studies using human CD34+ HSPCs have shown profound impairment of B-cell development from HSPCs4,11 because, in part, of the disruption of EBF1 and PAX5 signaling4; however, changes in gene expression during early differentiation of HSPCs, which may be responsible for these changes, have remained poorly understood.
In the present investigation, we sought to identify transcriptomic changes brought about by AHR activation in HSPCs in humans. Toward this end, we used our in vitro stromal cell–free model of hematopoietic differentiation using human umbilical cord blood–derived CD34+ HSPCs to investigate the role of AHR activation directly on HSPCs. To induce AHR activation for the duration of the study, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) was used. TCDD was the agonist of choice because it is a halogenated aromatic hydrocarbon with very high specificity and affinity for the receptor.12 TCDD is highly persistent and bioaccumulates with an estimated half-life in humans of ∼7 years.13,14 For identification of discrete cell populations that may develop as HSPCs differentiate in vitro, and to discern transcriptomic changes brought about by TCDD treatment of HSPCs, we used single-cell RNA-sequencing (scRNA-seq) to characterize the gene expression changes across different cell types at the single-cell level. We then, confirmed findings from scRNA-seq analysis with flow cytometry. Our studies demonstrate that this model allows development of multiple immune cell types within a single system, and modulation of AHR signaling in HSPCs by AHR activation in this system shifts differentiation of human HSPCs primarily toward monocyte and granulocyte lineages by disrupting gene expression programs that regulate lymphoid-myeloid balance.
Methods
Chemicals
TCDD was obtained from AccuStandard (New Haven, CT). Dimethyl sulfoxide (DMSO) and AHR antagonist CH223191 were obtained from Sigma Aldrich (St. Louis, MO).
In vitro culture of human CD34+ HSPCs
Human CD34+ HSPCs isolated from umbilical cord blood from mixed donors were purchased in cryopreserved state from AllCells LLC (Alameda, CA). 104 cells per well were plated and cultured over a 28-day period in a 96-well cell culture plate in media enriched with cytokines and growth factors (Figure 1A) in the same manner as performed in a previous study from our laboratory.4 The media comprised RPMI-1640 media (Life Technologies) supplemented with 5% human AB serum (Valley Biomedical), 100 U/mL of penicillin (Life Technologies), 100 μg/mL of streptomycin (Life Technologies), and 50 μM 2-mercaptoethanol. Stem cell factor (SCF), fms-like tyrosine kinase 3 ligand (Flt3L) and interleukin-6 (IL-6) (each at 25 ng/mL; Miltenyi Biotec) were added on day 0 to promote proliferation of HSPCs, and cells were treated with either TCDD (1 nM) or control vehicle (0.02% DMSO). On day 7, half of the cell culture medium was replaced with fresh medium supplanted with SCF (25 ng/mL), Flt3L (25 ng/mL), and IL-7 (20 ng/mL; Miltenyi Biotec). Interleukin-7 (IL-7) was added to promote development of lymphoid progenitors and B cells. For remainder of the study, cytokine-free fresh media was used to replace half of the medium every 7 days. Because TCDD is highly lipophilic and partitions poorly into aqueous compartments, accurate estimation of TCDD remaining in the system after replacement of media is difficult. To ensure that more TCDD was not being added to the system than that being taken out, no further addition of TCDD was made.
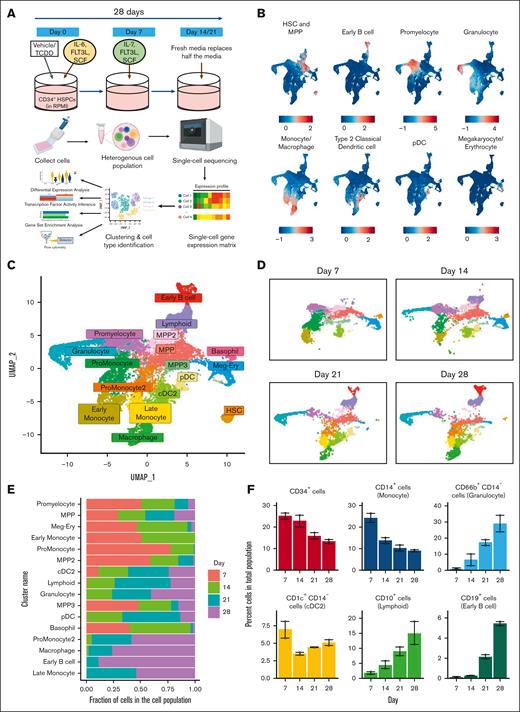
Single-cell transcriptomic analysis reveals development of multiple hematopoietic cell types in an in vitro model of human hematopoietic differentiation. (A) Experimental design: in vitro culture system of hematopoiesis from human cord blood HSPCs and schema of sample processing and data analysis. (B) Different cell types were identified based on expression of a suite of lineage-specific genes (refer to “Methods”). (C) Representation using UMAP of all cells from control group collected over the 28-day period. (D) Development of cell clusters over time is shown. (E) Proportion of cells of each cluster across time. (F) Percent of cells belonging to major cell types that develop from untreated HSPCs were identified using flow cytometry. Error bars show mean ± standard error of the mean from 2 independent experiments. HSC, hematopoietic stem cells; MPP, multipotent progenitors; cDC2, type 2 classical dendritic cells; pDC, plasmacytoid dendritic cells; Meg-Ery, megakaryocyte-erythroid progenitors.
Single-cell transcriptomic analysis reveals development of multiple hematopoietic cell types in an in vitro model of human hematopoietic differentiation. (A) Experimental design: in vitro culture system of hematopoiesis from human cord blood HSPCs and schema of sample processing and data analysis. (B) Different cell types were identified based on expression of a suite of lineage-specific genes (refer to “Methods”). (C) Representation using UMAP of all cells from control group collected over the 28-day period. (D) Development of cell clusters over time is shown. (E) Proportion of cells of each cluster across time. (F) Percent of cells belonging to major cell types that develop from untreated HSPCs were identified using flow cytometry. Error bars show mean ± standard error of the mean from 2 independent experiments. HSC, hematopoietic stem cells; MPP, multipotent progenitors; cDC2, type 2 classical dendritic cells; pDC, plasmacytoid dendritic cells; Meg-Ery, megakaryocyte-erythroid progenitors.
For any treatment group, cells received the treatment only on day 0. DMSO (0.02%) was used as the solvent for TCDD and AHR antagonist CH223191 (10 μM). For treatments involving both TCDD (1 nM) and the AHR antagonist CH223191 (10 μM), cells were treated with the antagonist 30 minutes before the addition of TCDD.
scRNA-seq and processing of cells
Cells were harvested on day 0 from untreated HSPCs and every 7 days from 3 pooled 96-wells from both vehicle- and TCDD-treated groups. Cell suspensions containing approximately 104 cells from each group were then processed using the 3′ messenger RNA chemistry-based gel beads-in-emulsion technology from 10X Chromium Single Cell Gene Expression platform v3.2 (10X Genomics, Pleasanton, CA) to generate complementary DNA (cDNA) libraries as per company protocols. cDNA libraries were subjected to quality control analysis including a double-sided size selection. DNA fragments were selected with a size range between 400 and 1000 base pairs and analyzed using Agilent Bioanalyzer DNA1000 chip. cDNA libraries were sequenced on an Illumina HiSeq platform at an average sequencing depth of 50 000 paired end reads per cell. Reads were processed by CellRanger software (v 2.1) from 10X Genomics.
Gene expression data for each cell across days were concatenated into a single matrix which was analyzed with the scRNA-seq analysis package Seurat (v 4.0).15 After quality control of the single-cell gene expression matrix (refer to supplemental Methods), data were normalized using the SCTransform method. In total, 3000 variable genes were used for dimension reduction, and cells were represented in 2-dimensional space using Uniform Manifold Approximation and Projection (UMAP). Unsupervised clustering (resolution = 0.5) of cells generated cell clusters, which were annotated based on enrichment of hematopoietic lineage–specific gene signatures.
Developmental trajectory analysis
Cell clusters associated with the lineage of interest were selected and dimensionality reduction and projection in UMAP space were redone. This was followed by trajectory analysis and pseudotime calculations for cells using the R package “slingshot”16 (v 3.14).
Analysis of differentially expressed genes (DEGs)
DEGs analysis between vehicle- and TCDD-treated groups was carried out using “FindMarkers” function of Seurat (testing method called upon the MAST package and assay used was "RNA"). Log2 fold-change (log2FC) threshold was set at 0.25 unless otherwise stated. Adjusted P-value threshold of 0.05 based on Bonferroni multiple testing correction was used as a cutoff to select significant DEGs. For gene and cluster dot-plots in the figures, an equal number of upregulated and downregulated significantly DEGs were plotted based on fold-change in expression among groups. Top DEGs in each category were selected and shown in figures.
Inference of transcription factor (TF) activity
TF activity was estimated using the R-language–based package “SCENIC”17 (refer to supplemental Methods).
Gene set enrichment analysis (GSEA)
Seurat’s “FindMarkers” function was used to calculate DEGs but without any threshold for log2 fold-change (log2FC = 0). The genes were ranked based on fold-change. The R package “fgsea” (v 3.14)18 was used to carry out GSEA using the preranked list of genes and Hallmark gene sets from human MSigDB database.
Statistical analysis
Statistical analysis for flow cytometry data was performed using GraphPad Prism 9.5.0 (GraphPad Software, San Diego, CA). For flow cytometry data involving comparison of 2 groups at a particular timepoint, a paired, 2-tailed student t test was used to determine significance of the results. For comparison of multiple groups at any time, a repeated measures 1-way analysis of variance was used, followed by a Dunnett multiple comparisons test. Regarding SCENIC analysis and M2 macrophage score comparisons, statistical analysis for differences in TF activity and M2 macrophage scores among groups was done using Wilcoxon rank sum test (“wilcox.test” function in R).
Flow cytometric analysis
On days of cell collection, cells were harvested and washed using Hanks' balanced salt solution (pH 7.4, Invitrogen). Cell surface Fc receptors were blocked by incubating cells with human AB serum. For cell surface staining, cells were incubated with antibodies in fluorescence-activated cell sorter (FACS) buffer (1× Hanks' balanced salt solution containing 1% bovine serum albumin and 0.1% sodium azide, pH 7.4-7.6) for 30 minutes, washed and fixed using Cytofix fixation buffer (BD Biosciences) for 20 minutes (refer to supplemental Methods for details of cell surface antibodies). For intracellular staining, fixed cells were permeabilized by incubating in eBioscience Foxp3/TF permeabilization buffer (Invitrogen) for 20 minutes and incubated with relevant antibodies (antihuman Ctip-1 [Fluorophore PE, clone NB600-261; Novus Biologicals], antihuman IRF8 [Fluorophore APC, clone V3GYWCH; Thermo Fisher Scientific] and antihuman AHR [Fluorophore PE-Cy7 / PE, clone FF3399; Thermo Fisher Scientific]) for 60 minutes. This was followed by washing in the presence of the buffer. Finally, cells were resuspended in FACS buffer. Flow cytometric analysis was performed using BD FACSCanto-II cell analyzer (BD Biosciences) with FACSDiva software (BD Biosciences) or Cytek Northern Lights full spectral flow cytometer (Freemont, CA). Flow cytometry data were analyzed using FlowJo (version 10.1; Treestar Software, Ashland, OR). Please refer to supplemental Figures 5 and 6 for gating strategies.
Results
Single-cell transcriptomic analysis reveals development of multiple hematopoietic cell types in an in vitro model of human hematopoietic cell differentiation
To study the role of AHR signaling during differentiation of human HSPCs, we developed an in vitro stromal-cell–free model (Figure 1A; refer to "Methods"). HSPCs were initially treated with either the control vehicle (0.02% DMSO) or TCDD (1 nM). For the scRNA-seq study, cells from both groups were harvested at multiple time points for sequencing. Upon processing of sequenced reads, exploratory gene expression analysis in cells suggested the existence of diverse hematopoietic lineages in the system. Based on expression of multiple genes associated with distinct hematopoietic lineages, numerous hematopoietic cell types were identified (Figures 1B; supplemental Figure1A-B, supplemental Methods). To characterize the natural development of hematopoietic progenitors in absence of TCDD, we selected only cells from Day 0 and that of the vehicle group. UMAP projection of different cell clusters reflected a natural progression of development of different hematopoietic lineages from early progenitors (Figure 1C-E). Automated annotation against a dataset of human bone marrow–derived cells was largely consistent with clustering performed under our supervision (supplemental Figure 2A). To confirm the formation of bona fide hematopoietic lineages in our system, flow cytometry was used using antibodies directed against markers for major hematopoietic lineages. The markers selected were informed through both literature and scRNA-seq analysis and were highly and uniquely expressed in the population of interest (supplemental Figure 2B). Flow cytometric analysis of cells developed from untreated HSPCs confirmed our scRNA-seq–based findings regarding the formation of several major immune cell types (Figure 1F).
AHR activation by TCDD alters the development of several hematopoietic lineages
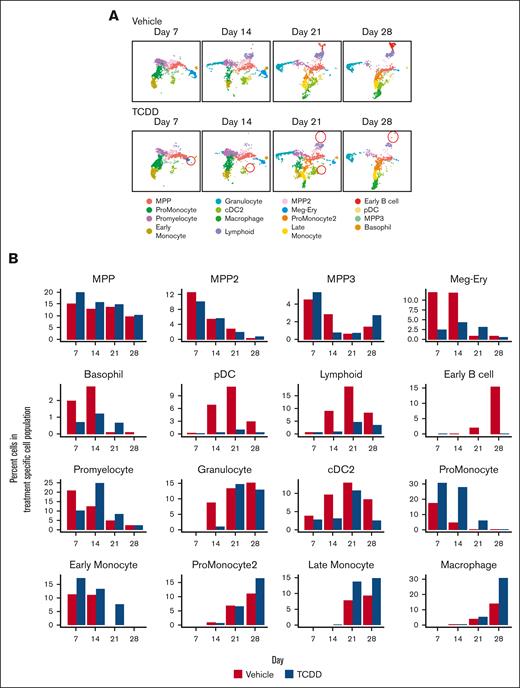
In the presence of TCDD, we observed multiple changes in the hematopoietic landscape. A time and treatment-dependent split of cell clusters (scRNA-seq data) demonstrated the chronological development of different cell types and their modulation by TCDD (Figure 2A-B). With TCDD treatment, on day 7, the largest change is seen in the megakaryocyte (MK)-erythroid cluster with fewer cells being present in this cluster in the TCDD-treated group, relative to the vehicle group. Concomitantly, we see an increase in the proportion of cells in the promonocyte and early monocyte clusters. This trend continued till day 14. Cells belonging to the lymphoid and plasmacytoid dendritic-cell (pDC) clusters emerge on day 14, and a reduced proportion of cells belonging to these clusters was observed in the TCDD group. This was suggestive of a skewing of cells toward the monocyte lineage at the expense of other lineages owing to TCDD treatment and is in agreement with previous findings reported in the mouse.9 By day 21, promonocyte and early monocyte clusters give rise to promonocyte-2, late monocyte and macrophage clusters. In contrast to the vehicle group, cells belonging to the early monocyte cluster are still observed in the TCDD-treated group, suggesting that TCDD may delay maturation of the earlier monocytes. A substantial number of early B cell cluster cells appear by day 21 and progressively increase in number as seen on day 28 in the vehicle group, which was almost nonexistent in the TCDD group in agreement with our previous study.4 Our previous studies with TCDD had demonstrated that reduction in cell number in the presence of TCDD was not attributable to cell death by apoptosis or necroptosis.19 A similar decrease is also observed for the pDC cluster with TCDD. GSEA did not point to changes in cell cycle–mediated effects that could explain the above phenomena (supplemental Figure 4A).
AHR activation by TCDD alters the development of several hematopoietic lineages. (A) Changes in cell clusters from vehicle and TCDD groups across the 28-day developmental period. Some of these changes are highlighted with red circles. (B) Percent of cells belonging to each scRNA-seq data associated cell cluster for Vehicle and TCDD-treated groups across the 28-day developmental period. (C) Representative UMAP plots of cells captured by flow cytometry on day 21, with major cell populations identified through expression of different lineage markers. Comparison of UMAP plots of an equal number of cells (30,000) from both vehicle and TCDD-treated groups; (D) Percent of cells belonging to major hematopoietic lineages captured using flow cytometry for vehicle and TCDD-treated groups across the 28-day developmental period. Data presented are composite of 6 independent experiments. Statistical significance of differences in percentage of cells between treatments at any time point was calculated using a 2-tailed paired t test. ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001. Error bars show mean ± standard error of the mean. Refer to supplemental Figures 5 and 6 for gating strategy. cDC2, type 2 classical dendritic cells; CLP, common lymphoid progenitor.
AHR activation by TCDD alters the development of several hematopoietic lineages. (A) Changes in cell clusters from vehicle and TCDD groups across the 28-day developmental period. Some of these changes are highlighted with red circles. (B) Percent of cells belonging to each scRNA-seq data associated cell cluster for Vehicle and TCDD-treated groups across the 28-day developmental period. (C) Representative UMAP plots of cells captured by flow cytometry on day 21, with major cell populations identified through expression of different lineage markers. Comparison of UMAP plots of an equal number of cells (30,000) from both vehicle and TCDD-treated groups; (D) Percent of cells belonging to major hematopoietic lineages captured using flow cytometry for vehicle and TCDD-treated groups across the 28-day developmental period. Data presented are composite of 6 independent experiments. Statistical significance of differences in percentage of cells between treatments at any time point was calculated using a 2-tailed paired t test. ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001. Error bars show mean ± standard error of the mean. Refer to supplemental Figures 5 and 6 for gating strategy. cDC2, type 2 classical dendritic cells; CLP, common lymphoid progenitor.
We next verified the changes in cell population induced by AHR activation with TCDD as observed from scRNA-seq analysis using flow cytometry. With antibodies directed against cell markers expressed on the major hematopoietic cell populations, changes in the number of different cell types with TCDD treatment were tracked over 28 days. The percent of CD66b+ CD14– cells, representing granulocytes, and CD14+ cells (monocytes/macrophages) were significantly greater in the TCDD treatment group relative to the vehicle group on days 14, 21, and 28 of the study (Figure 2C-D). Because eosinophils and basophils constitute a minor proportion of hematopoietic populations physiologically as well as in our system as inferred from scRNA-seq analysis, we assumed that CD66b+ CD14– cells largely represented the neutrophil population. CD10+ cells represent the lymphoid population which can give rise to B, T, and NK-cell populations. The number of CD10+ cells decreased significantly with TCDD treatment. There was a significant trend toward a decrease in CD1c+ CD14– cells (type 2 classical dendritic cells) across most days upon TCDD treatment.
TCDD is known to exert its toxic effects primarily through activation of AHR.20 To determine whether the effects on the hematopoietic profile that were observed with TCDD were mediated through the AHR, we treated HSPCs with TCDD in presence of an AHR antagonist CH223191 (supplemental Figure 3A). CH223191 was initially developed to antagonize the binding of TCDD to AHR and its translocation to the nucleus, and its structure-activity relationship regarding AHR activity has been extensively investigated.21,22 A treatment group in which cells were treated with CH223191 alone was also included. supplemental Figure 3A shows that the aberrant hematopoietic profile induced with TCDD treatment was abrogated in presence of the AHR antagonist, demonstrating complete antagonism of TCDD. In fact, HSPCs treated with TCDD and CH223191 had lower monocyte and granulocyte development than the vehicle group, suggestive of antagonism of endogenous AHR activation by the antagonist. HSPCs that differentiated in presence of the AHR antagonist alone demonstrated a greater skewed development along the lymphoid lineage compared to the vehicle group, suggesting again antagonism of endogenous AHR activity. These experiments demonstrated that TCDD-mediated effects on HSPCs were through AHR activation.
We used other cell markers for detecting multipotent progenitors (MPPs) (CD34), B cells (CD19), megakaryocytic cells (CD41), erythroid cells, (CD235a) and pDCs (CD303). Similar to our previous studies,19 we observed strong reductions in CD19+ cells and CD34+ cells with TCDD treatment (supplemental Figure 3B). AHR activation also resulted in a significant decrease in MK populations (supplemental Figure 3B). CD235a is a specific marker for erythroid committed cells. A trend toward a slight increase in CD235a+ cells could be detected in the TCDD group. pDC population development seemed to be highly HSPC lot–specific and could not always be reliably detected. However, there was a trend toward a decrease in CD303+ cells in 2 independent experiments (supplemental Figure 3B), consistent with findings from the scRNA-seq analysis (Figure 2B).
GSEA of major immune cell lineages showed upregulation of oxidative phosphorylation pathway with AHR activation across almost all cell types (supplemental Figure 4A-C). Even though they are on distinct hematopoietic trajectories, both granulocytes and lymphoid progenitors that develop by day 28 in the presence of TCDD had a reduced interferon-alpha signature (supplemental Figure 4A-B). Further studies are warranted to elucidate the role of AHR in orchestrating this response.
In summary, AHR activation in HSPCs led to an increase in monocytes and granulocytes and a decrease in lymphoid, dendritic, and MK populations.
AHR activation suppresses critical genes and key TFs involved in B cell and dendritic cell development
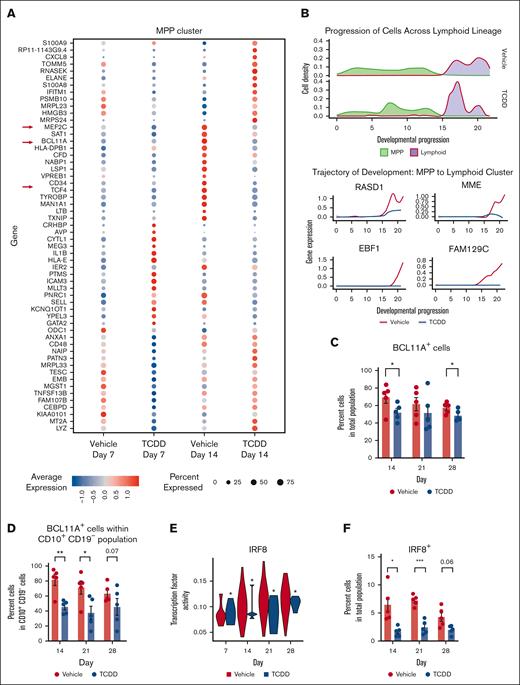
Apart from the hematopoietic stem cell cluster, MPP cluster is an early forming cluster which is not lineage-specified compared with other progenitor clusters. Early gene changes induced by AHR activation with TCDD that lead to persistent alterations in the hematopoietic profile may be gleaned from transcriptomic changes specific to this cluster. We focused upon the MPP cluster on day 14 because this was when IL-7 was present in culture to allow lymphoid cell proliferation and when lymphoid lineages were starting to develop. We found, among the most DEGs on day 14 in the MPP cluster, genes that are crucial for lymphoid and dendritic lineages, such as TFs BCL11A, MEF2C, and TCF4,23,24 were downregulated with TCDD treatment (Figure 3A). Suppression of these genes may explain AHR activation–mediated reduction in lymphoid cells and the pDC cluster. Next, we focused on the lymphoid lineage to identify genes dysregulated by AHR activation, which may further explain the reduced number of cells within lymphoid and early B cell clusters with TCDD treatment. We found expression of genes essential to lymphoid lineage and B cell lineage specification (MME and EBF1) to be significantly downregulated along the lymphoid trajectory as well as that of B cell–associated genes that have not previously been linked to early B cell development, such as RASD1 and FAM129C (Figure 3B). As mentioned above, among the top downregulated DEGs in MPP cluster cells on day 14, owing to AHR activation by TCDD, was BCL11A, a TF that is known to regulate EBF1 essential for B cell specification of lymphoid progenitors.25 Mouse embryos with mutant Bcl11a do not develop B cells, and fetal liver hematopoietic progenitors show absence of Ebf1, Pax5, and IL-7Rα transcripts in Bcl11a−/− embryos,25 highlighting the importance of Bcl11a in B cell development. Flow cytometric analysis confirmed a reduction in percent of BCL11A+ cells in the overall population with TCDD treatment across multiple days (Figure 3C). There was a significant and greater reduction in BCL11A+ cells with AHR activation within the CD10+ CD19– cells (common lymphoid progenitors) on days 14 and 21 (Figure 3D). A decrease in BCL11A expression in lymphoid progenitors owing to TCDD could thus contribute to the reduction in early B cell formation, which was seen with TCDD treatment.
AHR activation suppresses critical genes and key transcription factors involved in B cell and dendritic cell development. (A) Dot plot of the average gene expression of TCDD-treatment induced differentially expressed genes in MPP (multipotent progenitor) cluster on days 7 and 14. All DEGs are associated with a Bonferroni adjusted P value <.05. (B) Density of cells along MPP to lymphoid cells trajectory and expression of the top 4 highly variable genes that are differentially expressed by TCDD treatment and are involved in the development of lymphoid cells. (C) Percent BCL11A protein–expressing cells in overall population from 5 independent experiments. (D) Percent BCL11A protein–expressing cells in CD10+ CD19– cells of vehicle and TCDD-treated groups across days from 5 independent experiments. (E) Transcription factor activity of IRF8 (analyzed by SCENIC) in pDC (plasmacytoid dendritic cell) cluster. Statistical significance of differences in TF activity between treatments at any time point was calculated using a Wilcoxon rank sum test. ∗P < .05. (F) Percent IRF8 protein–expressing cells in overall population from 5 independent experiments. (C-D,F) Protein expression was measured using flow cytometry. Error bars show mean ± standard error of the mean. Statistical significance of differences in percentage of cells between treatments at any time point was calculated using a 2-tailed paired t test. ∗ P < .05, ∗∗ P < .01, ∗∗∗ P < .001.
AHR activation suppresses critical genes and key transcription factors involved in B cell and dendritic cell development. (A) Dot plot of the average gene expression of TCDD-treatment induced differentially expressed genes in MPP (multipotent progenitor) cluster on days 7 and 14. All DEGs are associated with a Bonferroni adjusted P value <.05. (B) Density of cells along MPP to lymphoid cells trajectory and expression of the top 4 highly variable genes that are differentially expressed by TCDD treatment and are involved in the development of lymphoid cells. (C) Percent BCL11A protein–expressing cells in overall population from 5 independent experiments. (D) Percent BCL11A protein–expressing cells in CD10+ CD19– cells of vehicle and TCDD-treated groups across days from 5 independent experiments. (E) Transcription factor activity of IRF8 (analyzed by SCENIC) in pDC (plasmacytoid dendritic cell) cluster. Statistical significance of differences in TF activity between treatments at any time point was calculated using a Wilcoxon rank sum test. ∗P < .05. (F) Percent IRF8 protein–expressing cells in overall population from 5 independent experiments. (C-D,F) Protein expression was measured using flow cytometry. Error bars show mean ± standard error of the mean. Statistical significance of differences in percentage of cells between treatments at any time point was calculated using a 2-tailed paired t test. ∗ P < .05, ∗∗ P < .01, ∗∗∗ P < .001.
We also observed a reduction in TF activity (calculated with SCENIC) of IRF8 in the pDC cluster for the TCDD group (Figure 3E). IRF8 plays an important role in the development of several hematopoietic lineages with increased expression being linked to development of lymphoid, pDCs, classical dendritic cells, and in macrophage maturation, whereas its absence is associated with an increase in granulocyte populations.26 Using flow cytometry, we observed that percent IRF8 protein–expressing cells was also significantly reduced in the TCDD-treated group on days 14 and 21 of the study (Figure 3F). The changes in IRF8 with TCDD treatment are thus consistent with the reduction in CD10+ cells and an increase in CD66b+ CD14– cells. Overall, we identified that AHR activation is associated with a reduction of multiple lymphoid and dendritic cell–associated genes at different time points and specifically demonstrated a reduction in BCL11A and IRF8 at both transcriptomic and protein levels that may contribute to the observed phenomena.
Single-cell transcriptomic analysis shows that the macrophage cluster cells that develop in presence of TCDD have a reduced M2 macrophage signature
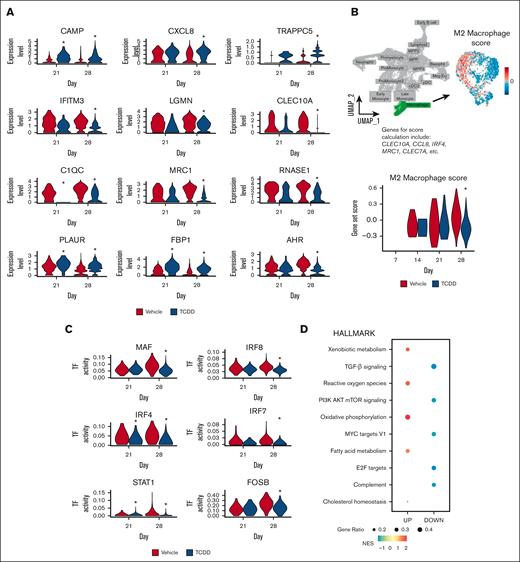
Although there is an increase in proportion of cells of the macrophage cluster with TCDD treatment (Figures 2B), cells in this cluster that developed in the presence of TCDD showed reduced expression of several markers that are usually associated with an M2 macrophage phenotype (Figure 4A; supplemental Figure 4B). These include genes that encode complement proteins, such as C1QA, C1QC, M2 macrophage associated marker MRC1, and lectins, such as CLEC10A (Figure 4A; supplemental Figure 4D).27,28 A decreased M2 macrophage gene set score, calculated based on curated M2 macrophage markers (refer to supplemental Methods), in macrophages of the TCDD-treated group (Figure 4B) was also consistent with these observations. Upon performing calculation of TF activity with SCENIC, we observed a reduction in TF activity of IRF4, MEF2A, MAF, and IRF8 among others in cells of the macrophage cluster (Figure 4C). IRF4 and MAF are known to promote M2 polarization of macrophages, whereas studies in mice have suggested that IRF8 can also play important regulatory roles in macrophage polarization.29,30 GSEA demonstrated, within the macrophage cluster, an upregulation in pathways known to be modulated by AHR activation, such as xenobiotic metabolism, oxidative phosphorylation, and fatty acid metabolism (Figure 4D). More interestingly, to our knowledge, we identified for the first time in cells of the TCDD group, downregulation of transforming growth factor-β signaling, MYC signaling, and PI3K-AKT-mTOR pathway. These downregulated pathways have been associated with M2 macrophage development.31,32 Overall, this suggests that AHR activation might preclude maturation of monocytes into M2 macrophages. Interestingly, a previous study using a macrophage cell line cocultured with endometrial cells had reported that TCDD in combination with 17β-estradiol drives macrophages toward an M2 phenotype33; although, the effect was not observed with TCDD alone, suggesting that this occurrence may have been a result of possible cross talk in signaling between AHR and estrogen receptors.
Single-cell transcriptomic analysis shows that the macrophage cluster cells that develop in presence of TCDD have a reduced M2 macrophage signature. (A) Expression of select TCDD-treatment induced differentially expressed genes in macrophages on days 21 and 28; ∗Bonferroni adjusted P value <.05. (B) Macrophage cluster was selected, and an M2 macrophage score was calculated based on expression of several genes (refer to “supplemental Methods”). M2 macrophage score within the macrophage cluster, and across time for vehicle and TCDD-treated groups is shown. Statistical significance of differences in M2 macrophage score between treatments at any time point was calculated using a Wilcoxon rank sum test. ∗P < .05. (C) TF activity (calculated with SCENIC) in the macrophage cluster for selected TFs across time is shown. Statistical significance of differences in TF activity between treatments at any time point was calculated using a Wilcoxon rank sum test. ∗P < .05. (D) GSEA using the Hallmark pathways database shows significantly enriched pathways in cells of the macrophage cluster owing to TCDD treatment at day 28. All enriched pathways have a Benjamini-Hochberg adjusted P value <.05. DOWN, downregulated pathways; NES, normalized enrichment score; UP, upregulated pathways.
Single-cell transcriptomic analysis shows that the macrophage cluster cells that develop in presence of TCDD have a reduced M2 macrophage signature. (A) Expression of select TCDD-treatment induced differentially expressed genes in macrophages on days 21 and 28; ∗Bonferroni adjusted P value <.05. (B) Macrophage cluster was selected, and an M2 macrophage score was calculated based on expression of several genes (refer to “supplemental Methods”). M2 macrophage score within the macrophage cluster, and across time for vehicle and TCDD-treated groups is shown. Statistical significance of differences in M2 macrophage score between treatments at any time point was calculated using a Wilcoxon rank sum test. ∗P < .05. (C) TF activity (calculated with SCENIC) in the macrophage cluster for selected TFs across time is shown. Statistical significance of differences in TF activity between treatments at any time point was calculated using a Wilcoxon rank sum test. ∗P < .05. (D) GSEA using the Hallmark pathways database shows significantly enriched pathways in cells of the macrophage cluster owing to TCDD treatment at day 28. All enriched pathways have a Benjamini-Hochberg adjusted P value <.05. DOWN, downregulated pathways; NES, normalized enrichment score; UP, upregulated pathways.
Discussion
In this study, to our knowledge, we report for the first time, the use of single-cell transcriptomics to investigate the role of AHR activation in human hematopoiesis using a novel in vitro stromal cell–free model system of human hematopoietic differentiation. Earlier studies have reported the use of cytokines or growth factors SCF, FLT3L, and IL-6 to facilitate in vitro HSPC proliferation and differentiation with a myeloid bias.1,34 IL-7, which we add to our system on day 7, has been known to facilitate expansion and differentiation of B cells.35,36 Studies by Kraus et al have used culture conditions similar to our studies but have mainly focused on the study of B cell development.3 Early MK and erythroid progenitors also develop in the culture. Because this model was established to identify effects of perturbations on early hematopoietic differentiation and not for full differentiation of all progenitors, we refrained from addition of thrombopoietin and erythropoietin to avoid further differentiation bias toward the MK/erythroid branch at the expense of other major immune cell lineages. The current model as demonstrated through its characterization in this study, thus allows differentiation of proliferating CD34+ HSPCs and development of almost a complete spectrum of lymphoid and myeloid lineage–specified cells. Our single-cell transcriptomic analysis of differentiating HSPCs collected at different time points enabled identification of distinct cell clusters associated with multiple hematopoietic lineages and allowed us to follow their chronological development. Flow cytometric analysis confirmed existence of major cell types as identified using scRNA-seq analysis, with the exception of T cells, which are known to require Notch ligand engagement of lymphoid progenitors.37 Collectively, our results suggest that this in vitro model may be especially useful in studies of early human hematopoiesis and in identification of agents that can alter the human developing immune system.
Results from flow cytometry–based analysis and scRNA-seq analysis were highly concordant with occasional differences. There was 1 discrepancy in the case of erythroid progenitors. scRNA-seq analysis suggested a decrease in erythroid cells in the TCDD-treated group relative to that of the control. However, flow cytometry–derived results did not show a decrease in erythroid populations in the TCDD-treated group. This disparity might reflect the formation of an overall small number of erythroid populations in vitro and failure to capture these cells during processing of TCDD-treated cells for scRNA-sequencing.
Our findings that AHR activation by TCDD elicits a decrease in early B cells and MK populations is in agreement with previous experimental or epidemiological studies.4,38 In contrast to previous studies, we also demonstrated that AHR activation in HSPCs increased development of both monocyte and granulocyte populations from HSPCs. A previous study using human cord blood–derived CD34+ cells had reported impaired development of both monocytes and dendritic cells with AHR agonists, including TCDD as well as no change in neutrophils population.39 This discrepancy between our findings and the study could be because of differences in culture conditions, because the study used diverse myeloid lineage–promoting cytokines, such as granulocyte colony–stimulating factor, granulocyte-macrophage colony–stimulating factor, and IL-4. AHR signaling is highly context specific,40 and the influence of different cytokines could modify AHR signaling, giving rise to the observed differences.
scRNA-seq analysis of progenitor clusters allowed identification of key genes that putatively govern the increase in monocyte and granulocyte populations at the expense of other lineages with AHR activation. We observed TCDD-mediated reduction in BCL11A and IRF8, which are key TFs that regulate lymphoid-myeloid balance with increased expression of these TFs being linked to increased output of lymphoid cells and pDCs. It is noteworthy that regulation of hematopoietic cell fate results from the combined influence of multiple TFs to orchestrate lineage–specific gene transcription over time.41 Modulation of a few TFs by AHR signaling could alter expression and activity of other TFs. Further experimental evidence is warranted to ascertain the exact mechanisms. In addition, we identified decreased levels of key maturation markers in the macrophage cluster (MRC1, CLEC10A, etc). This is in agreement with previous reports of attenuation of monocyte and macrophage maturation with AHR activation by xenobiotics.40 Our transcriptomic analysis also identified decreased activity with TCDD treatment of several critical TFs involved in macrophage maturation, such as MAF, that have not been previously known to be modulated by AHR signaling in macrophages.
Collectively, using scRNA-seq analysis and flow cytometry, we characterized a model of human hematopoiesis and identified gene expression programs perturbed by AHR activation during hematopoietic multilineage specification, thus providing a better understanding of the molecular mechanisms by which AHR signaling may impair human hematopoietic differentiation. Importantly, the model system offers an opportunity to study concurrent multilineage human hematopoiesis and can also be used to evaluate the effects of other small molecules or genetic perturbations on processes governing HSPC to lineage commitment. In conclusion, our study has provided important insights into the role of AHR in human hematopoiesis.
Acknowledgments
The authors thank Jinpeng Li and Rance Nault for suggestions regarding data analysis, and Lance Blevins for comments on the manuscript. The graphical abstract was created using BioRender.com.
This work was funded by National Institutes of Health grant P42ES004911.
Authorship
Contribution: D.M.I.O.K., P.W.F.K., and N.E.K. conceptualized the study and wrote and reviewed the manuscript; D.M.I.O.K. and A.B. performed sample processing for scRNA-sequencing; D.M.I.O.K. and R.B.C. carried out sample processing and sample run for flow cytometry; D.M.I.O.K. and P.W.F.K. provided methodology for scRNA-sequencing analysis; D.M.I.O.K. performed all other experimentation and scRNA-seq data analysis, and wrote and edited the original draft of the manuscript; and N.E.K. acquired funding and provided overall supervision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Norbert E. Kaminski, Michigan State University, 1129 Farm Ln, East Lansing, MI 48824; e-mail: kamins11@msu.edu.
References
Author notes
The sequencing data provided in this study has been submitted to National Center for Biotechnology Information’s Gene Expression Omnibus repository accession number GSE224172. Other relevant data are available on request from the corresponding author, Norbert E. Kaminski (kamins11@msu.edu).
The full-text version of this article contains a data supplement.