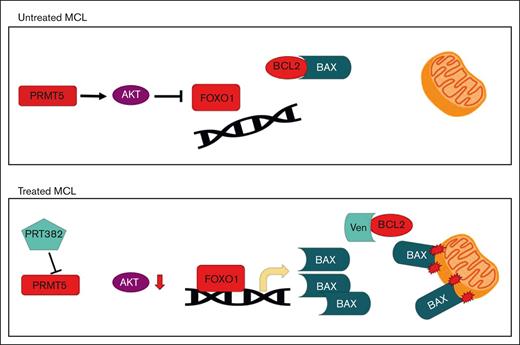
Key Points
PRMT5 inhibition induces a FOXO1 driven, proapoptotic program in MCL.
Combination therapy with PRMT5 and BCL-2 inhibition is synergistic in preclinical MCL models, including those with ibrutinib resistance.
Abstract
Mantle cell lymphoma (MCL) is an incurable B-cell malignancy that comprises up to 6% of non-Hodgkin lymphomas diagnosed annually and is associated with a poor prognosis. The average overall survival of patients with MCL is 5 years, and for most patients who progress on targeted agents, survival remains at a dismal 3 to 8 months. There is a major unmet need to identify new therapeutic approaches that are well tolerated to improve treatment outcomes and quality of life. The protein arginine methyltransferase 5 (PRMT5) enzyme is overexpressed in MCL and promotes growth and survival. Inhibition of PRMT5 drives antitumor activity in MCL cell lines and preclinical murine models. PRMT5 inhibition reduced the activity of prosurvival AKT signaling, which led to the nuclear translocation of FOXO1 and modulation of its transcriptional activity. Chromatin immunoprecipitation and sequencing identified multiple proapoptotic BCL-2 family members as FOXO1-bound genomic loci. We identified BAX as a direct transcriptional target of FOXO1 and demonstrated its critical role in the synergy observed between the selective PRMT5 inhibitor, PRT382, and the BCL-2 inhibitor, venetoclax. Single-agent and combination treatments were performed in 9 MCL lines. Loewe synergy scores showed significant levels of synergy in most MCL lines tested. Preclinical, in vivo evaluation of this strategy in multiple MCL models showed therapeutic synergy with combination venetoclax/PRT382 treatment with an increased survival advantage in 2 patient-derived xenograft models (P ≤ .0001, P ≤ .0001). Our results provide mechanistic rationale for the combination of PRMT5 inhibition and venetoclax to treat patients with MCL.
Introduction
Mantle cell lymphoma (MCL) is a CD5+/CD19+ B-cell non-Hodgkin lymphoma, defined by the t(11;14) translocation juxtaposing CCND1 downstream of the IgH promoter; this results in cyclin D1 overexpression and cell cycle dysregulation. MCL comprises up to 6% of non-Hodgkin lymphoma cases diagnosed annually1 and is associated with an overall poor prognosis due to multiple factors, including advanced stage of disease at diagnosis, resistance to standard immunochemotherapy regimens, and clinical factors.2 Because of the late median age of diagnosis (∼70 years of age3), aggressive chemotherapy and stem cell transplantation are often not realistic options.4 Without stem cell transplant, the average overall survival of patients with MCL is ∼6 years,5 and for most of the patients who progressed on targeted agents like ibrutinib before the recent Food and Drug Administration approval of brexucabtagene autoleucel chimeric antigen receptor T-cell therapy,6 survival remained very poor.7 Short of salvage immunochemotherapy followed by a stem cell transplant, relapse is virtually universal, and for the most part, MCL is considered incurable.8 Thus, there is a major unmet need to identify new therapeutic strategies that are well tolerated by less fit patients to improve prognosis and quality of life.3
Protein arginine methyltransferase 5 (PRMT5) is a type II PRMT enzyme that modulates the activity of a wide range of proteins through symmetric dimethylation of arginine residues (sDMA).9 PRMT5 is required for normal B-cell development and the formation of germinal centers via direct and indirect modulation of P53 and the spliceosome.10 We and others have documented the overexpression of PRMT5 and its oncogenic activity in promoting the growth and survival of MCL and other lymphoid malignancies.11-18 The sDMA activity of PRMT5 regulates many cellular functions, including alternative splicing, epigenetic control of gene expression, and survival/growth and death pathways orchestrated by P53,19,20 NF-κB/p65,11,21 BCL-6,22 and E2F1.23-25 Inhibition of PRMT5 leads to reduced cancer cell growth,26,27 abrogation of a stem cell phenotype,28,29 and increased survival in in vivo models.13,14,26,30 These observations have led to the development of several unique classes of small-molecule PRMT5 inhibitors11,14,31-33 that are currently being explored in clinical trials (#NCT03886831, #NCT04089449, #NCT05245500, #NCT05094336, and others).
Previous work has shown that PRMT5 promotes the survival of lymphoma cells by epigenetically suppressing AXIN2 and WIF1, supporting the WNT-β-CATENIN pathway, and enhancing AKT activity.34 AKT phosphorylates protein and lipid kinases, cell cycle regulators, and transcription factors, among others.35 AKT is also known to provide progrowth and survival signals through several pathways, including DNA damage repair,36 cell cycling,37 degradation of p53,38 and receptor tyrosine kinase signal modulation.39
One of the direct targets of AKT is the forkhead box protein O1 (FOXO1), a transcription factor canonically known as a tumor suppressor40-42 and critical for normal B-cell development.43,44 FOXO1 has been shown to be essential for pro-B cells to advance to pre-B cells, peripheral blood B cells to traffic to lymph nodes, and to support immunoglobulin class switching to drive efficient antibody memory responses.43 In cancer, FOXO1 regulates cell cycle,45,46 autophagy,47,48 and has been correlated with prognosis in multiple types of cancer.49-51 These functions are suppressed by AKT activity, where AKT phosphorylates FOXO1, preventing FOXO1’s transcriptional activity and triggering export from the nucleus.41 In lymphomas, PRMT5 supports the activity of AKT through the sDMA of R391 of AKT.34,52,53 We hypothesized that PRMT5 inhibition may lead to interruption of this pathway and restore tumor suppressor activity of downstream targets like FOXO1.
Here, we show how PRMT5 inhibition leads to the dissociation of AKT and FOXO1, followed by the nuclear translocation of FOXO1 and recruitment to the promoter regions of target genes, including members of the proapoptotic BCL-2 family. After PRMT5 inhibition, FOXO1 directly binds to the promoter region of BCL-2–associated X protein (BAX), leading to its increased expression and decreased apoptotic threshold in MCL cells. We hypothesized this would drive a therapeutic vulnerability to BCL-2 inhibition and demonstrated that treatment with the BCL-2 inhibitor, venetoclax, and the PRMT5 inhibitor, PRT382 leads to synergistic cell death of MCL cells in both in vitro and in vivo preclinical models. Basal expression of BCL-2 was found to correlate with the synergistic antitumor activity of this combinatorial strategy. This study provides support for combining PRMT5 and BCL-2 inhibition in clinical trials for patients with MCL.
Materials and methods
Cell culture, measurement of antitumor activity, and synergy
Nine cell lines were used in this work: Jeko, Rec-1, SP53, UPN-1, CCMCL1, Z-138, Mino, Maver-1, and Granta-519. All lines were cultured at 37°C, 5% CO2, in RPMI 1640 supplemented with 10% FBS, 1% glutamax, and 1% penicillin/streptomycin. Cell lines were validated by short tandam repeats (STR) sequencing. Mycoplasma testing was performed monthly. PRT382 was supplied by Prelude Therapeutics (Wilmington, DE). Venetoclax (ABT-199) was purchased from MedChemExpress. Inhibitory concentration (IC50s), defined as a 50% reduction in the percentage of live cells, were measured with annexin V/Propidium iodide (PI) staining and flow cytometry. IC50s were measured at day 9 for PRT382 and day 3 for venetoclax. Synergy was measured via 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay at day 9 after 6 days of PRT382 pretreatment and 3 days of combination treatment. Synergy scores and plots were calculated with the Loewe model via Combenefit.54
Chromatin immunoprecipitation sequencing (ChIP-seq) and quantitative polymerase chain reaction (q-PCR)
Sample preparation, library construction, and ChIP-seq were performed as described previously.55 Briefly, cells treated with and without PRT382 were harvested after 48 hours and fixed. Nuclei were harvested and chromatin sheared via sonication before immunoprecipitation with 10 μg of anti-FOXO1 (custom-raised rabbit polyclonal) was performed at 4°C overnight. The preparation was cleaned with RNase and proteinase K. DNA was reverse-crosslinked and extracted via NucleoSpin Gel and a PCR cleanup DNA extraction kit. Libraries were generated using the KAPA Hyper Prep kit and 8-cycle PCR amplified, followed by purification using 1X SPRI beads. Sequencing and postprocessing of the raw data were performed at the Genomics Core facility at Weill Cornell Medicine.
Reverse transcription was carried out on 200 ng of total RNA using the RevertAid RT kit. reverse transcription qPCR was performed on complementary DNA samples using the PowerUp SYBR Green Master Mix on the 7500 Fast Real-time PCR system. The messenger RNA (mRNA) level of each sample was normalized to that of beta actin (ACTB) mRNA. The relative mRNA level was presented as unit values of 2ˆdCt (=Ct of ACTB-Ct of gene).
Western blotting and immunofluorescence
Cells were treated with small-molecule inhibitors for up to 9 days, with media changed completely every 3 days. Doses are listed in supplemental Table 1 and were chosen to maintain viability above 70% at the time of collection. Cells were harvested by pelleting at 300g for 10 minutes, washed with ice-cold phosphate-buffered saline, and pelleted at 300g for 8 minutes at 4°C. Lysates were made using radioimmunopercipitation assay (RIPA) buffer with phosphatase and protease inhibition cocktails. Western blots were run with 20 to 30μg of protein on 4% to 20% sodium dodecyl sulfate- polyacrylamide gel electrophoresis gels before being transferred to Polyvinylidene fluoride (PVDF) using the Turbo Transfer System. Blots were blocked, probed, washed, and imaged according to LiCor protocols. For immunofluorescence, cells were fixed with paraformaldehyde and permeabilized with 0.2% TX100. Incubation with the primary antibody was performed overnight at 4°C and imaging was completed with an Alexa488-conjugated donkey anti-rabbit immunoglobulin G (IgG) secondary. FOXO1 localization was determined by quantification of cells with FOXO1-enriched nuclei by view field. Additional primary and secondary antibodies are listed in the supplemental Methods.
Knockdown cell lines
BAX and BAK1 knockdown cell lines were created using 2 short hairpin RNA (shRNA) plasmids (Mission shRNA, Sigma) for each gene. Briefly, glycerol bacterial stocks were expanded and harvested for plasmid. This was transduced with a packaging and envelope plasmid into Lenti X 293T cell. Virus was produced, collected, and used to transduce cells of interest. Successfully transduced cells were selected with puromycin and knockdown was confirmed via reverse transcription PCR and western blot. A pLKO.1 empty plasmid SHC001 was used as a control. PRMT5 knockdown cell line details can be found in supplemental Methods.
In vivo studies
Two patient-derived xenograft (PDX) and 2 cell line–derived xenograft (CDX) models were used in this work. The CCMCL1 CDX, PDX.AA.MCL, and PDX.IR.96069 studies were performed at the Ohio State University (OSU) under protocol 2009A0094-R4 and institution animal care and use comittee (IACUC) approval. PDX.AA.MCL was developed by the OSU lymphoma group from an ibrutinib-resistant patient sample,56 whereas PDX.IR.96069 was obtained from PROXE57 and tested for continued ibrutinib resistance. The Granta-519 CDX flank model was performed at Crown Bioscience on behalf of Prelude Therapeutics under their ethical guidelines. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) or NOD.Cg-Prkdcscid/J (NOD SCID) mice (OSU ULAR or LC Shanghai Lingchang Bio-Technology co, LTD) were engrafted either via tail vein or on the flank with 10e6 cells. Disease burden was monitored via flow cytometry or measurement of tumor size. Mice were dosed variably with the CCMCL1 CDX (refer to supplemental Methods), 4 days on, 3 days off (PDX.AA.MCL and PDX.IR.96069), or daily (Granta-519 CDX) via oral gavage.
Statistics
Data were analyzed with a two-way analysis of variance, Student t test, Spearman correlation, or log-rank (Mantel-Cox) test, as applicable. To compare changes in disease burden over time, we used generalized estimating equations with autoregressive correlation structure to test the differences in slopes between groups. For this exploratory preclinical study, P values were not adjusted for potential multiple comparisons. Error bars show the standard deviation of the data. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.
Results
Selective inhibition of PRMT5 with PRT382 is cytotoxic against MCL cells in vitro and in vivo
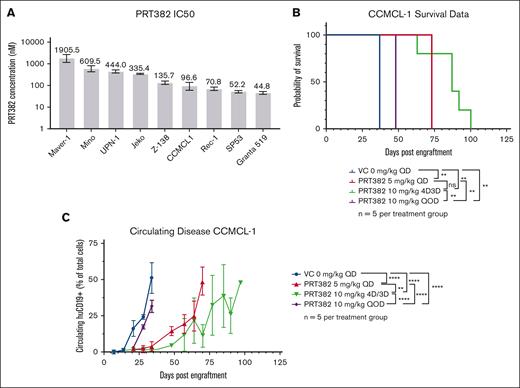
To selectively target PRMT5 activity in MCL, we used PRT382 (Prelude Therapeutics), a novel S-adenosyl methionine competitive, selective small-molecule inhibitor of PRMT5 enzymatic activity58 (supplemental Figure 1A). In vitro anti-MCL activity was demonstrated in 9 cell lines, which showed IC50s ranging from 44.8 to 1905.5 nM, with the maximal effect occurring on day 9 (Figure 1A; supplemental Figure 1B). This compound was found to be more potent than EPZ015666 (GSK3235025), a well-described PRMT5 inhibitor58 (supplemental Figure 1C). In vivo, PRT382 demonstrated a favorable oral bioavailability and pharmacokinetic profile in mice (area under curve, 1175 h∗kg∗ng/mL per mg at 10 mg/kg) (supplemental Figure 1D). The human MCL CDX, CCMCL1, was used to evaluate a range of doses and schedules for evaluation of toxicity and antitumor activity. A dose of 10 mg/kg every other day showed the lowest efficacy compared with a dose of 5 mg/kg daily or 10 mg/kg 4 days on, 3 days off (4D/3D), demonstrating the importance of a dosing schedule for this compound (Figure 1B-C). PRT382 delivered on a dose and schedule of 10 mg/kg 4D/3D off achieved a prolonged significant reduction in circulating disease and extended median survival from 37 to 87 days (Figure 1B-C; P < .01). This schedule avoided dose-limiting toxicities, defined as greater than 10% body weight loss in a week (supplemental Figure 1E), and provided robust antitumor activity. Despite the significant survival advantage, all treated mice eventually reached early removal criteria (ERC) because of the MCL disease burden, prompting consideration of combination strategies.
PRMT5 inhibition via PRT382 is effective at killing MCL in vitro and in vivo. (A) IC50 of 9 MCL cell lines measured by annexin V/PI staining and flow cytometry on day 9. (B) Survival of CCMCL1-engrafted NSG mice treated with PRT382 at varying doses and frequencies. PRT382 10 mg/kg 4D3D showed the greatest survival advantage (P < .01). QD – daily, 4D3D – 4 days on treatment, 3 days off QOD – every other day. (C) Circulating huCD19+ cells in CCMCL1-engrafted NSG mice. PRT382 treatment significantly delayed disease progression. A log-rank test with significance was used for panel B. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Error bars show standard deviation of the data.
PRMT5 inhibition via PRT382 is effective at killing MCL in vitro and in vivo. (A) IC50 of 9 MCL cell lines measured by annexin V/PI staining and flow cytometry on day 9. (B) Survival of CCMCL1-engrafted NSG mice treated with PRT382 at varying doses and frequencies. PRT382 10 mg/kg 4D3D showed the greatest survival advantage (P < .01). QD – daily, 4D3D – 4 days on treatment, 3 days off QOD – every other day. (C) Circulating huCD19+ cells in CCMCL1-engrafted NSG mice. PRT382 treatment significantly delayed disease progression. A log-rank test with significance was used for panel B. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Error bars show standard deviation of the data.
PRMT5 inhibition promotes reactivation and relocalization of FOXO1 in MCL
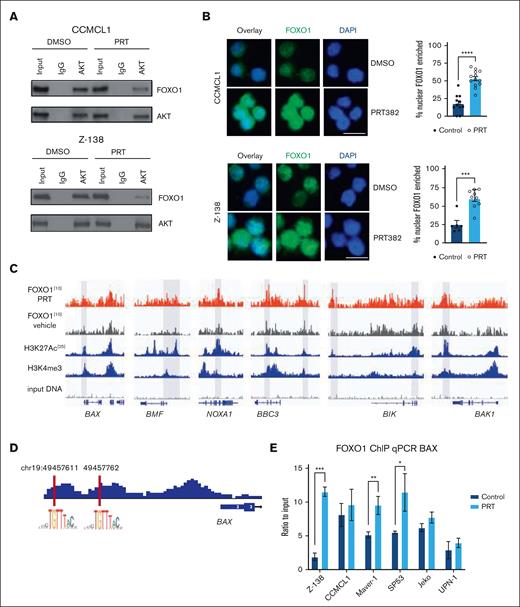
We and others have previously reported that PRMT5 directly and indirectly supports AKT activity.34,53 We hypothesized that the reduced AKT activity occurring because of PRMT5 inhibition would lead to perturbation of the AKT:FOXO1 interaction, FOXO1 nuclear translocation, and modulation of genes with tumor suppressor activity. Using CCMCL1 and Z-138 as representative MCL cell lines, we confirmed that the PRMT5 inhibition disrupted the physical interaction between AKT and FOXO1 (Figure 2A). We evaluated the nuclear localization of FOXO1 in CCMCL1 and Z-138 with immunofluorescence, comparing control and PRMT5-inhibited cells. Within the nuclear compartment, we saw increased levels of FOXO1 after PRMT5 inhibition (both P < .001) (Figure 2B). This observation led us to explore FOXO1 recruitment among potential target promoters.
PRMT5 inhibition activates the FOXO1-dependent transcription program in MCL. (A) Immunoprecipitation of AKT in CCMCL1 and Z-138 cells shows the decrease in interaction between AKT and FOXO1 with treatment of 100 or 150 nM, respectively, of PRT382 for 6 days. (B) Immunofluorescence of CCMCL1 and Z-138 cell lines looking at the localization of FOXO1 after 72 hours of treatment with 100nM of PRT382 or dimethyl sulfoxide (DMSO). Cells are stained with FOXO1 primary antibody and alexafluor488-conjugated donkey anti-rabbit secondary. Cells were also stained with 4′,6-diamidino-2-phenylindole (DAPI). Images taken on an EVOS FL Cell Auto Imaging system at 40× original magnification. The number of cells with FOXO1 enriched in the nucleus was quantified and graphed. Scale bar, 10 μM. (C) Traces of ChIP sequencing performed on FOXO1 in CCMCL1 cells treated for 48 hours with 100 nM of PRT382 or DMSO as a control. Selected BCL-2 family proteins are shown here. (D) FOXO1 consensus sequence was confirmed in the promoter sequences of BAX upstream of the gene body. (E) FOXO1 ChIP qPCR of BAX showing significant enrichment in Z-138, Maver-1, and SP53 as well as a trend in CCMCL1, Jeko, and UPN-1 after 72 hours of PRT382 treatment. A student t test was performed to show significance for panels B,E. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Error bars show standard deviation of the data.
PRMT5 inhibition activates the FOXO1-dependent transcription program in MCL. (A) Immunoprecipitation of AKT in CCMCL1 and Z-138 cells shows the decrease in interaction between AKT and FOXO1 with treatment of 100 or 150 nM, respectively, of PRT382 for 6 days. (B) Immunofluorescence of CCMCL1 and Z-138 cell lines looking at the localization of FOXO1 after 72 hours of treatment with 100nM of PRT382 or dimethyl sulfoxide (DMSO). Cells are stained with FOXO1 primary antibody and alexafluor488-conjugated donkey anti-rabbit secondary. Cells were also stained with 4′,6-diamidino-2-phenylindole (DAPI). Images taken on an EVOS FL Cell Auto Imaging system at 40× original magnification. The number of cells with FOXO1 enriched in the nucleus was quantified and graphed. Scale bar, 10 μM. (C) Traces of ChIP sequencing performed on FOXO1 in CCMCL1 cells treated for 48 hours with 100 nM of PRT382 or DMSO as a control. Selected BCL-2 family proteins are shown here. (D) FOXO1 consensus sequence was confirmed in the promoter sequences of BAX upstream of the gene body. (E) FOXO1 ChIP qPCR of BAX showing significant enrichment in Z-138, Maver-1, and SP53 as well as a trend in CCMCL1, Jeko, and UPN-1 after 72 hours of PRT382 treatment. A student t test was performed to show significance for panels B,E. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Error bars show standard deviation of the data.
FOXO1 reactivation promotes the expression of proapoptotic BCL-2 family proteins
To determine the downstream response of FOXO1 activation, CCMCL1 cells were treated with PRT382 for 48 hours and then processed for ChIP sequencing. Among those genes associated with FOXO1 peaks and sites of active transcription, genes in the BCL-2 family were of particular interest (Figure 2C; supplemental Table 2). This collection of proteins containing a BH3 motif includes both proapoptotic and prosurvival proteins, where the balance of concentrations and interactions determines whether a cell enters intrinsic apoptosis. We found FOXO1 to be associated with the proapoptotic genes BAX, BAK1, BIK, and BBC3 (Figure 2C). These genes either produce direct effectors of apoptosis, as in the case of BAX and BAK1, or mediate apoptotic activity, as in the case of BIK and BBC3. In support of the ChIP-seq data, we identified the presence of a FOXO1 consensus–binding motif (5′-GTAAA(T/C)A-3’)59 in the BAX gene promoter (Figure 2D). ChIP q-PCR confirmed that FOXO1 was significantly enriched on the BAX gene promoter in Z-138, Maver-1, and SP53 cell lines after PRT382 treatment (Figure 2E). ChIP qPCR on Z-138, CCMCL1, and Maver-1 cell lines also confirmed increased enrichment for FOXO1 binding to the active regulatory regions of BAK1, BIK, or NOXA1 when PRMT5 was inhibited (supplemental Figure 2A-C).
Supporting the relevance of FOXO1 enrichment on these genes, qPCR showed several proapoptotic BH3 family members were upregulated on a transcript level after PRMT5 inhibition (Figure 3A-C). Western blotting showed that BAX, BAK1, and BBC3 protein levels were all upregulated in multiple MCL cell lines after 6 days of PRMT5 inhibition (Figure 3D-G). BAX was the most frequently upregulated and had the greatest fold increase across all cell lines with PRMT5 inhibition (Figure 3D). This increase was confirmed to be an on-target effect of PRMT5 inhibition and FOXO1 activity through genetic knockdown (supplemental Figure 2D-E). These data show that proapoptotic proteins are upregulated with PRMT5 inhibition and suggest that FOXO1 regulates the expression of the key proapoptotic protein BAX.
PRMT5 inhibition promotes apoptosis through the transcription and translation of proapoptotic genes. qPCR measurement of the transcripts of select BCL-2 family genes in (A) Z-138, (B) CCMCL1, and (C) Maver-1 after 72 hours of 100 nM PRT382 treatment. Quantification of (D) BAX, (E) BAK1, and (F) BBC3 (Puma) protein levels with 6 days of PRT382 treatment. (G) Representative western blot of 6 MCL cell lines after treatment with 6 days of PRT382 or DMSO showing the levels of BAX, BAK1, and BBC3 including their cleaved forms. PRT382 doses are as follows: Z-138 150 nM, CCMCL1 100 nM, Granta 519 50 nM, Maver-1 1 μM, Mino 450 nM, and Jeko 300 nM. Note: BBC3 expression in CCMCL1 is extremely low compared with the other cell lines; an overexposed blot with CCMCL1 bands can be found in supplemental Figure 9. A student t test was used to determine significance for panels A-F. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
PRMT5 inhibition promotes apoptosis through the transcription and translation of proapoptotic genes. qPCR measurement of the transcripts of select BCL-2 family genes in (A) Z-138, (B) CCMCL1, and (C) Maver-1 after 72 hours of 100 nM PRT382 treatment. Quantification of (D) BAX, (E) BAK1, and (F) BBC3 (Puma) protein levels with 6 days of PRT382 treatment. (G) Representative western blot of 6 MCL cell lines after treatment with 6 days of PRT382 or DMSO showing the levels of BAX, BAK1, and BBC3 including their cleaved forms. PRT382 doses are as follows: Z-138 150 nM, CCMCL1 100 nM, Granta 519 50 nM, Maver-1 1 μM, Mino 450 nM, and Jeko 300 nM. Note: BBC3 expression in CCMCL1 is extremely low compared with the other cell lines; an overexposed blot with CCMCL1 bands can be found in supplemental Figure 9. A student t test was used to determine significance for panels A-F. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
PRMT5 and BCL-2 inhibition drive synergistic MCL cell death
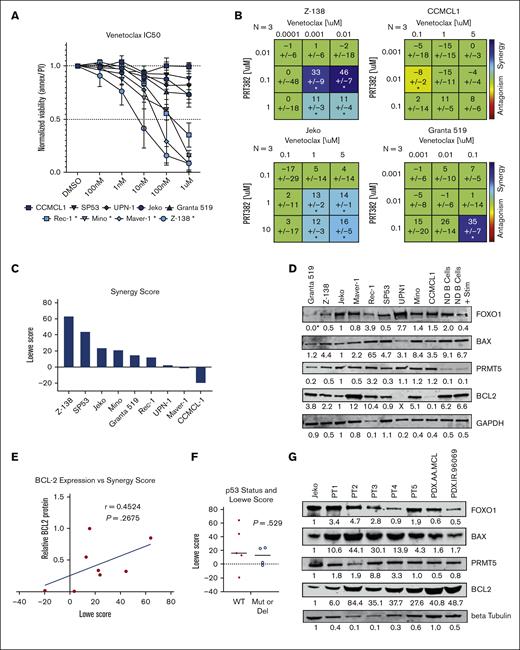
With the upregulation of proapoptotic proteins, including BAX with PRMT5 inhibition, we looked for an agent that could capitalize on this shift in intrinsic apoptotic signaling. The protein BCL-2 binds to and blocks the activity of BAX so we chose to test venetoclax, a BCL-2 inhibitor approved for the treatment of chronic lymphocytic leukemia.60 As a single agent, venetoclax produced IC50 values below 1 μM in 4 of the MCL lines tested (Figure 4A). We evaluated the synergy of venetoclax in combination with PRTM5 inhibition via PRT382 using MTS and the Loewe Model of Synergy computed by Combenefit54,61 (Figure 4B-C; supplemental Figure 3). Z-138 was found to be the most sensitive to the combination treatment, with synergy scores reaching as high as 63.9 (Figure 4C), whereas other lines such as Mino and Jeko were found to be moderately sensitive. The range of sensitivities across 9 MCL cell lines, as shown in Figure 4C, shows significant synergy in 6 of the 9 cell lines tested (synergy score, 12.8-63.9). This led us to explore why 3 lines showed resistance to this combination and whether we could determine a biomarker correlative to the degree of synergy.
Six of 9 MCL cell lines show synergistic killing with treatment with PRT382 and BCL-2 inhibition, venetoclax. (A) IC50 of 9 MCL cell lines measured with annexin V/PI and flow cytometry after 72 hours of treatment with venetoclax. Cell lines with an IC50 below 1 μM are starred and considered sensitive. (B) Synergy matrices calculated through Combenefit54 using the Lowe model of synergy. Values below −10 are antagonistic, −10 to 10 are additive, and values over 10 are synergistic. Significance is shown by stars at the bottom of each box. ∗P < .05; ∗∗P < .01; ∗∗∗ P < .001. (C) Single synergy values calculated from at least 3 separate replicates for each cell line. The same levels of synergy are used as with the synergy matrices. (D) Western blot showing the baseline levels of key proteins in MCL cell lines and normal donor B Cells (ND B cells) without and with IgG stimulation. Values are corrected by the loading control and normalized to Jeko. (E) The correlation between the baseline level of BCL-2 protein and the Lowe score seen across the MCL cell lines (Maver-1 was censored as an outlier). (F) Correlation between P53 status and synergy score. (G) Western blot showing the baseline levels of key proteins in Jeko, 5 primary patient peripheral blood mononuclear cells (PBMCS), and 2 PDX murine models of MCL. Values are corrected by the loading control and normalized to Jeko. A Spearman correlation (E) or Student t test (F) was used to determine significance P < .05; ∗∗P < .01 ∗∗∗P < .001.
Six of 9 MCL cell lines show synergistic killing with treatment with PRT382 and BCL-2 inhibition, venetoclax. (A) IC50 of 9 MCL cell lines measured with annexin V/PI and flow cytometry after 72 hours of treatment with venetoclax. Cell lines with an IC50 below 1 μM are starred and considered sensitive. (B) Synergy matrices calculated through Combenefit54 using the Lowe model of synergy. Values below −10 are antagonistic, −10 to 10 are additive, and values over 10 are synergistic. Significance is shown by stars at the bottom of each box. ∗P < .05; ∗∗P < .01; ∗∗∗ P < .001. (C) Single synergy values calculated from at least 3 separate replicates for each cell line. The same levels of synergy are used as with the synergy matrices. (D) Western blot showing the baseline levels of key proteins in MCL cell lines and normal donor B Cells (ND B cells) without and with IgG stimulation. Values are corrected by the loading control and normalized to Jeko. (E) The correlation between the baseline level of BCL-2 protein and the Lowe score seen across the MCL cell lines (Maver-1 was censored as an outlier). (F) Correlation between P53 status and synergy score. (G) Western blot showing the baseline levels of key proteins in Jeko, 5 primary patient peripheral blood mononuclear cells (PBMCS), and 2 PDX murine models of MCL. Values are corrected by the loading control and normalized to Jeko. A Spearman correlation (E) or Student t test (F) was used to determine significance P < .05; ∗∗P < .01 ∗∗∗P < .001.
BCL2 expression is a biomarker for synergy response in MCL cells
We explored the basal expression of key proteins to determine if a correlative pattern of expression was associated with the level of antitumor synergy observed in cell lines (Figure 4D). Baseline levels of BCL-2, BAX, FOXO1, and PRMT5 and the ratio of BAX to BCL-2 expression were all correlated to Loewe synergy scores. BCL-2 expression, with Maver-1 censored as an outlier, provided the strongest correlation (Spearman r = 0.4524, P = .2675), and higher expression of BCL-2 resulted in higher synergy scores (Figure 4E). FOXO1 and PRMT5 expression had negative correlations with synergy score, whereas BAX expression and the ratio of BAX to BCL-2 resulted in positive correlations (supplemental Figure 4A-D). Clustering the cell lines by p53 status (wild type or mutant) showed no difference in synergy scores (Figure 4F, P = .529). Five patient samples and 2 MCL PDX model cells were also analyzed for basal expression of BAX, BCL-2, FOXO1, and PRMT5 (Figure 4G). BCL-2 was found to be highly expressed in all samples tested, suggesting a high level of synergy could be achieved.
PRMT5 inhibition in combination with venetoclax induces intrinsic apoptosis dependent on BAX expression
To determine the mechanism driving cell death with drug treatment, caspase 3, 8, and 9 expression was examined in cells treated with DMSO, PRT382 only, venetoclax only, or the combination. The cleavage of caspase 9, indicating intrinsic apoptosis, was seen as early as day 2 (supplemental Figure 5), whereas caspase 8 remained relatively unchanged, showing little to no extrinsic apoptosis signaling. Corresponding with viability measurements, the greatest cleavage of caspases 3 and 9 was seen in the combo treatment cohort on day 6 (Figure 5A).
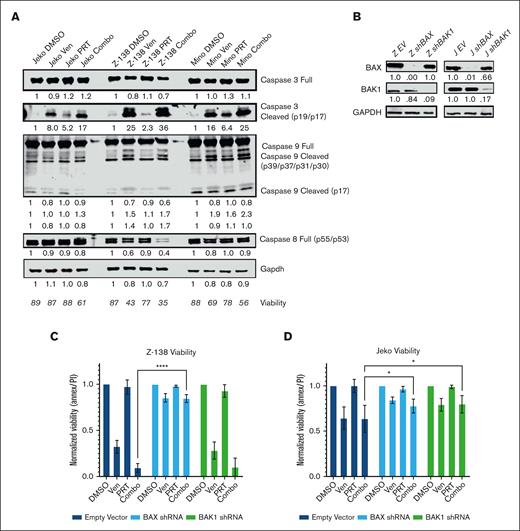
Synergistic intrinsic apoptosis is dependent on the BAX protein. (A) Western blot showing caspase activity with control, single agent, and combo-treated Jeko, Z-138, and Mino MCL cell lines (4 days of exposure). Jeko doses PRT382 at 300 nM, venetoclax at 1 μM, Z-138 doses PRT382 at 150 nM, venetoclax at 10 nM, Mino doses PRT382 at 450 nM, venetoclax at 10 nM. Caspase cleavage was used as an indication of activity, with caspase 8 being indicative of extrinsic apoptosis, caspase 9 of intrinsic apoptosis, and caspase 3 of general apoptosis. (B) Western blots showing the knockdown of BAX and BAK1 proteins after transfection with shRNA against these transcripts in Z-138 and Jeko cells. Values are adjusted by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and normalized to the empty vector control. (C) Viability of Z-138 and (D) Jeko knockdown variants with control, single agent, or combo treatment for 4 days. Doses as in panel A. At least 3 replicates were completed, and data were measured with annexin V/PI staining and flow cytometry. A 2-way analysis of variance with multiple comparisons was used to determine statistical significance for panels C-D. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Synergistic intrinsic apoptosis is dependent on the BAX protein. (A) Western blot showing caspase activity with control, single agent, and combo-treated Jeko, Z-138, and Mino MCL cell lines (4 days of exposure). Jeko doses PRT382 at 300 nM, venetoclax at 1 μM, Z-138 doses PRT382 at 150 nM, venetoclax at 10 nM, Mino doses PRT382 at 450 nM, venetoclax at 10 nM. Caspase cleavage was used as an indication of activity, with caspase 8 being indicative of extrinsic apoptosis, caspase 9 of intrinsic apoptosis, and caspase 3 of general apoptosis. (B) Western blots showing the knockdown of BAX and BAK1 proteins after transfection with shRNA against these transcripts in Z-138 and Jeko cells. Values are adjusted by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and normalized to the empty vector control. (C) Viability of Z-138 and (D) Jeko knockdown variants with control, single agent, or combo treatment for 4 days. Doses as in panel A. At least 3 replicates were completed, and data were measured with annexin V/PI staining and flow cytometry. A 2-way analysis of variance with multiple comparisons was used to determine statistical significance for panels C-D. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
From our western blots, we determined that BAX was the most commonly and significantly upregulated protein among the proapoptotic BCL-2 family. To examine the importance of BAX for venetoclax activity and the synergistic response with combination treatment, we created BAX knockdown lines with Z-138, Jeko, Granta-519, and Maver-1 using shRNA (Figure 5B; supplemental Figure 6). As BAK1 is also capable of triggering the mitochondrial depolarization that leads to intrinsic apoptosis, we also created shRNA knockdowns using the same 4 cell lines (Figure 5B; supplemental Figure 6). Each line was treated with DMSO, PRT382 only, venetoclax only, or the combination for 4 days. Annexin V/PI staining with flow cytometry was used to measure the viability of each treatment. As seen in Figure 5C, BAX knockdown was protective in Z-138 (P < .0001), whereas both knockdowns of BAX and BAK1 were protective in Jeko (P = .0285, P = .0102) (Figure 5D). Granta-519 also showed a trend toward rescue with BAX knockdown (P = .223) (supplemental Figure 6B), whereas neither protein knockdown rescued Maver-1 cells (supplemental Figure 6D).
PRMT5 and BCL-2 inhibition is synergistic in vivo reducing disease burden and improving survival
One CDX and 2 MCL PDX models were used to test the combination of venetoclax and a PRMT5 inhibitor. The CDX was a flank model using Granta-519 cells engrafted subcutaneously. The combination of subtherapeutic PRT543, the clinical PRMT5 inhibitor for which PRT382 is the tool compound, and subtherapeutic venetoclax showed decreased tumor burden (supplemental Figure 7) compared with progressive disease in the single-agent cohorts.
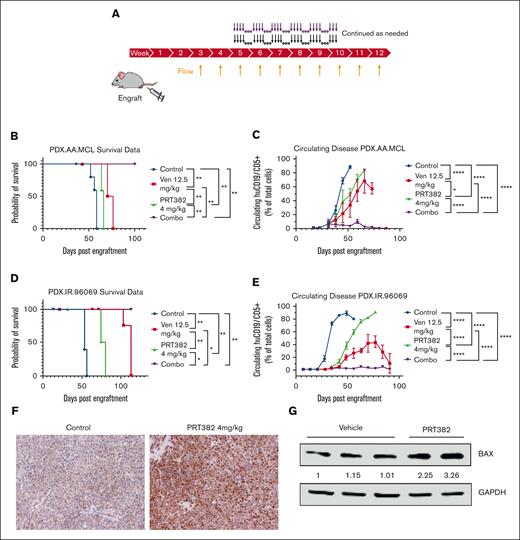
This led us to test the combination in 2 systemic PDX MCL models: PDX.AA.MCL developed from an ibrutinib-resistant patient sample by the OSU lymphoma research group,56 and PDX.IR.96069, an ibrutinib-resistant model obtained from PRoXe.57 NSG mice were engrafted with the respective cells and monitored weekly by flow cytometry for circulating huCD19+/huCD5+ cells. Once disease was detectable by flow cytometry, treatment began, 4 days on, 3 days off for both drugs (Figure 6A; supplemental Methods). Disease burden continued to be monitored weekly by flow cytometry and the examination of mice. Body weight was maintained during the course of treatment (supplemental Figure 8). The control and PRT-only cohorts reached a median survival of 58 days and 66 days in the AA model and 53 and 77 days in the 96069 model (Figure 6B,D). The venetoclax only cohort in the AA model had also reached ERC with a median survival of 73 days (Figure 6B), whereas a median survival was not reached in the 96069 model (Figure 6D). During the course of the experiment, the combination treatment reduced the tumor burden in the peripheral blood below the level of detection via flow cytometry (Figure 6C,E), which translated into a statistically significant survival advantage as no mice had reached ERC by the end of the experiment (days 100 and 104) (Figure 6B,D). FOXO1 expression was found to be increased in PRT382-treated mice (Figure 6F), supporting the role of FOXO1 in the PRMT5 inhibitor response. As with the in vitro results, BAX expression was upregulated in PRT382-treated mice, supporting the role of this protein in the synergistic response (Figure 6G). These results show significant synergy between PRMT5 and BCL-2 inhibition, reducing systemic disease burden and improving survival in 2 MCL PDX models.
Combination treatment with PRT382 and venetoclax is synergistic in vivo. (A) Schematic showing the setup of in vivo experiments. Mice were engrafted on day 0, and weekly bleeds with flow cytometry were used to assess the circulating disease burden. Once ∼1% of the circulating lymphocytes were MCL cells (huCD5+/huCD19+), treatment began. Mice were treated with each single agent or the combination for 4 days on, 3 days off. (B) Kaplan-Meier curve showing survival of the PDX.AA.MCL model. Median survival was 58 days for vehicle, 63 days for PRT, 73 days for venetoclax, and the combo did not reach a median survival by the experiment’s end of 101 days P < .0001. (C) A graph of circulating disease burden over time in the PDX.AA.MCL model is measured as a percentage of huCD5+/huCd19+ cells in the lymphoid compartment. (D) Kaplan-Meier curve showing the survival of the PDX.IR.96069 model. The median survival was 53 days for vehicle and 77 days for PRT. The venetoclax and combo groups did not reach a median survival by the experiment’s end at 104 days. (E) A graph of circulating disease burden over time in the PDX.IR.96069 model measured as a percentage of huCD5+/huCd19+ cells in the lymphoid compartment. (F) Immunohistochemistry of FOXO1 (brown) of a control and PRT382-treated mouse spleen. Scale bars, 100 μm. (G) Western blot showing BAX expression of cells from vehicle or PRT382-treated mice. Values are adjusted by GAPDH loading control and normalized against lane 1. A log-rank test for trend was performed on the survival data in panels B,D. Generalized estimating equations with an autoregressive correlation structure were used to compare the disease burden over time in panels C,E. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Combination treatment with PRT382 and venetoclax is synergistic in vivo. (A) Schematic showing the setup of in vivo experiments. Mice were engrafted on day 0, and weekly bleeds with flow cytometry were used to assess the circulating disease burden. Once ∼1% of the circulating lymphocytes were MCL cells (huCD5+/huCD19+), treatment began. Mice were treated with each single agent or the combination for 4 days on, 3 days off. (B) Kaplan-Meier curve showing survival of the PDX.AA.MCL model. Median survival was 58 days for vehicle, 63 days for PRT, 73 days for venetoclax, and the combo did not reach a median survival by the experiment’s end of 101 days P < .0001. (C) A graph of circulating disease burden over time in the PDX.AA.MCL model is measured as a percentage of huCD5+/huCd19+ cells in the lymphoid compartment. (D) Kaplan-Meier curve showing the survival of the PDX.IR.96069 model. The median survival was 53 days for vehicle and 77 days for PRT. The venetoclax and combo groups did not reach a median survival by the experiment’s end at 104 days. (E) A graph of circulating disease burden over time in the PDX.IR.96069 model measured as a percentage of huCD5+/huCd19+ cells in the lymphoid compartment. (F) Immunohistochemistry of FOXO1 (brown) of a control and PRT382-treated mouse spleen. Scale bars, 100 μm. (G) Western blot showing BAX expression of cells from vehicle or PRT382-treated mice. Values are adjusted by GAPDH loading control and normalized against lane 1. A log-rank test for trend was performed on the survival data in panels B,D. Generalized estimating equations with an autoregressive correlation structure were used to compare the disease burden over time in panels C,E. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Discussion
The discovery of new therapeutic strategies is essential for patients with MCL because this disease is currently incurable. Although targeted therapies such as ibrutinib have improved outcomes for patients with MCL over the last 10 years, patients almost uniformly develop progressive, resistant disease and have very poor prognoses.62,63 Here, we describe novel mechanistic data and provide rationale for combining PRMT5 and BCL-2 inhibition for treating patients with MCL. After treatment with a PRMT5 inhibitor, we observed a physical dissociation of FOXO1 from AKT, freeing it from inhibition and facilitating its nuclear translocation, where it promotes the transcription of a key BCL-2 family gene, BAX. This novel PRMT5-AKT-FOXO1-BCL-2 family axis was observed in multiple MCL cell lines. Modulation of this axis through the inhibition of PRMT5 created a sensitivity to the BCL-2 inhibitor venetoclax, as shown by synergistic cell death with combination treatment in vitro and in vivo.
Currently, patients who relapse on ibrutinib have few available lines of treatment and a very poor prognosis.63,64 Of the 5 ibrutinib-resistant cell lines assessed, 3 lines show synergistic cell death with PRMT5 and BCL-2 inhibition. The 3 murine models used for the combination studies are also ibrutinib-resistant, as previously determined by in vitro testing for Granta-519 and in vivo dosing for the PDX.AA.MCL and PDX.IR.96069 models.56 Our preclinical data show strong evidence in support of using PRMT5 inhibition in combination with venetoclax, especially in the setting of ibrutinib resistance.
With an increased survival advantage and undetectable disease burden, our in vivo models showed impressive responses to combination treatment. This work should be continued to optimize treatment strategies and determine what, if any, remaining disease burden exists in the combination cohort. We are exploring additional treatment regimens, including increasing the dose of PRT382 and venetoclax to therapeutic levels, using alternative PRMT5 inhibitors, and creating a reduced maintenance dosing regimen for long-term survival studies.
Here, we focused on the BH3 family genes that showed enrichment for FOXO1 recruitment after PRMT5 inhibition. FOXO1 ChIP sequencing after PRMT5 inhibition revealed over 2000 potential targets, providing an opportunity to better understand the biologic relevance of FOXO1 to the pathogenesis of MCL. Interestingly, attempts to create FOXO1 knockdown cell lines or use a small-molecule inhibitor resulted in cytotoxicity in all MCL cell lines, a counterintuitive finding suggesting that FOXO1 is relevant for MCL survival. FOXO1 likely plays a complex role in MCL, acting as an oncogene necessary for maintaining the B lymphocyte lineage transcriptional program to promote MCL survival65 while, in the context of PRMT5 inhibition, acting as a tumor suppressor.45,46 A similar dichotomy has been described by Zhao et al in colon cancer and HeLa cervical cancer cells, in which cytosolic FOXO1 is essential for stress-induced autophagy but also has the potential to induce apoptosis.47 Our data suggest that PRMT5 inhibition reprograms FOXO1 from an oncogene to a tumor suppressor through its transcriptional program. Further studies are needed to elucidate the context-dependent cellular roles of FOXO1 in PRMT5-driven tumors.
Although we found FOXO1 to play a role in the expression of BAX, it may not be the only player at work, as multiple proapoptotic BCL-2 family proteins were upregulated with PRMT5 inhibition and functioned independently of FOXO1. For example, the Z-138 cell line treated with PRT382 showed induction of BAX, BAK1, BIK, and BBC3 at the transcript and protein levels, whereas only the BAX promotor was enriched for FOXO1 interaction. P53 activity, although not correlative to synergy, could play a role as it is known as both a target of PRMT5 and a regulator of apoptosis.30 KLF4 has also been shown to be supported by PRMT5 activity and to repress BAX expression.66 The increased expression seen here is likely due to promotion from FOXO1 as well as release from inhibitors like KLF4.
In addition to AKT and FOXO1, there are numerous other targets of PRMT5; at the time of publication, the NCBI showed PRMT5 directly associates with over 300 proteins, from which there are numerous downstream targets.67 As we continue to unravel how PRMT5 orchestrates a malignant, resistant, and stem phenotype in cancers, following direct targets such as p53 or smD3 to their effectors will be important. This study shows that PRMT5 inhibition continues to be a promising target in cancer and supports ongoing PRMT5 inhibitor clinical trials. Because of the drug-resistant nature of MCL, combining these novel compounds with secondary therapeutics that can take advantage of an exposed vulnerability may prove crucial in the treatment of MCL.
Acknowledgments
The authors thank Reena Shakya and ULAR staff for help with the animal studies conducted at The Ohio State University (OSU). Prelude Therapeutics supported this study by providing PRMT5 selective inhibitors, scientific correspondence, and preliminary data.
This study was supported by funding from the National Institutes of Health (grant 5P01CA214274-03), Leukemia and Lymphoma Society (longitudinal functional genomics in mantle cell lymphoma therapy and drug resistance), and the OSU Comprehensive Cancer Center. The authors are grateful for the patients and healthy volunteers who provided tissue samples for these studies, to the OSU Comprehensive Cancer Center Leukemia Tissue Bank Shared Resource (supported by NCIP30 CA016058) for sample procurement.
F.B.-B. was supported by the OSU Biomedical Sciences Graduate Program Systems and Integrative Biology Training Fellowship, the Pelotonia Graduate Fellowship, and the OSU Presidential Fellowship during the course of this work.
Authorship
Contribution: F.B.-B., Y.Z., P.G., J.P., and R.A.B. conceptualized and designed the study; F.B.-B., S. Sloan, Y.Y., W.K.C., J.-H.C., Y.Z., J.P., and R.A.B. developed the methodology; F.B.-B., I.H., C.H., J.H.-M., A.P., J.-H.C., Y.Z., and S. Singh acquired the data; F.B.-B., I.H., S. Sloan, C.H., Z.C., M.L., Y.Z., O.E., R.L., J.P., and R.A.B. performed analysis and interpretation of data (eg, statistical analysis, biostatistics, computational and analysis); F.B.-B., C.H., S. Sloan, M.L., L.A., P.S., R.L., J.P., and R.A.B. wrote the manuscript; J.H.-M., W.K.C., A.P., N.B., J.-H.C., K.V., S.C.-K., M.D., O.E., L.S., L.A., P.S., R.L., J.P., and R.A.B. provided administrative, technical, or material support (ie, reporting or organizing data, selection of compounds, constructing databases); and J.P., and R.A.B. supervised the study.
Conflict-of-interest disclosure: Y.Z., K.V., P.S., and N.B. are employees of Prelude Therapeutics. Y.Z., K.V., P.S., and N.B. own stocks of Prelude Therapeutics. R.A.B. received research support from Prelude Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Robert A. Baiocchi, Division of Hematology, The Ohio State University, 400 W 12th Ave, 481B Wiseman CCC, Columbus, OH 43210; e-mail: robert.baiocch@osumc.edu; and Jihye Paik, Department of Pathology and Laboratory Medicine, Weill Cornell Medicine, 1300 York Ave, Room C336, New York, NY 10065; e-mail: jep2025@med.cornell.edu.
References
Author notes
∗J.P. and R.A.B. contributed equally to this study.
ChIP-seq data are available on the Gene Expression Omnibus database with accession number GSE182689.
Data are available on request from the corresponding author, Robert Baiocchi (robert.baiocch@osumc.edu).
The full-text version of this article contains a data supplement.