Key Points
CRISPR/Cas9–mediated editing in HSPC models of cancer development revealed Pbrm1 as a regulator of myeloid leukemia progression.
Pbrm1 loss leads to leukemic progression through immune regulation and interferon signaling.
Abstract
CRISPR/Cas9 screening approaches are powerful tool for identifying in vivo cancer dependencies. Hematopoietic malignancies are genetically complex disorders in which the sequential acquisition of somatic mutations generates clonal diversity. Over time, additional cooperating mutations may drive disease progression. Using an in vivo pooled gene editing screen of epigenetic factors in primary murine hematopoietic stem and progenitor cells (HSPCs), we sought to uncover unrecognized genes that contribute to leukemia progression. We, first, modeled myeloid leukemia in mice by functionally abrogating both Tet2 and Tet3 in HSPCs, followed by transplantation. We, then, performed pooled CRISPR/Cas9 editing of genes encoding epigenetic factors and identified Pbrm1/Baf180, a subunit of the polybromo BRG1/BRM-associated factor SWItch/Sucrose Non-Fermenting chromatin–remodeling complex, as a negative driver of disease progression. We found that Pbrm1 loss promoted leukemogenesis with a significantly shortened latency. Pbrm1-deficient leukemia cells were less immunogenic and were characterized by attenuated interferon signaling and reduced major histocompatibility complex class II (MHC II) expression. We explored the potential relevance to human leukemia by assessing the involvement of PBRM1 in the control of interferon pathway components and found that PBRM1 binds to the promoters of a subset of these genes, most notably IRF1, which in turn regulates MHC II expression. Our findings revealed a novel role for Pbrm1 in leukemia progression. More generally, CRISPR/Cas9 screening coupled with phenotypic readouts in vivo has helped identify a pathway by which transcriptional control of interferon signaling influences leukemia cell interactions with the immune system.
Introduction
Cancers are characterized by multiple genetic and epigenetic alterations in oncogenes and tumor suppressor genes. Although genomic studies have contributed to the discovery of cancer-associated genes, many genes are “passengers” rather than “drivers.”1 Hematological malignancies result from the sequential acquisition of driver mutations in a hematopoietic stem cell and its descendants. Over time, cooperating mutations in a subclone(s) may culminate in an overt malignancy.2,3 Large-scale sequencing of samples from patients with acute myeloid leukemia (AML) has provided a set of driver genes.4 Of these, mutations in genes encoding epigenetic modifiers and transcription factors are among the most frequent.5
The use of mouse models of human myeloid malignancies supports the detailed and focused investigation of selected driver mutations through various genetic manipulations and represent powerful tools in the study of these disorders.6 Advances in gene editing technologies, particularly CRISPR/Cas9, have facilitated disease modeling through the generation of specific genetic lesions, with 1 or multiple mutations in primary hematopoietic stem and progenitor cells (HSPCs) and subsequent transplantation into a suitable host. For the most part, such transplantation studies have focused on modifying 1 gene, several genes, or translocations using CRISPR/Cas9 and relied on the manipulation of previously recognized leukemia-associated genes rather than unbiased searches for new candidates.7-10
CRISPR screens enable high throughput interrogation of gene functions in diverse tumors, which have, to date, been performed ex vivo or in transplantation models. To recapitulate cancer development more faithfully, CRISPR screens have recently been used to modify genes within tissues in vivo and to identify drivers of tumorigenesis in native microenvironments, such as glioma and liver tumors.11,12 Compared with ex vivo or transplantation-based approaches, screens that target tissues in vivo have the advantages of retaining the native microenvironment and interactions with the immune system.13
Here, we performed a pooled CRISPR/Cas9 gene editing screen of primary HSPCs with the goal of identifying factors that participate in leukemogenesis in vivo in the context of the bone marrow environment. We hypothesized that in vivo screening might reveal previously unknown genes that cooperate with driver mutations to accelerate disease progression. We, first, demonstrated that in vivo CRISPR/Cas9 targeting of Tet2 and Tet3 leads to myeloid leukemia. With mutation of Tet2 as a baseline, we performed an unbiased screening of genes encoding epigenetic factors. Pooled single guide RNAs (sgRNAs) were introduced into HSPCs ex vivo, followed by Cas9 induction and transplantation to identify genes that accelerate leukemia in vivo.
Material and methods
Pooled CRISPR/Cas9 screening
For epigenetic pooled screening, mice with doxycycline-inducible Cas9 allele (KH2/iCas9; The Jackson Laboratory, stock number 029415) of 8 to 12 weeks old were euthanized, and a total of 1 million Lin–Sca-1+c-Kit+ (LSK) cells were sorted and transduced ex vivo with lentivirus containing the epigenetic sgRNA library (supplemental Table 1)14 and then transplanted into B6/129 recipient mice (see supplemental Materials for additional details). Briefly, LSK cells were cultured in 96-well round-bottom microplates in StemSpan Serum-Free Expansion Medium (StemCell Technologies) supplemented with 100 ng/mL recombinant murine stem cell factor (rmSCF), 10 ng/mL rm interleukin-11 (IL-11), and 5 ng/mL rmFlt3l (R&D Systems) and cotransduced at a low multiplicity of infection, with lentivirus expressing Tet2 sgRNA (Tet2_e10.1; refer to supplemental Table 1 for full sequence) and an epigenetic sgRNA library to achieve ∼50% infection efficiency. After 24 hours, the cultured cells were then pooled, washed extensively, and injected intravenously into 17 lethally irradiated B6/129 recipient mice.
Mice were assessed periodically for disease onset and progression and euthanized when moribund. Peripheral blood was collected periodically via submandibular bleeding into EDTA-containing tubes at the indicated times, stained with antibodies as indicated, and analyzed using BD Accuri (Becton, Dickinson and Company). All animal protocols were approved by the Institutional Animal Care and Use Committee at Boston Children’s Hospital.
Readout of the pooled screen, sgRNAs, or edited alleles
To identify sgRNAs present in leukemia cells, genomic DNA from leukemic cells was harvested using QuickExtract DNA (Epicenter), and sgRNA cassettes were amplified via polymerase chain reaction, using the respective primers (supplemental Table 1). Because of the largely clonal nature of transformed leukemia cells, only a limited number of sgRNAs were present; TOPO TA cloning (Qiagen) followed by Sanger sequencing of each individual clone was used to identify the respective sgRNAs. To confirm the clonal nature of the transformed leukemia cells, bulk leukemia cells were plated onto M3434 methylcellulose (Stem Cell Technologies), and each colony was subjected to additional rounds of polymerase chain reaction identification for sgRNAs or edited alleles.
To identify each edited allele, the locus of Tet2, Tet3, or Pbrm1 that was targeted by each sgRNA was firstly amplified using the respective primers (supplemental Table 1) and then subjected to Sanger sequencing. The sequencing products were aligned against wild-type alleles to identify the corresponding mutations. Sanger sequencing with multiple peaks was deconvoluted using DECODR.15
RNA sequencing
Total RNA was collected from either mouse primary leukemic cells or HAP1 cells using the RNeasy Mini Kit (Qiagen) following the manufacturer’s recommendations, and genomic DNA was removed using the TURBO DNA-free Kit (Invitrogen). RNA was then enriched using the polyA enrichment method, and libraries were prepared and sequenced on an Illumina NovaSeq to a minimum of 40 million 150 bp read pairs per sample. RNA sequencing (RNA-seq) data were then analyzed using DESeq216 (see supplemental Materials for additional details).
ChIP sequencing
Chromatin immunoprecipitation (ChIP) was performed as described.17 Briefly, between 10 and 30 million cells were crosslinked with 1% formaldehyde for 12 minutes at room temperature, followed by quenching with 125 mM glycine for 10 minutes, washed with phosphate-buffered saline, pelleted, and flash-frozen. Nuclei were isolated, rinsed, and sonicated using an E220 Covaris sonicator. The supernatant was used for immunoprecipitation using the indicated antibody. The antibodies used were anti-Pbrm1 (Bethyl, catalog no. A301-590A and Novus Biologicals, catalog no. NB100-79832). ChIP-seq libraries were prepared using automated Swift 2S ligation chemistry according to standard protocols and sequenced to a minimum of 40 million 150 bp read pairs per sample. ChIP-seq data were analyzed using deepTools (v3.0.2)18 (see supplemental Materials for additional details).
Gene set enrichment analysis
Statistical analysis
For group comparison, Student t test was used unless otherwise specified in the legends. For survival analysis, log-rank (Mantel-Cox) tests were used.
Results
CRISPR/Cas9 targeting of Tet2 and Tet3 in vivo leads to myeloid leukemia
We used mice with an inducible Cas9 allele to perform in vivo targeted gene editing. sgRNAs were introduced into HSPCs ex vivo, and doxycycline was added ex vivo and administered to mice after transplantation to allow for persistent gene editing. Epigenetic factors have been implicated in clonal hematopoiesis and the knockout of these genes has demonstrated their involvement in myeloid malignancies.20 Ten-eleven translocation (TET) family of dioxygenases, which initiate the removal of 5-methylcytosine through successive oxidization of 5mC to 5-hydroxymethylcytosine (5hmC), are frequently mutated in human AML, largely by frameshift or nonsense mutations.21Tet2–/– mice contain an increased LSK cell pool and both heterozygous and homozygous Tet2 loss results in the development of myeloid malignancies.22,23
We first assessed whether the generation of loss-of-function alleles of Tet2 using a domain-targeting sgRNA led to leukemia after the induction of Cas9 in transplanted HSPCs (Figure 1A). Tet2 loss-of-function generated by an sgRNA targeting the catalytic domain failed to elicit leukemia after 6 months (Figure 1B). Because previous studies suggested that Tet3 might compensate for Tet2 loss and combined knockout of Tet2 and Tet3 accelerated leukemic development compared with single knockouts,24 we inactivated both Tet2 and Tet3 by multiplexing domain-targeting sgRNAs (Figure 1A). Mice with Tet2/Tet3-edited HSPCs developed aggressive myeloid malignancies with a median latency of 112 days (Figure 1B), whereas single editing of either Tet2 or Tet3 did not result in disease. The bone marrow of leukemic mice contained >90% green fluorescent protein–positive (GFP+) donor-derived cells, mainly Mac-1+, and large blasts were visible in the blood and bone marrow (Figure 1C). Widespread infiltration of myeloid cells into the hematopoietic system, including the spleen and lymph nodes, was evident (Figure 1C).
Modeling of myeloid leukemia using CRISPR/Cas9 and sgRNA targeting Tet2 and/or Tet3. (A) Schematic diagram of in vivo transformation of primary HSPCs. (B) Kaplan-Meier survival curve of mice that received transplantation with HSPCs infected with empty control, Tet2 sgRNA only, Tet3 sgRNA only, and Tet2/Tet3 sgRNAs. n = 7 per group; ∗∗∗P < .0001. (C) Characterization of myeloid leukemia in various hematopoietic organs. Representative fluorescence-activated cell sorting (FACS) plot of various hematopoietic organs, picture showing splenomegaly and enlarged lymph node, histology section of the bone marrow, and cytospin of leukemic cells. (D) Allelic examples of dominating leukemia clones with Tet2 and Tet3 loss-of-function in different biological replicates. Indels, insertion/deletions.
Modeling of myeloid leukemia using CRISPR/Cas9 and sgRNA targeting Tet2 and/or Tet3. (A) Schematic diagram of in vivo transformation of primary HSPCs. (B) Kaplan-Meier survival curve of mice that received transplantation with HSPCs infected with empty control, Tet2 sgRNA only, Tet3 sgRNA only, and Tet2/Tet3 sgRNAs. n = 7 per group; ∗∗∗P < .0001. (C) Characterization of myeloid leukemia in various hematopoietic organs. Representative fluorescence-activated cell sorting (FACS) plot of various hematopoietic organs, picture showing splenomegaly and enlarged lymph node, histology section of the bone marrow, and cytospin of leukemic cells. (D) Allelic examples of dominating leukemia clones with Tet2 and Tet3 loss-of-function in different biological replicates. Indels, insertion/deletions.
Editing of both Tet2 and Tet3 alleles was confirmed using bulk leukemic cells isolated from the bone marrow, and both sustained homozygous loss of the enzymatic domains in Tet2 and Tet3 through either simple frameshift or large deletions (Figure 1D). Notably, most of the bulk GFP+ cells in each mouse exhibited the same editing at both the Tet2 and Tet3 alleles, suggesting that the leukemic blasts were largely clonal. This finding was confirmed by plating leukemia cells derived from the bone marrow with methylcellulose and genotyping single colonies (data not shown). Together, these data demonstrate that the combined inactivation of Tet2 and Tet3 leads to the malignant transformation of HSPCs into myeloid leukemia in transplanted mice.
Unbiased pooled epigenetic screen identifies Pbrm1 as a cooperating gene in Tet2/Tet3 leukemia
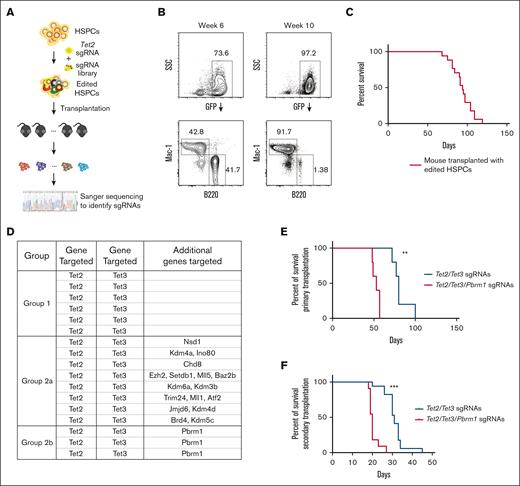
Because the mutation of Tet2 alone was insufficient to generate leukemia and an additional mutation of Tet3 induced disease, we reasoned that we might identify cooperating genes for leukemogenesis in a pooled screen initiated on a background of Tet2-edited HSPCs. As a library for this screen, we chose sgRNAs targeting the domains of epigenetic factors,14 which consisted of 1032 sgRNAs targeting 192 genes (supplemental Table 1). At a minimum, we anticipated identifying Tet3 as a cooperating factor. HSPCs carrying the inducible Cas9 allele were isolated from KH2/iCas9 mice, transduced ex vivo with Tet2 sgRNA and the sgRNA library, and transplanted into syngeneic hosts (Figure 2A). After the induction of Cas9, the mice were examined for signs of leukemia. At week 6, the recipients appeared healthy and exhibited normal lineage distribution of donor cells. At week 10 and beyond, some recipients showed a significant myeloid bias of transplanted cells in the peripheral blood (Figure 2B). Four months after transplantation, all mice showed signs of disease and were euthanized (Figure 2C). Leukemic cells were GFP+ and strongly Mac-1+, consistent with a myeloid-lineage origin (supplemental Figure 1), and splenomegaly was evident in all mice that received transplantation with the edited HSPCs (data not shown).
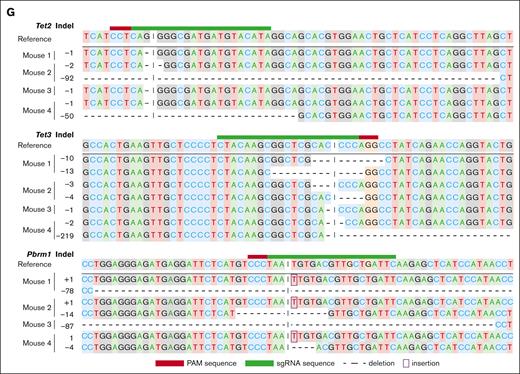
Pooled genetic screen using a domain-targeting epigenetic sgRNA library together with Tet2 sgRNA. (A) Schematic diagram of pooled genetic screen of HSPCs. (B) Representative FACS plot with normal lineage contribution at week 6 and disease manifestation at week 10 in the same biological replicate. (C) Kaplan-Meier survival curve of mice that received transplantation with primary HSPCs edited with an epigenetic sgRNA library. (D) Group summary of gene targeted in each leukemia mouse in the genetic screen. Targeted genes were identified via genomic polymerase chain reaction of sgRNA loci from total bone marrow cells at the terminal point (see “Material and methods” for details). Group 1 includes mice that had only Tet2/Tet3 targeted by sgRNAs. Group 2a and 2b include mice that had additional gene(s) targeted besides Tet2 and Tet3. Group 2b includes mice that had all Tet2/Tet3/Pbrm1 alleles targeted with different sgRNAs (see supplemental Figure 2 for the full sgRNA list). (E) Kaplan-Meier survival curve of mice that received transplantation with primary HSPCs edited with either Tet2/Tet3-targeting or Tet2/Tet3/Pbrm1-targeting sgRNAs. n = 5 per group; ∗∗P = .0027. (F) Kaplan-Meier survival curve of secondary recipients that received transplantation with leukemic cells isolated from primary recipients in panel E. n = 17 and 11 from 4 and 3 primary donors in each group, respectively; ∗∗∗P < .0001. (G) Allelic examples of dominating leukemia clones showing Tet2, Tet3, and Pbrm1 loss-of-function in different biological replicates. SSC, side scatter.
Pooled genetic screen using a domain-targeting epigenetic sgRNA library together with Tet2 sgRNA. (A) Schematic diagram of pooled genetic screen of HSPCs. (B) Representative FACS plot with normal lineage contribution at week 6 and disease manifestation at week 10 in the same biological replicate. (C) Kaplan-Meier survival curve of mice that received transplantation with primary HSPCs edited with an epigenetic sgRNA library. (D) Group summary of gene targeted in each leukemia mouse in the genetic screen. Targeted genes were identified via genomic polymerase chain reaction of sgRNA loci from total bone marrow cells at the terminal point (see “Material and methods” for details). Group 1 includes mice that had only Tet2/Tet3 targeted by sgRNAs. Group 2a and 2b include mice that had additional gene(s) targeted besides Tet2 and Tet3. Group 2b includes mice that had all Tet2/Tet3/Pbrm1 alleles targeted with different sgRNAs (see supplemental Figure 2 for the full sgRNA list). (E) Kaplan-Meier survival curve of mice that received transplantation with primary HSPCs edited with either Tet2/Tet3-targeting or Tet2/Tet3/Pbrm1-targeting sgRNAs. n = 5 per group; ∗∗P = .0027. (F) Kaplan-Meier survival curve of secondary recipients that received transplantation with leukemic cells isolated from primary recipients in panel E. n = 17 and 11 from 4 and 3 primary donors in each group, respectively; ∗∗∗P < .0001. (G) Allelic examples of dominating leukemia clones showing Tet2, Tet3, and Pbrm1 loss-of-function in different biological replicates. SSC, side scatter.
Genomic DNA of bulk leukemic cells from the bone marrow identified sgRNAs that were overrepresented. All mice examined (17/17) contained sgRNAs targeting both Tet2 and Tet3 genes (Figure 2D; supplemental Figure 2A-B), confirming that Tet3 loss-of-function alone cooperated with Tet2 loss to drive leukemia. Additional sgRNAs targeting different genes were also selectively identified (Figure 2D; supplemental Figure 2A-B). Given the largely clonal nature of these leukemic cells from each mouse, these sgRNAs were either acquired by chance together with Tet2/Tet3 sgRNAs or contributed to an aggressive phenotype. The latter possibility was suggested by the presence of independent sgRNAs targeting the bromodomains of Pbrm1 in 3 of 17 mice (P = 8.93e-5; Figure 2D; supplemental Figure S2A-B). Furthermore, within the cohort, mice with Pbrm1 sgRNA tended to exhibit a shorter disease latency (supplemental Figure 2C).
The presence of independent sgRNAs targeting Pbrm1 and a more aggressive phenotype provided the initial evidence that Pbrm1 cooperates with Tet loss during leukemogenesis. To test this hypothesis, we performed multiplexed gene editing and in vivo phenotype readout of HSPCs with Tet2 and Tet3 sgRNAs, with or without Pbrm1 sgRNA, as shown in Figure 1A. Targeting Pbrm1 resulted in a fully penetrant disease with latency reduced from 80 to 53 days (P = .0027; Figure 2E). Furthermore, the phenotypes were transplantable, and the secondary recipients showed a similar decrease in disease latency (from 31 to 20 days; P < .0001; Figure 2F). Molecular characterization demonstrated that the Pbrm1 alleles in this Pbrm1-edited cohort sustained loss-of-function alleles, in addition to Tet2 and Tet3 loss-of-function (Figure 2G). The immunophenotype was similar to that of Tet2/Tet3 leukemic mice (supplemental Figure 3A-B), and colony formation in methylcellulose was comparable (supplemental Figure 3C). Taken together, these data provide evidence that loss of Pbrm1 leads to a more aggressive form of Tet2/Tet3-deficient AML.
Leukemic cells lacking Pbrm1 are less immunogenic
PBRM1, a component of the multisubunit switch/sucrose nonfermenting (SWI/SNF) chromatin–remodeling complex, is often deleted or mutated in human malignancies, most prominently in renal cancers.25 However, its role in hematological malignancies has not yet been investigated. To explore how Pbrm1 contributes to hematological disease progression, we compared the RNA-seqs of bulk GFP+ leukemic cells from the bone marrow cells of Tet2/Tet3/Pbrm1-targeted and Tet2/Tet3-targeted mice. Transcripts of several genes were differentially expressed (supplemental Figure 4A; supplemental Table 2). Of particular interest, we observed that a subset of major histocompatibility complex (MHC) genes, mainly MHC class II (MHC II), was selectively downregulated in Pbrm1-deficient leukemic cells at the RNA and protein level (Figure 3A; supplemental Figure 4B-C).
Pbrm1-deficient AML cells are less immunogenic, with attenuated IFN signaling and reduced MHC expression. (A) Volcano plot of RNA-seq data from mouse AML cells, comparing Pbrm1-intact and Pbrm1-deficient AML cells. Red dots denote MHC II–related genes. (B) GSEA showing IFN pathways, including IFN-⍺ and IFN-γ, and signaling pathways involved in IFN signaling (TNF-⍺) were significantly downregulated in Pbrm1-deficient AML cells (see supplemental Figure 4D-E for the full list). Normalized enrichment score (NES) and false discovery rate (FDR) are as indicated. (C) Experimental design for secondary transplantation into NSG mice (see “Material and methods” for details). (D) Kaplan-Meier survival analysis of secondary transplantation in NSG mice. n = 20 and 16 from 5 primary donors in each group; P = .4752. AML was modeled based on Tet2/Tet3 deletions in mice.
Pbrm1-deficient AML cells are less immunogenic, with attenuated IFN signaling and reduced MHC expression. (A) Volcano plot of RNA-seq data from mouse AML cells, comparing Pbrm1-intact and Pbrm1-deficient AML cells. Red dots denote MHC II–related genes. (B) GSEA showing IFN pathways, including IFN-⍺ and IFN-γ, and signaling pathways involved in IFN signaling (TNF-⍺) were significantly downregulated in Pbrm1-deficient AML cells (see supplemental Figure 4D-E for the full list). Normalized enrichment score (NES) and false discovery rate (FDR) are as indicated. (C) Experimental design for secondary transplantation into NSG mice (see “Material and methods” for details). (D) Kaplan-Meier survival analysis of secondary transplantation in NSG mice. n = 20 and 16 from 5 primary donors in each group; P = .4752. AML was modeled based on Tet2/Tet3 deletions in mice.
MHC molecules are controlled by a variety of signaling pathways.26 To assess the signaling pathways that could potentially contribute to MHC downregulation, we performed GSEA to compare Tet2/Tet3/Pbrm1-targeted and Tet2/Tet3-targeted cells (supplemental Figure 4D-E). GSEA revealed that several interferon (IFN) pathways, including IFN-⍺ and IFN-γ pathways, were downregulated most significantly (Figure 3B). Signaling pathways involved in IFN signaling (TNF-⍺, IL-2, and IL-6) were also significantly downregulated in the Pbrm1-deficient AML cells (Figure 3B; supplemental Figure 4E).
Interferon pathways and MHC molecules, particularly MHC II, have been implicated in leukemogenesis.27-30 The shorter latency of leukemia observed upon targeting Pbrm1 could be cell-intrinsic or reflect interactions with the host immune system. To distinguish between these possibilities, we induced leukemia in vivo by targeting Tet2 and Tet3 with or without Pbrm1 and transplanted primary leukemic cells into secondary immunodeficient NSG mice (Figure 3C), which lack B, T, and natural killer cells. Notably, Pbrm1-intact and Pbrm1-deficient AML cells exhibited similar latencies after secondary transplantation into NSG mice (Figure 3D). Taken together, our findings indicate that the more aggressive leukemic phenotype seen upon Pbrm1-loss in immunologically competent mice was cell extrinsic and dependent on the host immune system. Because we did not observe any difference in the level of expression of MHC I components (supplemental Figure 4B), these data suggested that Pbrm1-deficient AML cells are less immunogenic, perhaps because of attenuated interferon signaling and reduced MHC II expression, which together contribute to immune escape and a more aggressive phenotype in mice.
PBRM1 controls IFN signaling and HLA class II levels in human leukemic cells
Before exploring the mechanistic relationships between Pbrm1 with IFN signaling and MHC II expression, we sought to determine the relevance of our findings in mice in the human context. We surveyed 15 human leukemic cell lines and identified 2 cell lines HAP1 and SHI-1, in which CRISPR/Cas9 inactivation of PBRM1 reduced HLA class II upregulation upon IFN-γ stimulation (Figure 4A; supplemental Figure 5A-B). Notably, these 2 cell lines have no known deleterious TET2 or TET3 mutations,31,32 suggesting that these effects of PBRM1 loss are independent of the TET2 or TET3 allele status. We performed bulk RNA-seq and assessed global gene expression changes in PBRM1-intact and PBRM1-deficient HAP1 cells with or without IFN-γ stimulation. As expected, we observed strong upregulation of HLA upon IFN treatment in wild-type HAP1 cells (Figure 4B; supplemental Table 3). Consistent with observations at the protein level (Figure 4A), HLA upregulation was also blunted at the messenger RNA level upon PBRM1 knockout (Figure 4B).
PBRM1 regulates IFN signaling and HLA class II levels in human leukemic cells. (A) Attenuated HLA class II upregulation upon treatment with the indicated concentrations of IFN-γ in PBRM1 knockout HAP1 cells (n = 3 for each treatment; see “Material and methods” for details). (B) Dot plot of gene counts of HLA class II genes in PBRM1-deficient HAP1 cells compared with those of PBRM1-intact HAP1 cells with or without IFN-γ treatment. Each dot represent 1 biological replicate. Light colors represent gene expression in untreated cells, and dark colors represent gene expression in IFN-γ–treated cells. Red represents knockout and blue represents control. (C) GSEA analysis showing IFN pathways, including IFN-⍺ and IFN-γ, and other signaling pathways, including TNF-⍺ and hypoxia pathways, were significantly downregulated in PBRM1-deficient HAP1 cells treated with IFN-γ (see supplemental Figure 5F-G for the full list). NES and FDR are as indicated. (D) Dot plot of gene counts of all IRFs in PBRM1-intact or PBRM1-deficient HAP1 cells with or without IFN-γ treatment. (E) HLA expression after IFN-γ treatment of HAP1 cells with IRF1 knockout via sgRNAs compared with the empty vector control. (F-H) PBRM1 binding profile in untreated and IFN-γ-treated wild-type HAP1 cells across the gene body of all genes (F), of top interferon-induced genes (G), and of top PBRM1-regulated interferon-induced genes (H) (see “Material and methods” for analysis details). ChIP-seq was performed using antibodies against PBRM1 (antibody 1: Bethyl, catalog no. A301-590A and antibody 2: Novus Biologicals, catalog no. NB100-79832). (I) ChIP-seq profiles of PBRM1 binding at the indicated loci, including IRF1, CIITA, and HLA class II, in untreated and IFN-γ–treated wild-type HAP1 cells.
PBRM1 regulates IFN signaling and HLA class II levels in human leukemic cells. (A) Attenuated HLA class II upregulation upon treatment with the indicated concentrations of IFN-γ in PBRM1 knockout HAP1 cells (n = 3 for each treatment; see “Material and methods” for details). (B) Dot plot of gene counts of HLA class II genes in PBRM1-deficient HAP1 cells compared with those of PBRM1-intact HAP1 cells with or without IFN-γ treatment. Each dot represent 1 biological replicate. Light colors represent gene expression in untreated cells, and dark colors represent gene expression in IFN-γ–treated cells. Red represents knockout and blue represents control. (C) GSEA analysis showing IFN pathways, including IFN-⍺ and IFN-γ, and other signaling pathways, including TNF-⍺ and hypoxia pathways, were significantly downregulated in PBRM1-deficient HAP1 cells treated with IFN-γ (see supplemental Figure 5F-G for the full list). NES and FDR are as indicated. (D) Dot plot of gene counts of all IRFs in PBRM1-intact or PBRM1-deficient HAP1 cells with or without IFN-γ treatment. (E) HLA expression after IFN-γ treatment of HAP1 cells with IRF1 knockout via sgRNAs compared with the empty vector control. (F-H) PBRM1 binding profile in untreated and IFN-γ-treated wild-type HAP1 cells across the gene body of all genes (F), of top interferon-induced genes (G), and of top PBRM1-regulated interferon-induced genes (H) (see “Material and methods” for analysis details). ChIP-seq was performed using antibodies against PBRM1 (antibody 1: Bethyl, catalog no. A301-590A and antibody 2: Novus Biologicals, catalog no. NB100-79832). (I) ChIP-seq profiles of PBRM1 binding at the indicated loci, including IRF1, CIITA, and HLA class II, in untreated and IFN-γ–treated wild-type HAP1 cells.
Transcripts of several genes were differentially expressed (supplemental Figure 5C). Further hierarchical clustering analysis of RNA-seq data revealed several distinct clusters, 1 of which consisted principally of genes induced by IFN-γ in wild-type HAP1 cells but misregulated in PBRM1-deficient cells (supplemental Figure 5D). This cluster included downstream effectors produced in response to IFN-γ, such as the transcription factor IRF1 and the accumulation of the activated lymphocyte chemokines CXCL9 and CXCL11 (supplemental Figures 5E; supplemental Table 3). Additional analysis using GSEA revealed that IFN-γ and IFN-⍺ as well as TNF-⍺ and IL-2 pathway were most significantly downregulated in PBRM1-deficient HAP1 cells (Figure 4C; supplemental Figure 5F-G). These data demonstrate that PBRM1 controls HLA class II and IFN signaling pathways in human leukemic cells, consistent with the findings in mice (Figure 3A-B; supplemental Figure 4E).
IFN-γ activates type 2 IFN signaling, which in turn induces the expression of MHC II genes in a variety of cell types,33 and the IFN regulatory factor (IRF) family of transcription factors acts as important activators and repressors of the IFN signaling process.34 Notably, the transcription factor IRF1 is critical for the IFN signaling pathway and is essential for the activation of the MHC II transactivator Class II Major Histocompatibility Complex Transactivator by IFN-γ.35 To evaluate the significance of each transcription factor IRF in HAP1 cells, we first compared the gene expression patterns of all IRFs and found that among all IRFs, IRF1 was particularly strongly induced in IFN-γ-treated HAP1 cells (Figure 4D). More importantly, the upregulation of IRF1 was also blunted in IFN-γ-treated PBRM1-deficient cells at both the messenger RNA and protein level (Figure 4D; supplemental Figure 6A). These data raise the possibility that the observed defects in HLA upregulation in PBRM1-deficient cells were secondary to defective IFN signaling due to reduced IRF1. Consistent with this hypothesis, knockout of IRF1 in wild-type HAP1 using different sgRNAs (supplemental Figure 6B) blunted HLA upregulation in IFN-γ–stimulated, IRF1-deficient HAP1 cells (Figure 4E; supplemental Figure 6C).
To determine whether the effects of PBRM1 on IFN and HLA genes might be direct, we examined the chromatin occupancy of PBRM1 genome-wide by performing ChIP-seq in wild-type HAP1 cells before and after IFN-γ stimulation. Consistent with previous literature and its role in transcriptional control,36 we found that PBRM1 was located mainly at gene promoters (Figure 4F). Treatment with IFN-γ resulted in a small change in the global binding of PBRM1 (Figure 4F; fold change of 1.10, with P = 1e-300). Next, we assessed the binding profile of PBRM1 at the IFN genes. Although treatment with IFN-γ induced striking changes in the gene expression of IFN-induced genes (Figure 4B; supplemental Figure 5D-E), the binding of PBRM1 at IFN-induced genes was only modestly increased (Figure 4G; fold change of 1.263, with P = 2.22e-16). In contrast, the binding of PBRM1 to IFN-suppressed genes remained unchanged (supplemental Figure 6D; fold change of 1.037, with P = 0.0019). Furthermore, PBRM1 binding was also modestly elevated in PBRM1-regulated interferon-induced genes upon IFN-γ treatment (Figure 4H; fold change of 1.29, with P = .00727). These data suggest that PBRM1 regulates IFN genes by binding to these loci at the basal level as well as upon stimulation.
Next, we focused on IRF1 and HLA-related genes that were differentially expressed in PBRM1-deficient HAP1 cells (Figure 4B,D). In untreated cells, we detected PBRM1 binding at the promoter regions of IRF1 but not at the CIITA or HLA genes (Figure 4I). Upon stimulation with IFN-γ, a dramatic increase in PBRM1 occupancy was observed at IRF1, but not at gene loci of other downregulated genes (Figure 4I). Taken together, these data suggest that PBRM1 occupancy does not increase globally after IFN-γ treatment. Rather, PBRM1 occupancy was selectively increased in a subset of IFN-γ-regulated genes, such as IRF1, which is most consistent with the conclusion that PBRM1 acts as a positive regulator of IRF1 expression, which in turn controls HLA expression.
Discussion
We performed an unbiased pooled genetic screen to identify novel factors that contribute to leukemogenesis in the context of an intact microenvironment. We, first, generated AML with full penetrance in mice after in vivo induction of Cas9 within transplanted HSPCs transduced ex vivo with domain-targeting sgRNAs for Tet2 and Tet3 (Figure 1). By harnessing this experimental platform, we used a focused domain-targeting sgRNA library to screen for epigenetic factors that cooperate with Tet loss to drive leukemia. In this manner, we discovered that the loss of Pbrm1/Baf180, in the setting of the Tet2/Tet3 mutation, created a more aggressive AML, characterized by a shortened latency of disease onset (Figure 2). Pbrm1-deficient AML cells exhibited attenuated IFN signaling pathways and reduced MHC II expression, rendering cells less immunogenic and more competent to escape immune surveillance (Figure 3). Our study has important implications because PBRM1 regulates IFN signaling in a subset of human myeloid leukemia cell lines (HAP1 and SHI-1), likely via the regulation of the expression of IFN pathway genes, such as IRF1 (Figure 4), to control the immunogenicity of leukemic cells.
PBRM11/BAF180 is a component of the polybromo BRG1/BRM-associated factor (PBAF) SWI/SNF chromatin–remodeling complex that acts at numerous genome-wide loci. Inactivating mutations of PBRM1 are found in multiple cancers, with an incidence of >40% in clear cell renal cell cancer (ccRCC).36,37 PBRM1 has been reported to participate in a variety of signaling processes, including hypoxia-inducible factor-α38 and IFN-γ.39 Recently, PBAF has been proposed as a biomarker for immune checkpoint therapy and immune modulation. However, these effects are not consistent from one cellular context to another.39-44 Patients with RCC treated with immune checkpoint blockade demonstrate shorter overall survival than those with intact PBRM1.40 However, in another cohort of patients with immune checkpoint blockade–treated ccRCC, the clinical benefit was associated with loss-of-function mutations in PBRM1, with prolonged overall survival and progression-free survival.43
IFNs are critical regulators of the immune response against tumors.45 IFNs display a range of antitumor activities, including induction of apoptosis, inhibition of angiogenesis and proliferation, cell terminal differentiation, and immune regulation. IFN-α, IFN-β, and IFN-γ directly upregulate the expression of surface tumor-associated antigens via the augmentation of MHC I and II molecules, thereby increasing the immunogenicity of tumor cells and rendering them vulnerable to destruction by the immune system. Understanding the immune regulatory mechanisms used by AML cells upon exposure to IFN-γ is critical for the development of immunotherapy and checkpoint blockade therapy approaches.27,29,30 Data obtained from our myeloid leukemia model suggested that Pbrm1 loss is associated with reduced expression of IFN-responsive genes (Figures 3 and 4).
In castrate-resistant prostate cancer, PBRM1 appears to be necessary for the IRF1-induced expression of several genes that suppress the immune tumor microenvironment and inhibit T-cell function; knockdown of PBRM1 results in reduced IFN-γ signaling.39 Likewise, in RCC, in which PBRM1 mutations are most frequent, PBRM1 loss defines a nonimmunogenic tumor phenotype characterized by coordinated downregulation of the immunomodulatory gene set relative to the PBRM1 intact group, and PBRM1 mutations are associated with a less immunogenic tumor microenvironment in human ccRCC.40 This scenario appears similar to that reported here in the context of myeloid leukemia. However, studies have presented more complex scenarios. Among ccRCC cell lines, PBRM1 exhibited context-dependent, tumor-promoting, or tumor-inhibiting effects, dependent on the hypoxia-inducible factor1A gene mutational status.46 In another study on renal cancer cells, the genes most strongly enriched in PBAF-deficient cell lines were immunostimulatory and PBRM1-deficient tumors exhibited increased hypoxia and IL-6/JAK-STAT3 gene set enrichment.43 In melanoma, the loss of Pbrm1 sensitized mouse B16F10 melanoma cells to killing by T cells.44 In colon cancer cells, PBRM1 appears to associate cancer stem cells with immune evasion by restricting the basal activity of the innate immune system; knockdown of PBRM1 upregulates IFN-related and inflammation-related gene signatures.47 Thus, PBRM1 appears to affect IFN signaling positively or negatively, depending on the cell or tumor type. The effects of PBRM1 loss on tumor cells could reflect distinct immune cell signatures that are determined by the different contexts of each cancer type48 and its effects on the immune microenvironment could vary based on the tumor lineage.40
Little is known regarding the involvement of PBRM1 in myeloid malignancies and primary hematopoiesis. Among the South Korean cohort of patients with AML, PBRM1, SMARCC1, and DPF3, were mutated at a frequency of 6.7% each.49 However, this association has not been highlighted in any other cohorts. In primary hematopoiesis, Pbrm1 regulates cellular senescence and hematopoietic stem cell homeostasis through p21.50 Our findings implicate Pbrm1 in immune surveillance in a subset of myeloid leukemia and provide evidence that PBRM1 acts through the regulation of IFN signaling and IRF1 and controls HLA class II expression (Figure 4). An appreciation of the impact of PBRM1 loss on leukemia progression merits further study within genetically defined subsets of patients with myeloid malignancies.
Generally, the experimental strategy we used to identify Pbrm1 as a regulator of the myeloid leukemia phenotype highlights the potential of agnostic genetic screens performed in the context of an intact microenvironment. Variations in this approach, perhaps by leveraging CRISPR interference or CRISPR activation, which repress or activate protein-coding genes or regulatory elements, respectively, could provide further insights into cancer dependencies.
Acknowledgments
The authors thank Huafeng Xie and Semir Beyaz for valuable discussion, Brikena Gjeci for technical support, the Molecular Biology Core Facilities at Dana-Farber Cancer Institute for next-generation sequencing, Flow Cytometry Core at Dana-Farber Cancer Institute and HSCI-BCH Flow Cytometry Core at Boston Children’s Hospital for cell sorting, and Rodent Histopathology Core at Dana-Farber/Harvard Cancer Center for histology. This work was supported by a National Heart, Lung, and Blood Institute grant (P01 HL131477).
Authorship
Contribution: B.E.L. and S.H.O. conceptualized and designed the study, interpreted the data, and wrote the manuscript, with input from all authors; B.E.L., G.Y.L., W.C., D.S., and Y.F. performed the research and analyzed the data; B.E.L. and Q.Z. analyzed and interpreted the next-generation sequencing data; and C.R.V. generated the CRISPR library and provided feedback on experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for W.C. is Regor Therapeutic Inc, Shanghai, China.
The current affiliations for Q.Z. are the Department of Molecular and Human Genetics and Lester & Sue Smith Breast Center, Baylor College of Medicine, Houston, TX.
The current affiliations for D.S. are the St. Anna Children's Cancer Research Institute and CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, Vienna, Austria.
Correspondence: Stuart H. Orkin, Children's Hospital Boston, Dana-Farber Cancer Institute, Howard Hughes Medical Institute, Harvard Medical School, 1 Blackfan Cir, Karp Family Research Bldg, Boston, MA 02115; e-mail: stuart_orkin@dfci.harvard.edu.
References
Author notes
The data generated in this study are deposited in the Gene Expression Omnibus database (accession number GSE211263).
Data are available on request from the corresponding author, Stuart Orkin (stuart_orkin@dfci.harvard.edu).
The full-text version of this article contains a data supplement.