Key Points
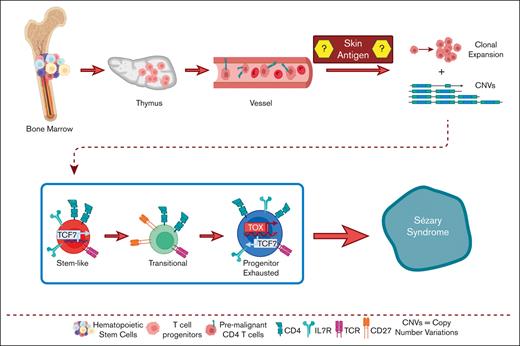
CTCL cells arise from mutated hematopoietic stem cells after thymic egression.
Clonally enriched CTCL cells carry these progenitor mutations.
Abstract
The pathogenesis of cutaneous T-cell lymphoma (CTCL) remains unclear. Using single-cell RNA or T-cell receptor (TCR) sequencing of 32 619 CD3+CD4+ and CD26+/CD7+ and 29 932 CD3+CD4+ and CD26−/CD7− lymphocytes from the peripheral blood of 7 patients with CTCL, coupled to single-cell ATAC-sequencing of 26,411 CD3+CD4+ and CD26+/CD7+ and 33 841 CD3+CD4+ and CD26−/CD7− lymphocytes, we show that tumor cells in Sézary syndrome and mycosis fungoides (MF) exhibit different phenotypes and trajectories of differentiation. When compared to MF, Sézary cells exhibit narrower repertoires of TCRs and exhibit clonal enrichment. Surprisingly, we identified ≥200 mutations in hematopoietic stem cells from multiple patients with Sézary syndrome. Mutations in key oncogenes were also present in peripheral Sézary cells, which also showed the hallmarks of recent thymic egression. Together our data suggest that CTCL arises from mutated lymphocyte progenitors that acquire TCRs in the thymus, which complete their malignant transformation in the periphery.
Introduction
Peripheral T-cell lymphomas account for ∼12% of lymphoid tumors worldwide.1 Among them, cutaneous T-cell lymphoma (CTCL) represents a heterogeneous group of diseases characterized by a cutaneous infiltration of malignant T cells. There are no curative therapies available for these diseases, because in part of limited understanding of the origin and pathogenesis of the disease.
The most common forms of CTCL are mycosis fungoides (MF) and its leukemic variant, Sézary syndrome (SS), which have an annual incidence of ∼0.5 per 100 000.2 The similarities and differences in the molecular alterations driving these heterogeneous diseases remain incompletely understood. Thus, although many mutations in common oncogenes are found in both MF and SS,3 recent studies have also identified differences in structural variants and deletions in tumor suppressor genes, which are much more common in the leukemic variant of the disease.3 For instance, recent studies have suggested a role for mutations in known suppressor genes, oncogenes, and epigenetic modifiers in SS, including TP53, ARID1A, CDKN2A, RB1, PTEN, PLCG1, ZEB1, DNMT3A, and the JAK-STAT pathway.3-6 Some authors have speculated that SS could evolve from MF,7 whereas other studies have pointed to a different cell of origin for these diseases, based on differential expression of markers of antigen experience. Based on new evidence on phenotypic heterogeneity among malignant cells in both variants of CTCL, the field has recently evolved to the prevailing view that different functional states and the magnitude of genetic alterations, rather than different cells of origin, determine the manifestation of CTCL in the form of MF vs SS.8
T cells egress from the thymus after selection and acquisition of T-cell receptors (TCRs) from migrating lymphoid precursors. Although age-related regression of the thymus is associated with a decline in T-cell output, there are T cells with hallmarks of recent thymic egression in individuals aged ≥80 years.9 Although the mechanism of initiation of CTCL also remains elusive,10 recent studies found significant heterogeneity in the TCR repertoire of CTCL malignant cells, suggesting that malignant transformation could take place during early T-cell development.11 This may explain why up to 7% of patients with SS show concomitant T-cell large granular lymphocytic (T-LGL) proliferation in the periphery, which is paradoxically associated with favorable prognosis.12
To gain insight into the pathogenesis of CTCL, in this study we aimed to define the origin of CTCL, as well as differences and similarities between MF and Sézary cells. Our results point to heavily altered hematopoiesis in patients with CTCL. Sézary T-cells sharing mutations in oncogenes with hematopoietic stem cells (HSCs) egress from the thymus with multiple TCRs as premalignant lymphocytes, which complete their malignant transformation in the periphery, resulting into heterogeneous clonotypes and phenotypes. These results have obvious implications for future therapies designed to prevent CTCL recurrence after temporary remission, because of replenishment of peripheral malignant cells from HSCs.
Methods
Human samples
Deidentified human peripheral blood samples, bone marrow aspirates, and skin biopsies were procured under a protocol approved by Moffitt’s Scientific Review Committee. Cells were thawed in complete RPMI (10% heat-inactivated fetal bovine serum [FBS], 0.5 mM sodium pyruvate [Gibco, 11360070], 2 mM L-glutamine [Sigma, G7513], 100 international unit per mL penicillin, 100 μg/mL streptomycin [Lonza, 17-602E]). CD4+CD7+/CD26+, CD4+CD7−/CD26−, and naïve T cells were isolated through sorting on the BD fluorescence activated cell sorting (FACS) Aria special order research product with methods for staining described in the subsequent section. Allogenic SS skin biopsies were either snap frozen for antigen or dissociated and cryopreserved for later use. Skin samples were minced with scalpel before dissociated with 2 mg/mL collagenase IV (Thermo Fisher), 0.1 mg/mL DNAase I (Roche), 0.1 mg/mL hyaluronidase (Worthington Biochemical) in cRPMI. Biopsies were incubated on plate for 1 hour at 37°C on plate in cell-culture incubator 5% CO2. Fragments were gently homogenized with an 18 g syringe and filtered through 70 μM filter before being frozen in FBS and 10% dimethyl sulfoxide. Skin biopsies yielded between 0.25 × 106 and 1 × 106 cells from erythroderma lesion biopsies. CD4+CD7+/CD26+ and CD4+CD7−/CD26− were also isolated with EasySep human CD4+ T-cell isolation kit (STEM CELL, 17952) and EasySep release human PE positive selection kit (STEM CELL, 17654) from peripheral blood of patients with SS and MF. Human HSC CD34+ cells from bone marrow aspirates using human CD34 MicroBead Kit UltraPure Kit (Miltenyi, 130-100-453) following manufacturing instructions. Removal of CD45+ cells for enrichment of human fibroblast from bone marrow aspirates was done using EasySep human CD45 depletion kit II (STEM CELL, 17898). Primary bone marrow fibroblasts were grown in fibroblast growth medium 2 (PromoCell, C-23020) according to manufacturing instructions. Human normal primary epidermal keratinocytes (ATCC, PCS-200-011) were grown according to manufacturing instructions using dermal cell basal medium (ATCC, PCS-200-030) supplemented with the keratinocyte growth kit (ATCC, PCS-200-040).
Antibodies and flow cytometry
We used the following antihuman antibodies from Biolegend; CD45 (Clone HI30), CD26 (Clone BA57), CD7 (CD7-6B7), CD69 (Clone FN50), CD3 (OKT3), CD8 (RPA-T8), and CD4 (Clone OKT4). Human γ-globulin (Sigma, G4386) was used for Fc receptor blockade. Staining included Zombie NIR (Biolegend, 423105) and 4′,6-diamidino-2-phenylindole (Invitrogen, D1306) were used as viability probe for all samples. For flow cytometry, all samples were run on a LSRII (BD) and sorted using a FACSAria special order research product. Human samples were prepared as single-cell suspensions and resuspended in 1% of FBS and 1 mM EDTA (Invitrogen; Cat# AM9260G) before staining. The following cell populations were sorted for the Sézary samples: (1) naïve CD45+CD3e+CD4+CD45RA+CD45ROnegCD62L+CD26+CD7+ and (2) naïve CD45+CD3e+CD4+CD45RA+CD45ROnegCD62L+CD26neg. For control peripherical blood samples from either healthy donors, or a patient with benign ovarian tumor and umbilical cord blood as well, we sorted naïve CD45+CD3+CD4+CD45RA+CD45ROnegCD26+CD7+. Cells were stained with fluorescent conjugated antibodies and their clone numbers were as follows: from BD Biosciences, BV786 mouse antihuman CD45 (HI30, Cat#563716); BUV395 mouse antihuman CD3e (SK7, Cat#564001); BUV737 mouse antihuman CD4 (SK3, Cat#612748); and BB515 mouse antihuman CD45RO (UCHL1, Cat#564529). Similarly, from Biolegend: Alexa Fluor 700 antihuman CD14 (63D3, Cat#367114); phycoerythrin (PE)/cyanine7 antihuman CD45RA (HI100, Cat#304126); antigen presenting cell (APC) antihuman CD62L (DREG-56, Cat#304810); PE antihuman CD26 antibody (BA5b, Cat#302706) and PerCP/cyanine5.5 antihuman CD7, (CD7-6B7, Cat#343116). Cell viability was assessed through staining with the Zombie NIR (Biolegend, Cat#423106) for 15 minutes in phosphate-buffered saline (PBS). Data were analyzed using FlowJo v10 software.
Cell trace violet assays
Cell-trace assays were performed using CellTrace violet (Invitrogen, C34571) following manufacture’s recommendations with 5 μM stock concentration resuspended in dimethyl sulfoxide starting with 106 cell per mL cell concentration in sterile PBS. Cells were incubated at 37°C for 20 minutes protected from light. Reaction was quenched using complete cell culture medium 5 times the reaction volume and incubated for 5 minutes, spun for 5 minutes 4°C at 1500 revolutions per minute or 453 relative centrifugal force and resuspended at cell concentration of 106 cells per mL in fresh warm media and cultured for 5 to 7 days before fluorescence-activated cell sorting analysis.
Antigen presentation
APCs were isolated by depleting T cells from autologous peripheral blood mononuclear cell (PBMCs) from aphaeresis samples using EasySep release human CD3 positive selection kit (STEM CELL, 17751). T-cell–depleted PBMCs were cultured for 4 days with human 160 ng/mL interleukin 4 (IL-4) (PeproTech, 200-04) and 80 ng/mL granulocyte-macrophage colony-stimulating factor (PeproTech, 500-P33) in complete medium added on days 0 and 3. Malignant Sézary or MF (ie, CD7−CD26−) T cells were isolated from PBMCs as previously mentioned. Lysates were generated from skin biopsies or human normal primary epidermal keratinocytes (ATCC, PCS-200-011) using 10 successive cycles of freezing and thawing with 10 μL of PBS. Lysates were centrifuged at 300g for 10 minutes at 4°C to remove particulate debris. The lysate supernatants were collected, and their protein concentrations were measured using Pierce bovine calf albumin protein assay kit (Thermo Scientific, 23225). T-cell–depleted PBMCs were plated in triplicates in concentrations ranging from ∼10 000 to 50 000 per 100 μL. Lysates were added to T-cell–depleted aphaeresis at a final concentration of 12 μg per 100 μL. Malignant T cells were mixed with skin or control tissue-pulsed autologous PBMCs (1:10). After ∼20 hours, the assay supernatants were collected, and enzyme-linked immunosorbent assay were performed using ELISA MAX standard set kits for human interferon γ (Biolegend, 430101) and IL-4 (Biolegend, 430301) following manufacturer’s protocol to assess cytokine concentration.
10× RNA, variable diversity joining (VDJ), and assay for transposase-accessible chromatin (ATAC) sequencing
Previously described sorted isolated cells were resuspended in recommended 1000 cells per μL concentration in PBS with 0.04% w/v bovine serum albumin before library preparation. The target cell number for encapsulation was 6000 to 10 000 cells for single-cell RNA-sequencing and VDJ sequencing. Libraries were prepared using chromium single cell 5′ and V(D)J enrichment reagent kits (v2.0 Chemistry) (10X Genomics, Inc, Pleasanton, CA) following manufacturer’s instructions and run on NovaSeq6000 S4 200 cycles (26 bp × 90 bp). Read depth was ∼40 000 reads for RNA sequencing and 5000 reads for VDJ sequencing per sample. Cells were prepared as previously described. Target nuclei number for encapsulation was from 6000 to 10 000 cells for single-cell ATAC-sequencing. Libraries were prepared with chromium single cell ATAC reagent kit (v2.0 Chemistry) (10X Genomics, Inc) and run on NovaSeq6000 S2 100 cycles (50 bp × 50 bp). Read depth was ∼25 000 reads per sample.
Whole-exome sequencing
Following the collection of patients’ cells, genomic DNA was extracted using the QIAgen QIAamp DNA blood mini kit following the manufacturer’s protocol (Qiagen, 51104), and the DNA’s quality was assessed using the Agilent Tapestation (Agilent Technologies, Inc, Santa Clara, CA). Two hundred nanogram of DNA was used as input to generate whole-exome sequencing libraries using the Agilent SureSelect XT clinical research exome kit (Agilent Technologies, Inc). In brief, genomic DNA libraries were constructed according to the manufacturer’s protocol and the size and quality of the library was evaluated using the Agilent BioAnalyzer (Agilent Technologies, Inc). Equimolar amounts of library DNA were used for a whole-exome enrichment using the Agilent capture baits, and final libraries were screened on an Agilent TapeStation (Agilent Technologies, Inc) and quantitated using quantitative polymerase chain reaction (qPCR) with the kapa library quantification kit (Roche Diagnostics, U.S., Indianapolis, IN). An average of 100 million 75-base paired-end reads per sample were generated using v2 chemistry on an Illumina NextSeq 500 sequencer to achieve an average sequencing depth of 140×.
Molecular cloning
Genomic DNA isolated from CD4+ T cells from normal donor was used to amplify signal joint TCR rearrangement excision circle (sjTREC) (TREC Forward: 5′-AAAGAGGGCAGCCCTCTCCAAGGC-3′ and TREC Reverse: 5′-GGCTGATCTTGTCTGACATTTGC-3′) and TRCAC (TRCAC-Spel Forward: 5′-GACTAGTATGAGACCGTGACTTGCCAG-3′ and TRCAC-Spel Reverse: 5′-GACTAGTGCTGTTGTTGAAGGCGTTTGC-3′) sequencing using primers. Fragments were run on a 1.2% agarose gel and purified using Wizard SV Gel and PCR Clean-Up System (Promega, A9282). Fragments were cloned using Zero Blunt TOPO PCR cloning kit (Invitrogen, K280020) into vector pCRBlunt II-TOPO vector. Colonies were grown in Luria-Bertani broth, Lennox (BD, 240230) with BD Bacto dehydrated agar (BD, DF0140-15-4) with 50 μg/mL kanamycin (Gibco, 11815-032) and isolated with QIAGEN plasmid maxi kit (QIAGEN, 12163).
qPCR
Real-time PCR amplification was carried out using qPCR for which the quantification of the sjTRECs has been already published.13 In brief, we used a CFX384 real-time thermal cycler (BIO-RAD) and a 384-well plate with TaqMan fast advanced master mix for qPCR (Applied Biosystems, Cat# 4444557). The reaction mix was prepared as follows: TaqMan universal PCR 2×: 6 μL from a stock 2× to get 1×. TaqMan Probe: 0.3 μL from a 10 μM stock for a final concentration of 250 nM. Primers (forward/reverse): 0.216 μL from a stock 50 μM for a final concentration of 900 nM each one. DNA template: 5 μL from a stock at 40 ng per μL for a final concentration of 16.66 ng/μL. Water: 0.268 μL. The total volume per reaction was 12 μL. Standard curves were generated for both sjTRECs and the reference TCR α constant gene, as reported by Sottini et al, from linearized pCR Blunt II-TOPO vector with restriction enzyme XhoI (New England Biolabs, R0146S) and clean up with QIAquick PCR purification kit (QIAGEN, 28106). Serial dilutions were performed in diethyl pyrocarbonate-treated water (Thermo Scientific, R0601), and the number of sjTRECs per 106 cells (#TREC) was approximately estimated by following the equation as: (#TREC) = ([quantity of TRECs]/[quantity of TCR α constant/2]) × 106.
Study approval
Study conformed to approvals granted by Moffitt’s research regulatory committees, including institutional review board approvals MCC 50312, MCC 20403, MCC 19672, and MCC 20032. MCC 50312, MCC 20403, and MCC 20032 for SS and MF peripheral blood samples were acquired with informed written consent protocol MCC 14690. MCC 19672 for peripheral blood and skin biopsies were acquired with informed written consent protocol MCC 14690.
Results
Unique differentiation trajectories in SS vs MF
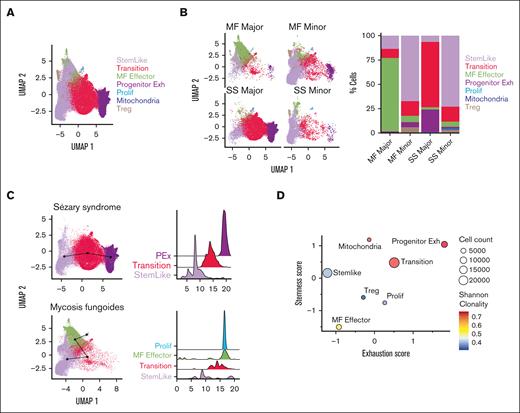
To identify CTCL-initiating cells with features of stemness, we first performed single-cell RNA sequencing, coupled to TCR VDJ sequencing, after sorting for CD4+CD26+ and CD4+CD26− T cells from patient peripheral blood aphaeresis from SS (n = 4) and MF (n = 3) (Table 1; supplemental Table 1). The gating strategy for sorting is represented in supplemental Figure 1A-B. Initial analysis yielded a total of 12 clusters for both diseases (supplemental Figure 2A-B). Seven groups emerged from gene expression analysis when compared to previously published data14,15 that were labeled as stem-like, transition, MF effector, proliferation, progenitor exhausted, mitochondrial activity, and T regulatory (Figure 1A; supplemental Table 2). Although the functionality of these cells may differ from their gene expression signatures, our primary aim was to define their trajectory of differentiation. We used determinants associated with T-cell phenotypes based on numerous studies which have already assessed the functional aspects of these genes.16-21 Interestingly, we found significant differences in clustering between MF and SS using our defined groups. The MF cells clustered in multiple groups with hallmarks of stemness, effector, and mitochondrial activity, and proliferation whereas Sézary cells predominately clustered in stem-like, transition, and progenitor-exhausted cells (supplemental Figure 2C-D). However, CD26 expression is not a definitive marker for malignancy with the majority of SS cells expressing the dominant clonal type.22 To further confirm the presence of malignancy in the clonal populations, we implemented inferCNV, which showed large copy number variations that were present (supplemental Figure 3A-B). Sézary is characterized by malignant T cells originating from dominant clones. Therefore, we defined malignant cells within our analysis independently of CD26 expression based on enriched clonal types in both SS and MF. Therefore, for each patient with Sézary, the CD26+ and CD26− cells in the most dominate clonotype were assigned as “SS major clones”; the cells in polyclonal clonotypes were assigned as “SS minor clones.” For each patient with MF, cells in small clones were assigned as “MF minor clones”; cells in other expanded larger clones were assigned as “MF major clones.” For both SS and MF, cells with no TCR identified were assigned as “Unknown” (supplemental Figure 4A). A similar method has been used in previous CTCL publications.23 The majority of CD26+ cells had smaller clones than the CD26− cells (supplemental Figure 4B-C). We examined the expression of markers of interest in these major and minor clones in MF and SS. MF-major clones highly expressed markers of effector cells (GNLY and PRF1), whereas Sézary major clones expressed stem-like markers (TCF7, SELL, and IL7R) as well as markers for exhaustion (TOX and TIGIT) (supplemental Figure 4D). We confirmed that the CD26− population is enriched with dominant clones compared to the CD26+ population, with a few clones shared between CD26− and CD26+ (supplemental Figure 4E). We then compared the major and minor clone populations of both SS and MF cells to their occupancy in our previously defined groups (Figure 1B, left). The majority of the MF-major clones were represented in the effector population whereas the major clones in SS were highly represented in the transition and progenitor exhausted group (Figure 1B, right). Trajectory and pseudotime analysis revealed a unique trajectory for patients with SS and MF. Sézary cells had a defined trajectory from the stem-like cells, transition, and then the progenitor exhausted cells (Figure 1C, top; supplemental Figure 2E, top). However, the MF cells started as stem-like, transition, effector, and lastly the proliferative population (Figure 1C, bottom; supplemental Figure 2E, bottom). In addition, a trajectory was constructed from “major” and “minor” clone cells. The trajectory was conducted without specifying the starting points and inferred developmental pseudotime using both SlingShot and Monocle methods. We plotted the inferred pseudotime of “major” clones vs “minor” clones (supplemental Figure 4F) and observed that the “major” clone cells are much more differentiated than the “minor” clone cells in both SS and MF samples. Using the combination of unsupervised trajectory analysis and pseudotime inference, we are able to establish a developmental trajectory from “minor” clone to “major” clone cells (supplemental Figure 4G). The progenitor exhausted T cells are “stem-like,” which are able to differentiate in terminally exhausted T cells.
Demographics table of patients with CTCL
| Characteristic . | Value, N = 11 . |
|---|---|
| Demographic and clinical features | |
| Sex, n (%) | |
| Male | 7 (64) |
| Female | 4 (36) |
| Age | |
| Mean, y (SD) | 70 (12) |
| Range, y | 46-82 |
| Race, n (%) | |
| White | 10 (91) |
| Black | 1 (9) |
| MF/SS diagnosis | |
| SS, n (%) | 8 (73) |
| MF, n (%) | 3 (27) |
| Stage at collection | |
| IA | 3 |
| IB | 1 |
| IIA | 1 |
| IIB | |
| III | |
| IV | 6 |
| MF/SS present in blood, # cells per μL | |
| SS, mean (range) | 2345 (484-57640) |
| MF, mean (range) | 107 (0-310) |
| Prior or coexisting neoplasm, # patients % | |
| Solid tumor | 1 (9) |
| Myeloid tumor | 2 (18) |
| Characteristic . | Value, N = 11 . |
|---|---|
| Demographic and clinical features | |
| Sex, n (%) | |
| Male | 7 (64) |
| Female | 4 (36) |
| Age | |
| Mean, y (SD) | 70 (12) |
| Range, y | 46-82 |
| Race, n (%) | |
| White | 10 (91) |
| Black | 1 (9) |
| MF/SS diagnosis | |
| SS, n (%) | 8 (73) |
| MF, n (%) | 3 (27) |
| Stage at collection | |
| IA | 3 |
| IB | 1 |
| IIA | 1 |
| IIB | |
| III | |
| IV | 6 |
| MF/SS present in blood, # cells per μL | |
| SS, mean (range) | 2345 (484-57640) |
| MF, mean (range) | 107 (0-310) |
| Prior or coexisting neoplasm, # patients % | |
| Solid tumor | 1 (9) |
| Myeloid tumor | 2 (18) |
Overall demographics of sex, age, race, diagnosis, stage, and prior or existing neoplasms independent of CTCL (n = 11).
Gene expression patterns in SS and MF. (A) Uniform manifold approximation and projection (UMAP) of combined analysis of single-cell RNA sequencing (scRNA-seq) from sorted CD4+CD26+ and CD4+CD26− isolated T cells from peripheral blood of patients with SS SS1-SS4 (n = 4) and patients with MF MF1-MF3 (n = 3) by group (n = 7). (B) UMAP plots of each MF and SS major and minor clones and occupation of cells in each group with bar graph (%) representative of percent of cells in each group. (C) Trajectory analysis and pseudotime (slingshot) analysis of SS cells (top) compared to MF cells (bottom). (D) Bubble plot of stemness score and exhaustion score colored by Shannon clonality index and sized by cell count of 7 groups. Bubble plot of single-cell ATAC-seq open chromatin regions and scRNA-seq gene expression patterns across phenotypes (stem-like, transition, effector, progenitor exhaustion, proliferation, mitochondrial activity, and T-regulatory) in SS (E) and MF (F). Color of each dot represents normalized average gene expression from high (red) to low (blue). Size of each dot represents the percentage of positive cells for each gene.
Gene expression patterns in SS and MF. (A) Uniform manifold approximation and projection (UMAP) of combined analysis of single-cell RNA sequencing (scRNA-seq) from sorted CD4+CD26+ and CD4+CD26− isolated T cells from peripheral blood of patients with SS SS1-SS4 (n = 4) and patients with MF MF1-MF3 (n = 3) by group (n = 7). (B) UMAP plots of each MF and SS major and minor clones and occupation of cells in each group with bar graph (%) representative of percent of cells in each group. (C) Trajectory analysis and pseudotime (slingshot) analysis of SS cells (top) compared to MF cells (bottom). (D) Bubble plot of stemness score and exhaustion score colored by Shannon clonality index and sized by cell count of 7 groups. Bubble plot of single-cell ATAC-seq open chromatin regions and scRNA-seq gene expression patterns across phenotypes (stem-like, transition, effector, progenitor exhaustion, proliferation, mitochondrial activity, and T-regulatory) in SS (E) and MF (F). Color of each dot represents normalized average gene expression from high (red) to low (blue). Size of each dot represents the percentage of positive cells for each gene.
When comparing the stemness and exhaustion scores of these defined groups, the progenitor exhausted group had the highest stem and exhausted score compared with all other groups (Figure 1D), which is consistent with previously described TCF7+TOX+ cells.20,24 Stem-like cells in both diseases differentiate into transitional populations with comparable transcriptional profiles, characterized by decreased coexpression of stem cell−like genes TCF7 and IL7R. The Sézary stem−like cells also gained the expression CD27 in the transitional group (Figure 1E-F). Sézary cells then accumulate a gene expression signature of progenitor exhausted cells with increased expression of stem gene TCF7 and exhaustion markers TOX and TIGIT (Figure 1E). However, only Sézary cells had attributes of increased mitochondrial activity (MT-ATP6, MT-CO1, MT-CYB, and MT-ND1) and T-regulatory cells (FOXP3) compared to MF (Figure 1E). In contrast, MF cells differentiated into a distinctive phenotype with features of effector activity, including the expression of IFNG, FGFBP2, GNLY, GZMB, GZMH, and PRF1; and downregulation of the stem cell marker TCF7 and the memory cell marker CD27 (Figure 1F). In addition, MF cells progressively acquired a proliferative phenotype, characterized by expression of cell cycle genes MKI67, CDK1, CCNA2, and CDCA2 and upregulation of exhaustion markers HLA-DRB1, HLA-DRA, and CD74 (Figure 1F).
We next analyzed notable transcription factors that have been previously reported to be critical in SS and MF development within these defined groups. As expected, we found that SATB1 expression decreases as cells differentiated from stem-like to more terminally differentiated phenotypes (supplemental Figure 2F). It has been previously reported that SATB1 expression is lost in SS and MF and is correlated with poor prognosis.25 In addition, increase in IKZF2 expression have been observed in SS.26 Interestingly, IKZF2 increases in transition group and progenitor exhausted group, but is minimally expressed in effector populations. ZEB2, another notable transcription factor in hematopoiesis and leukemic development,27 is increased in the effector population and proliferative population only (supplemental Figure 2F). SATB1 was also inversely correlated with PDCD1 expression, with increase in PDCD1 being predominately in proliferating Sézary cells rather than MF cells28 (supplemental Figure 2G). This inverse correlation was confirmed in CD4 and CD8 T cells from previous findings.28,29 Supplemental Figure 2H and I demonstrate an unbiased gene set enrichment analysis of previously reported T-cell phenotypes and their gene expression profiling, which correlated well to our groups14,15,20 (supplemental Table 3). Therefore, our findings indicated that SS and MF differentiate into expanded peripheral clonal cells with very distinct molecular attributes.
SS and MF are also different in terms of clonality and transcription factors driving malignant phenotypes
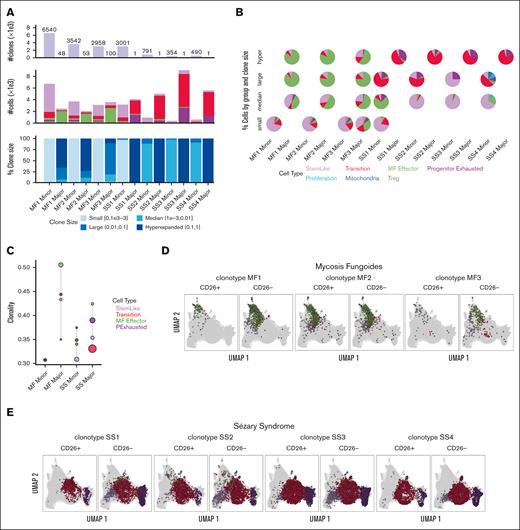
The single-cell TCR-sequencing of cells in both MF and SS showed a diverse range of clonotypes. However, MF cells exhibit much broader TCR repertoires than Sézary cells, which included larger clones (Figure 2A). MF-major clones were enriched in the MF effector group whereas SS-major clones increased in more differentiated populations, such as transition and progenitor exhaustion groups (Figure 2B and C). When comparing the most dominant clones in each sample from patient with MF and from those with SS between CD26+ and CD26− populations, we observe that the dominant clone for MF is within the expected effector group for either CD26+ or CD26− cells (Figure 2D), whereas the dominant SS clone was observed in the stem-like group only in the CD26− population (Figure 2E). Clonal enrichment in more differentiated malignant lymphocytes in MF and to an even greater extent, SS, suggests that disease progression is dependent on antigen stimulation.
TCR clonality patterns in SS and MF. (A) Quantification of number of clones, number of cells, and percent clone size in each sample for MF (MF1-MF3) and SS (SS1-SS4) major and minor clones from scRNA-seq and VDJ-seq. (B) Pie charts for composition of clones stratified by clone size, with color representing cells in different groups. (C) Shannon Clonality Index of TCR clones of MF and SS samples by group for major and minor clone T cells. UMAP representing the dominate clones for each MF (D) and SS (E) sample for CD4+CD26+ and CD4+CD26− T cells detailing the distribution of the clone in each group. Clonality was calculated for groups ≥50 cells.
TCR clonality patterns in SS and MF. (A) Quantification of number of clones, number of cells, and percent clone size in each sample for MF (MF1-MF3) and SS (SS1-SS4) major and minor clones from scRNA-seq and VDJ-seq. (B) Pie charts for composition of clones stratified by clone size, with color representing cells in different groups. (C) Shannon Clonality Index of TCR clones of MF and SS samples by group for major and minor clone T cells. UMAP representing the dominate clones for each MF (D) and SS (E) sample for CD4+CD26+ and CD4+CD26− T cells detailing the distribution of the clone in each group. Clonality was calculated for groups ≥50 cells.
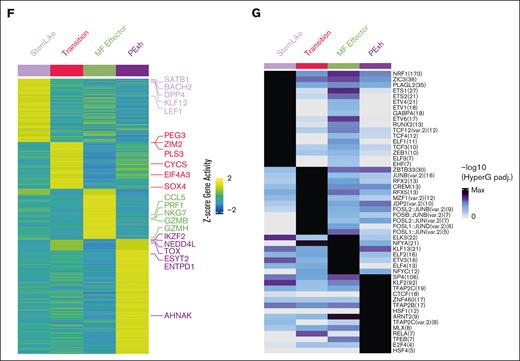
Equally important, single-cell ATAC-seq of sorted CD4+CD26+ and CD4+CD26− T cells of 4 patients with SS (SS1-SS4) and 1 with MF (MF3), confirmed that open or closed chromatin regions match expression patterns identified through single-cell RNA-seq (Figure 3A-B; supplemental Figure 5A-B). Although we were not able to define major clones and minor clones from single-cell ATAC-seq sequencing data, our previous findings (Figure 1B) support that the major clones for SS were enriched in the transition and progenitor exhaustion groups, whereas MF-major clones are enriched in the effector group. Owing to the differences in genes captured through single-cell ATAC-seq and RNA-seq, there was an expected absence of the mitochondrial genes. In addition, there was not a proliferative group present in the single-cell ATAC-seq data (Figure 3A-B). We examined the open chromatin regions near notable transcription factors and other genes to understand if the coordination and expression of these genes are potentially epigenetically regulated, and at what stage of differentiation this regulation occurs. Chromatin adjacent to SATB125,30,31 progressively shows a close conformation, because cells differentiate away from the stem-like clusters; particularly in SS. Similar to the single-cell RNA-seq data, ZEB2 has open chromatin regions in the effector population (Figure 3C,E; supplemental Figure 5C). In contrast, IKZF2 expression and open binding sites were present in transitional cells, as well as in progenitor exhausted cells compared to the stem-like population (Figure 3D-E). Open chromatin peaks near the promoter region of important transcription factors in CTCL, such as SATB1, IKZF2, ZEB1, RUNX3, and NOTCH1 were examined (Figure 3E). Coordinating the gene activity and transcription factor binding sites of genes, it appears that SATB1, KLF12, and LEF1 are highly expressed and have the potential of regulating the stem-like population. In the transition group, SOX4 is highly expressed with predicted binding in the transition group and maintains expression in the progenitor exhausted group. SOX4 has recently been attributed to being an important transcription factor in mediating the dysfunction signature in exhausted T cells.32 Lastly, IKZF2 is upregulated in the progenitor exhausted cells, indicating that it is important in establishing or maintaining the malignant state of SS cells (Figure 3F). In addition, single-cell ATAC-seq also revealed transcription factor motif enrichment for ZEB1, previously associated with tumor suppressor activity in SS,33 specifically in stem-like cells. Its loss in the subsequent differentiated groups supports its tumor suppression function (Figure 3G). Other transcription factors and genomic organizers previously associated with the pathophysiology of CTCL also showed distinctive (and matching) transcriptional and chromatin structure profiles. Together, these data suggest that MF and SS are distinct diseases in terms of diversity of clonotypes, clonal enrichment throughout malignant progression and chromatin structure. Furthermore, both diseases are driven by the expression of a distinctive pattern of transcription factors.
Chromatin confirmation patterns in SS and MF. (A) UMAP plot of single cell ATAC-seq from CD4+CD26+ and CD4+CD26− sorted T cells from peripheral blood of SS patients SS1-SS4 (n = 4) and MF patients MF3 (n = 1) by group (n = 5). (B) Individual UMAPs of each MF and SS CD4+CD26+ and CD4+CD26− T cells, similar to panel A. (C) Violin plots of single cell ATAC-seq open chromatin regions of notable CTCL and T-cell effector genes such as SATB1, TCF7, CD69, ITGAE, PDCD1, TXB21, ZNF638, and ZEB2. (D) Violin plots of single cell ATAC-seq open chromatin regions of notable CTCL and T-cell effector genes such as IKZF1, IKZF2, and IKZF3. (E) Chromatin peaks near notable CTCL genes and T-cell genes such as SATB1, IKF2, ZEB1, RUNX3, and NOTCH1. Heat maps of group-specific (F) gene activity and (G) transcription factor motif enriched in chromatin peaks genes. Columns present different cell groups. Rows present (F) gene showing different activity across group and (G) transcription factor (TF) motif enriched in differentially accessible peak regions. Color in panel E presents gene-wise z-scaled normalized average activity. Color in panel F presents normalized enrichment of the TF motif enriched within the differentially accessible peak regions.
Chromatin confirmation patterns in SS and MF. (A) UMAP plot of single cell ATAC-seq from CD4+CD26+ and CD4+CD26− sorted T cells from peripheral blood of SS patients SS1-SS4 (n = 4) and MF patients MF3 (n = 1) by group (n = 5). (B) Individual UMAPs of each MF and SS CD4+CD26+ and CD4+CD26− T cells, similar to panel A. (C) Violin plots of single cell ATAC-seq open chromatin regions of notable CTCL and T-cell effector genes such as SATB1, TCF7, CD69, ITGAE, PDCD1, TXB21, ZNF638, and ZEB2. (D) Violin plots of single cell ATAC-seq open chromatin regions of notable CTCL and T-cell effector genes such as IKZF1, IKZF2, and IKZF3. (E) Chromatin peaks near notable CTCL genes and T-cell genes such as SATB1, IKF2, ZEB1, RUNX3, and NOTCH1. Heat maps of group-specific (F) gene activity and (G) transcription factor motif enriched in chromatin peaks genes. Columns present different cell groups. Rows present (F) gene showing different activity across group and (G) transcription factor (TF) motif enriched in differentially accessible peak regions. Color in panel E presents gene-wise z-scaled normalized average activity. Color in panel F presents normalized enrichment of the TF motif enriched within the differentially accessible peak regions.
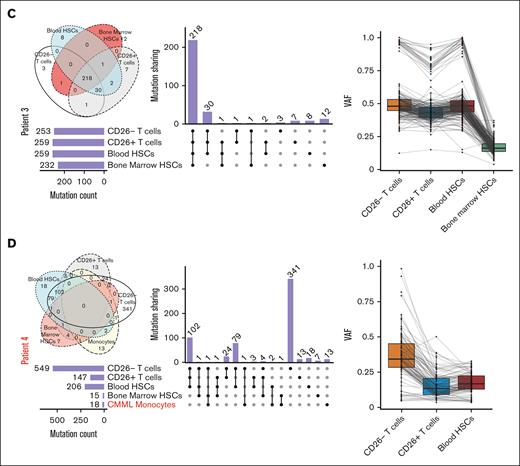
Heavily mutated hematopoietic progenitors share mutations in key oncogenes with malignant Sézary cells in all patients who were analyzed but not their normal T-cell counterparts in some patients
Given the expression of >200 nonsynonymous TCRs in malignant cells in all patients, we hypothesized that malignant T cells could arise from defective hematopoietic progenitors that acquire TCR expression in the thymus. To test this hypothesis, we conducted whole-exome sequencing on bead sorted CD34+ HSC extracted from bone marrow biopsies from 4 different patients with SS (Table 1; supplemental Table 4), as well as bead sorted autologous CD4+CD26− T cells, CD4+CD26+ T cells, and CD34+ HSCs from the aphaeresis of the same patients. Although we acknowledge that CD26− is not an explicit marker for malignancy, our previous data (supplemental Figure 2B-C) show that most of our CD26− population for SS contains the malignant expanded clone. To establish the baseline for calling true mutations without using hematopoietic cells, we cultured bone marrow fibroblasts from each patient. Unexpectedly, we found ≥200 mutations in CD34+ HSCs derived from either the bone marrow or peripheral blood in all patients (Figure 4). Supporting the validity of these results, some of these mutated genes in peripheral or bone marrow HSCs have been previously identified as key oncogenes. For instance, different mutations in the coding region of TP536,34,35 were present in autologous HSCs and peripheral Sézary cells in 2 patients, whereas a third patient showed a shared mutation in a noncoding region of TP53 (supplemental Table 4; supplemental Figure 6A). Analysis of each patient sample revealed TP53 (58%) as the most frequently mutated gene, which is supported by previous literature34-36 (supplemental Figure 6A). These mutations in TP53 as well as numerous other gene mutations occurred in either the peripheral blood or bone marrow HSCs samples as well as in peripheral T cells (Figure 4; supplemental Figure 6A). The exception was patient 1 who demonstrated a low number of overlapping mutations between progenitors and peripheral T cells despite harboring numerous mutations in bone marrow HSCs (Figure 4A). A number of patients with CTCL we observed in the clinic experienced co-occurring T-cell lymphomas12 or myeloid malignancies. An interesting point to explore was the differences in HSC mutational signatures in these patients with multiple malignancies (patient 4) compared to patients with only CTCL (patient 1-3). Patient 4 with co-occurring chronic myelomonocytic leukemia (CMML) and SS had the lowest mutation burden in the bone marrow HSC (Figure 4D; Table 1; supplemental Table 1). Given that previous studies in patients with CMML of similar age identified from not >10 to 15 mutations per exome in HSCs, this supports our findings.37,38 In addition, CMML has been reported to be driven by relatively few mutations with an especially low occurrence of TP53 mutations.39 Surprisingly, the majority of mutations shared with CD26− T cells in patient 4 were from the peripheral blood HSC rather than the bone marrow HSCs. Our results indicate that patients with SS have an unusually heavy mutational burden in hematopoietic precursors compared with those with CMML. In addition, this suggests that SS arise from different progenitors rather than co-occurring myeloid malignancies in the same patient. Previous publications claim that TP53 mutations occur in late phases of MF and are not typically detected in early-stage MF.6,40,41 However, because we were able to find these mutations in both the progenitor cells and peripheral T cells it is possible that there is a difference in primary and secondary patients with SS. The majority of our patients with SS did not have a prior history of MF, which might be a unique clinical feature of SS not previously reported, because we, to our knowledge, are the first to compare mutations from the progenitor cells of bone marrow and peripheral malignant cells of patients with SS (either primary or secondary). TP53 mutations were not present in every progenitor of the patients, which is expected given the rate of mutation detected in TP53 of patients with SS owing to the heterogeneity of the disease.6 In addition, patient 2 showed different mutations in the known oncogene ARID1A4,34 shared between HSCs and peripheral CD26− cells, whereas a shared mutation in the TNK2 tyrosine kinase gene, previously identified in acute myeloid leukemia and multiple other malignancies,42 was identified in another patient (Table 2).
Somatic mutations overlapping in HSCs and mature peripheral cells in SS. Upset plot of overlapping mutations across bone marrow HSCs, blood HSCs, CD26− cells, CD26+ T cells, and monocytes with corresponding Venn Diagrams and combined density plots with line graphs of variant allele frequency (VAFs) of mutations overlapping between progenitors and T cells in 4 patient samples (A-D).
Somatic mutations overlapping in HSCs and mature peripheral cells in SS. Upset plot of overlapping mutations across bone marrow HSCs, blood HSCs, CD26− cells, CD26+ T cells, and monocytes with corresponding Venn Diagrams and combined density plots with line graphs of variant allele frequency (VAFs) of mutations overlapping between progenitors and T cells in 4 patient samples (A-D).
Summary of repeated or oncogenic overlapping mutations across patients with SS
| Hugo symbol . | Patient 1 . | Patient 2 . | Patient 3 . | Patient 4 . |
|---|---|---|---|---|
| MAP6 | c.C1739T | c.C408A | ||
| PKD2 | c.C2384T | c.C2395T | ||
| UNC80∗ | c.C5415T | c.T7519A | ||
| TP53∗ | + | c.262delT | c.G42A | |
| GLI2 | + | c.C1506T | ||
| SCN2A | + | c.G1492A | ||
| TNK2∗ | + | + | ||
| FBN1 | + | + | ||
| CATSPERE | + | + | ||
| JPH3 | + | + | ||
| WDFY4 | + | + | ||
| RP1 | + | + | ||
| LAMA1 | + | + | ||
| CADPS | + | + | ||
| ARID1A∗ | c.C6382T | |||
| ARID1A∗ | c.C6381T | |||
| ARID1B∗ | c.C2722T |
| Hugo symbol . | Patient 1 . | Patient 2 . | Patient 3 . | Patient 4 . |
|---|---|---|---|---|
| MAP6 | c.C1739T | c.C408A | ||
| PKD2 | c.C2384T | c.C2395T | ||
| UNC80∗ | c.C5415T | c.T7519A | ||
| TP53∗ | + | c.262delT | c.G42A | |
| GLI2 | + | c.C1506T | ||
| SCN2A | + | c.G1492A | ||
| TNK2∗ | + | + | ||
| FBN1 | + | + | ||
| CATSPERE | + | + | ||
| JPH3 | + | + | ||
| WDFY4 | + | + | ||
| RP1 | + | + | ||
| LAMA1 | + | + | ||
| CADPS | + | + | ||
| ARID1A∗ | c.C6382T | |||
| ARID1A∗ | c.C6381T | |||
| ARID1B∗ | c.C2722T |
Whole-exome sequencing was performed on samples as previously described. Mutations which occurred in genes more than once across patient samples are shown or in oncogenes. +Intronic region.
Previously described oncogenes.
We identified specific mutations shared by autologous HSCs and malignant peripheral cells in every patient, ranging from 2 to 183 shared mutations (supplemental Table 4). Interestingly, in 2 out of 4 patients we found multiple mutations only shared between CD26− cells and HSCs, which were absent in autologous CD26+ T cells (supplemental Table 4), further supporting that CD26− T cells and at least a subset of CD26+ lymphocytes in these patients arose from different hematopoietic precursors. In addition, the low variant allele frequency (Figure 4) of these mutations in the HSCs suggests that there is a highly heterogenous population of progenitor cells in the bone marrow. Supporting this proposition and an obvious bias in lymphoid progenitors, monocytes from 3 samples from patient exhibited a low number of total mutations and even fewer overlapping mutations to any progenitor or mature T cells (supplemental Table 5). In addition, COSMIC mutation signatures for each patient in each sample for single bases substitutions (supplemental Figure 6B). The majority of these mutations were C>T transitions for patients 2 and 3, including UV signatures (SBS7A and SBS7B) shared between the HSC cells and mature CD26− T cells. Patient 1 had the most variation from the dominant signatures in the peripheral blood HSC being SBS6 (DNA mismatch repair associated with microsatellite instability tumors) and SBS7b (UV signature). SBS84 (activation-induced cytidine deaminase) was present in both blood and bone marrow HSCs. Peripheral CD26− T cells also contained signature SBS13 for activation-induced cytidine deaminase. Patient 4 with CMML had similar UV signatures in the peripheral blood apheresis HSCs and peripheral T cells. The signatures in bone marrow HSCs were different, likely because of the small number of mutations and sequencing artifacts. Similarly, the monocytes, unsurprisingly, have very different mutation signatures in all samples mostly relating to sequencing artifacts owing to the very small number of mutations. Considering the high number of mutations shared between CD26− T cells and HSCs and the diversity of TCRs in malignant cells, these results indicate that specific prethymic clones of mutated T-cell precursors with malignant potential acquire multiple TCRs in the thymus. Given the presence of some mutations in CD26+/CD7+ T cells, in addition to the clonal enrichment in differentiated malignant cells that lost stem-like attributes, these results further suggest that malignant transformation occurs in the periphery, postthymic selection, possibly in response to antigen.
CTCL cells show hallmarks of recent thymic egression but moderate response to skin antigens
Thymic activity greatly decreases with age, however the majority of patients with SS are older. To gain insight into thymic activity in patients with SS, we first quantified sjTRECs from naïve CD4+CD26− (n = 6) and CD4+ CD26+ T cells (n = 3) from peripheral blood of samples from patients with CTCL. Although the sjTRECs were lower in CD4+CD26− CTCL cells as previously reported43 compared to polyclonal T cells from age-matched healthy individuals, our purpose was to demonstrate that residue thymic activity exists in these patients with CTCL at an advanced age, and therefore these malignant cells from mutated progenitor HSCs have recently egressed from the thymus rather than simply acquiring mutations in the periphery. We have included a high control in the form of immature T cells from blood from cord blood. In addition, sorted CD4+ naïve T cells from the peripheral blood of benign ovarian tumor of advanced age were included as an additional control. We found that sjTREC signal is retained in Sézary cells, but not in control CD14+CD3− monocytes (n = 6) to show that patients with SS maintain thymic activity (Figure 5A-B). The gating strategy for sorting naïve T cells is shown for representative samples in supplemental Figure 7A-B.
Thymic egression of SS. (A) Schematic of recombination events in TCR rearrangement to generate signal joint TRECs and coding joint TRECs. (B) Quantification of sjTREC via real-time qPCR from naïve T cells from peripheral blood of CTCL patients that are CD4+CD26− (n = 6) or CD4+CD26+ (n = 3) compared to age-matched healthy donors (n = 6), cord blood, benign ovarian tumor peripheral patient blood, and sample matched CD14+CD3− monocytes (n = 6) using total DNA. Data are presented as mean ± standard error of the mean (SEM). ∗P < .05. Unpaired 2-tailed t test was used for calculating differences between means between experimental groups.
Thymic egression of SS. (A) Schematic of recombination events in TCR rearrangement to generate signal joint TRECs and coding joint TRECs. (B) Quantification of sjTREC via real-time qPCR from naïve T cells from peripheral blood of CTCL patients that are CD4+CD26− (n = 6) or CD4+CD26+ (n = 3) compared to age-matched healthy donors (n = 6), cord blood, benign ovarian tumor peripheral patient blood, and sample matched CD14+CD3− monocytes (n = 6) using total DNA. Data are presented as mean ± standard error of the mean (SEM). ∗P < .05. Unpaired 2-tailed t test was used for calculating differences between means between experimental groups.
To investigate how T cells with malignant potential egressing from the thymus complete their malignant transformation in the periphery, we tested whether MF and Sézary cells could react differently to skin antigens for clonal expansion. For that purpose, we procured skin biopsies from 2 patients with SS, produced lysates, and used them to pulse APCs generated from the peripheral blood of patients with MF and SS. As shown in Figure 6A, we observed only an increasing trend in IL-4 cytokine production when CD26− Sézary cells were incubated with autologous APCs pulsed with lysates from cultured (major histocompatibility complex-II+) keratinocytes, but not human skin lysates, in independent experiments in 2 of 3 patients. However, because this difference was not significant, other metrics of T-cell antigen response were explored. We did not observe significant proliferation in SS when exposed to lysates; however, there was a modest increase in surface expression of CD69, suggestive of possible reactivity from CD4+CD26− malignant cells to antigens contained with the primary skin sample, but not keratinocytes, compared to CD4+CD26+ cells (Figure 6B; supplemental Figure 8). CD69 is a well-established marker for activation44 with changes in CD69 expression indicating an effect elicited by possible antigen presentation from skin biopsies. However, the overall activation of these T cells throughout these experiments was modest.
Antigen presentation capacity in SS and MF. (A) Antigen pulsing of APCs with autologous Sézary peripheral blood CD4+CD26−CD7− cells (n = 3) against no antigen, keratinocyte lysate (12 μg), or allogenic Sézary skin biopsy lysate (12 μg). Positive control for proliferation was CD3/CD28 stimulated T cells. Enzyme-linked immunosorbent assay (ELISA) of supernatant from antigen presented cells for human IL-4 with a 1:1 dilution. Values are represented as fold change over APCs and T-cell negative control condition. Experiment was done in triplicate. (B) Quantification of fold change over APC and T cells for CD69 activated live CD4+CD26− T cells across keratinocyte, skin, APC and T cells only, and CD3/CD28 bead stimulated T cell conditions. Two-tailed Student t test: ∗P < .05; ∗∗P ≤ .01. (C) Conditions similar to panel A were performed in the peripheral blood MF samples CD4+CD26−/CD7− cells (n = 3) for the quantification of fold change of CD69 activated T cells. Two-tailed Student t test was performed. (D) Histogram displaying the FACs staining for CD69 (n = 3). n.s., nonsignificance.
Antigen presentation capacity in SS and MF. (A) Antigen pulsing of APCs with autologous Sézary peripheral blood CD4+CD26−CD7− cells (n = 3) against no antigen, keratinocyte lysate (12 μg), or allogenic Sézary skin biopsy lysate (12 μg). Positive control for proliferation was CD3/CD28 stimulated T cells. Enzyme-linked immunosorbent assay (ELISA) of supernatant from antigen presented cells for human IL-4 with a 1:1 dilution. Values are represented as fold change over APCs and T-cell negative control condition. Experiment was done in triplicate. (B) Quantification of fold change over APC and T cells for CD69 activated live CD4+CD26− T cells across keratinocyte, skin, APC and T cells only, and CD3/CD28 bead stimulated T cell conditions. Two-tailed Student t test: ∗P < .05; ∗∗P ≤ .01. (C) Conditions similar to panel A were performed in the peripheral blood MF samples CD4+CD26−/CD7− cells (n = 3) for the quantification of fold change of CD69 activated T cells. Two-tailed Student t test was performed. (D) Histogram displaying the FACs staining for CD69 (n = 3). n.s., nonsignificance.
Neither proliferation nor stimulation of CD69 was observed in malignant MF cells. We surmised that antigen recognition could be limited to a small subset of malignant lymphocytes contained within the peripheral blood, which is too narrow to detect proliferative responses23 (Figure 6C-D). Given the clonal diversity peripheral MF cells, these data suggest that skin-reactive clonotypes could be too diluted in the periphery to produce detectable antigen-specific responses. However, the clonal enrichment of TCRs and possible reactivity to skin antigens suggests Sézary cells malignant potential acquires before thymic development and complete their malignant transformation in the periphery.
Discussion
We report that Sézary and MF tumor–initiating cells have distinct phenotypes of differentiation and clonality. In addition, a major finding of our study is that HSCs in these patients share multiple mutations with malignant cells, including some in the coding region of known oncogenes, such as TP53 and ARID1A. Our results, therefore, not only confirm the recent questioning of the origin of mature T-cell lymphomas from memory T cells in the periphery,11 but also show that the etiology of CTCL is associated with a heavily mutated HSC compartment. Thus, using bone marrow fibroblasts as opposed to hematopoietic cells to establish the baseline, we found >200 mutations in HSCs of all patients with SS analyzed, which is significantly above ∼20 mutations per exome typically found even in patients with myelodysplasia,45 suggesting major alterations in hematopoiesis in these patients. In addition, we observed a higher proportion of mutations than show in the study by Choi et al34 and Wang et al5 likely because of fact that bone marrow fibroblasts, rather than skin fibroblast or monocytes, were used as a control to understand the mutation burden of T cells. This could help with understanding the propensity of developing other T-cell disorders in patients with CTCL, and in particular T-LGL in peripheral blood.12,46 Whether concurrent SS and T-LGL are driven by similar defects in HSCs differentiating into CD4+ vs CD8+ T cells in the thymus needs to be tested in future studies. Our conclusions are further supported by previous studies which have shown that CTCL can be transmitted through bone marrow transplantation.47,48 It has also been reported that the oligoclonality of CTCL49-51 and other T-cell lymphoma originate from immature precursors.52,53
The fact that CTCL originates from bone marrow precursors with malignant potential is further supported by the presence of multiple nonsynonymous TCRs in peripheral malignant cells, suggesting that malignant T cells egress from a thymus with residual activity. Accordingly, we found expression of sjTREC sequences in Sézary cells, indicative of recent thymic egression. Previous literature indicates that malignant CTCL cells have fewer TRECs than polyclonal lymphocytes.43 We have found the same result in our data, in which sjTRECS are lower than aged, matched healthy and autologous CD4+CD26+ T cells. However, sjTREC expression is still present in CTCL cells. Considering that CD4+CD26− CTCL cells, which according to our data show enriched malignant features, likely proliferate more and therefore dilute TREC sequences. Together, these results indicate that a mutated hematopoietic progenitor with malignant potential acquires different TCRs in the thymus and has the capacity to expand in the periphery. Interestingly, besides mutations shared between malignant cells and HSCs and absent in “normal” (CD26+CD7+) peripheral T cells, we also identified multiple mutations shared by “normal” and malignant T cells, suggesting that malignant transformation could be completed in the periphery through expansion and differentiation. This seems to occur in SS, with clonal enrichment of differentiated vs stem cells. Accordingly, peripheral blood T cells from SS modestly reacted against skin antigens compared to the MF samples, in which there was no detectable reactivity. Overall, despite transcriptional differences between SS vs MF, our results support that the degree of genomic alterations in the precursors that arrive to the thymus, along with the repertoire of TCRs that these cells acquire, could be the major determinants of the distinct pathogenesis of MF vs SS.
One limitation of the study was that the TCR repertoires from the malignant cells found in peripheral blood are likely different from those found in skin biopsies, and we were unable to collect matching skin samples. However, the novelty of the study focuses on the accumulation of mutations from the progenitor cells from bone marrow matching to the mutational profile of peripheral malignant cells. In addition, there are likely some small populations of nonmalignant CD26+ populations contaminating the bulk whole-exome sequencing, however, based on the clonality from VDJ sequencing and copy number variation (CNV) analysis, it is likely the majority of these CD26− cells are truly malignant cells. The variant allele frequencies of the mutations determine that these mutations are more prevalent in malignant cell population. Unfortunately, there were no nonmalignant, age-matched control bone marrow for this study. However, based on previous reports comparing normal aging of blood progenitors, in donors aged ∼60 years (similar to our patient cohort) there are 1000 base substitution per genome. In addition, this study used whole-genome sequencing whereas ours used whole-exome sequencing, therefore the number of mutations (∼200) isolated to exomes accounts for ∼20% of these mutation.54 It is possible that the more whole genome mutations are present within patients with SS.
Overall, our results identify altered hematopoietic progenitors as the likely origin of at least SS and identify important similarities and differences among the pathophysiology of the different manifestations of CTCL, which has obvious implications for future therapeutic interventions. It is tempting to speculate, for instance, that thymectomy could prevent malignant lymphocytes emerging from the thymus from replenishing the periphery after treatment. This could be supported, for instance, by future studies showing that malignant T cells are quickly replenished after peripheral depletion by lymphocytes with a totally different TCR repertoire. Whether lymphocyte progenitors would show their malignant potential through alternative differentiation into malignant B cells, for instance, would need to be clinically addressed, but our study supports that targeting the capacity of the bone marrow to replenish malignant T cells in the periphery is a requirement for findings curative interventions for patients with CTCL.
Acknowledgments
The authors thank all the patients, staff, and physicians who have participated in this study and sample collection. The authors are especially grateful to the Flow Cytometry Core of Moffitt Cancer Center and the Flow Cytometry Shared Resource from Duke Cancer Center. The authors thank Johana Melendez, Jonathan Semidey-Hurtado, Bethany Carter, and Jodi L. Kroeger for technical assistance.
This study was supported by National Institutes of Health grants R01CA240434, R01CA157664, and R01CA124515. S.B. was supported by National Institutes of Health grant 1K99CA266947-01. P.I. was supported by an Ann Schreiber Fellowship from the Ovarian Cancer Research Alliance. This work has been supported in part by the Flow Cytometry Core and Molecular Genomics Core at the H. Lee Moffitt Cancer Center & Research Institute, an National Cancer Institute-designated Comprehensive Cancer Center (P30-CA076292).
Authorship
Contribution: C.M.H. designed, performed, and analyzed most of the experiments and cowrote the manuscript; X.Y. performed all biological and statistical analysis for molecular genomic experiments; R.A.C. performed the sorting and experiments for sjTREC quantification; L.S. contributed to the design of the study, patient samples, and provided intellectual support; J.R.C.-G. oversaw and designed the study and experiments, analyzed data, and cowrote the manuscript; and K.B.S., J.J.P., P.I., C.M.A, Y.Z., S.B., G.M., J.A.M., C.C., M.Z.N., A.L.M., K.F.H., G.J.B., P.-L.C., and J.P.-I. contributed to the overall data interpretation, provided intellectual input, and approved the final manuscript.
Conflict-of-interest disclosure: J.P.-I. reports funding from TG Therapeutics, MEI, and Viracta, and consulting fees from Janssen, AbbVie, TG Therapeutics, Novartis, Takeda, and AstraZeneca. L.S. reports consulting fees from Dren Bio Inc; is an adviser for Kyowa-Kirin, Inc, Daiichi-Sankyo, and Kymera Therapeutics; and reports clinical research funding from Kyowa Kirin and EUSA Pharma, LLC. J.R.C.-G. reports funding and consulting fees from Anixa Biosciences; consulting fees from Alloy Therapeutics; and stock options from Compass Therapeutics, Anixa Biosciences, and Alloy Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Jose R Conejo-Garcia, Department of Immunology, Duke School of Medicine, Jones Bldg 302/305, 207 Research Dr, Durham, NC 27710; e-mail: jose.conejo-garcia@duke.edu; Lubomir Sokol, Malignant Hematology, H. Lee Moffitt Cancer Center & Research Institute, CSB 7TH 7133, 12902 Magnolia Dr, Tampa, FL 33612; e-mail: lubomir.sokol@moffitt.org; and Xiaoqing Yu, Biostatistics and Bioinformatics, H. Lee Moffitt Cancer Center & Research Institute, MRC Bldg, 12902 Magnolia Dr, Tampa, FL 33612; e-mail: xiaoqing.yu@moffitt.org.
References
Author notes
Raw and processed single-cell RNA-seq/VDJ, and single-cell ATAC-seq can be obtained from Gene Expression Omnibus (GEO) with accession numbers of GEO: GSE207678 and GSE20767, respectively. WES data of matched tumor and normal samples can be obtained from dbGaP under accession phs003158.v1.p1.
Data are available on request from the corresponding author, Xiaoqing Yu (xiaoqing.yu@moffitt.org).
The full-text version of this article contains a data supplement.