Key Points
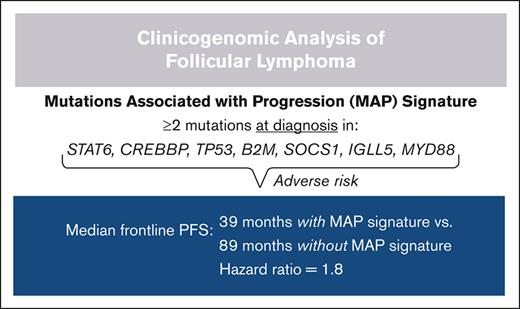
MAP (CREBBP, STAT6, TP53, IGLL5, B2M, SOCS1, and MYD88) confer adverse risk when found at FL diagnosis.
Low-risk m7-FLIPI is predictive of prolonged remissions with standard rituximab plus chemotherapy in newly diagnosed FL.
Abstract
Follicular lymphoma (FL) is clinically heterogeneous, with select patients tolerating extended watch-and-wait, whereas others require prompt treatment, suffer progression of disease within 24 months of treatment (POD24), and/or experience aggressive histologic transformation (t-FL). Because our understanding of the relationship between genetic alterations in FL and patient outcomes remains limited, we conducted a clinicogenomic analysis of 370 patients with FL or t-FL (from Cancer and Leukemia Group B/Alliance trials 50402/50701/50803, or real-world cohorts from Washington University School of Medicine, Cleveland Clinic, or University of Miami). FL subsets by grade, stage, watch-and-wait, or POD24 status did not differ by mutation burden, whereas mutation burden was significantly higher in relapsed/refractory (rel/ref) FL and t-FL than in newly diagnosed (dx) FL. Nonetheless, mutation burden in dx FL was not associated with frontline progression-free survival (PFS). CREBBP was the only gene more commonly mutated in FL than in t-FL yet mutated CREBBP was associated with shorter frontline PFS in FL. Mutations in 20 genes were more common in rel/ref FL or t-FL than in dx FL, including 6 significantly mutated genes (SMGs): STAT6, TP53, IGLL5, B2M, SOCS1, and MYD88. We defined a mutations associated with progression (MAP) signature as ≥2 mutations in these 7 genes (6 rel/ref FL or t-FL SMGs plus CREBBP). Patients with dx FL possessing a MAP signature had shorter frontline PFS, revealing a 7-gene set offering insight into FL progression risk potentially more generalizable than the m7–Follicular Lymphoma International Prognostic Index (m7-FLIPI), which had modest prognostic value in our cohort. Future studies are warranted to validate the poor prognosis associated with a MAP signature in dx FL, potentially facilitating novel trials specifically in this high-risk subset of patients.
Introduction
Follicular lymphoma (FL) is a common low-grade B-cell lymphoma of which >85% of cases possess t(14;18)(q32;q21), placing the antiapoptotic oncogene BCL2 under control of IGH regulatory elements.1,2 This event is sporadically found in rare cells from healthy individuals, most of whom do not develop FL,3,4 suggesting it is not sufficient for lymphomagenesis. Several studies suggest that premalignant B cells with IGH::BCL2 cyclically reengage germinal center (GC) reactions, permitting multihit lymphomagenesis through acquisition of cooperating mutations, promoting developmental arrest and proliferation.5-8
Initial FL presentations are heterogeneous. Some patients have symptomatic lymphadenopathy requiring timely intervention; others may be diagnosed incidentally and/or tolerate extended watch-and-wait.9-11 Most patients’ disease responds to frontline treatment12-15; however, their subsequent courses are highly variable. The majority of patients have prolonged remissions and low-grade recurrences but others suffer progression of disease within 24 months of treatment (POD24) and have poor outcomes despite receiving subsequent therapy.16,17 Furthermore, ∼30% of patients with FL experience aggressive histologic transformation (t-FL), which is enriched in patients experiencing POD24 and contributes to most FL-related deaths.18-21
Regarding pathobiology, the FL mutational landscape highlights the mechanisms driving lymphomagenesis.22,23 More than three-quarters of FL tumors possess mutations in genes encoding key histone modifiers (KMT2D, EZH2, CREBBP, and EP300),24-27 which control GC B-cell transcriptional programs. H1 linker histone mutations also cooperate with dysregulated BCL2 to promote FL.28,29 Other affected pathways include B-cell receptor, NF-κB, and JAK-STAT signaling.30-33 Several studies demonstrate that FL and t-FL often arise from divergent evolution of a common mutated precursor. Histone modifier alterations typically occur early in the common precursor, whereas transformation involves diverse factors including TP53 loss, immune evasion, MYC activation, and somatic hypermutation (SHM).34-37
Despite a growing understanding of FL pathogenesis, the translation of these insights into biology-informed clinical care has been slow. No predictive model exists to guide FL treatment, although the Groupe d'Etude des Lymphomes Folliculaires criteria differentiate patients with low vs high disease burden.38,39 The Follicular Lymphoma International Prognostic Index (FLIPI) includes 5 adverse clinical risk factors (age >60 years, advanced stage disease, hemoglobin level <12 g/dL, >4 involved nodal areas, and elevated lactate dehydrogenase).40 Although commonly applied in practice, the FLIPI rarely influences decisions of initial watch-and-wait vs treatment, or treatment selection.
Two strategies applying FL pathobiology to prognostication include DNA sequencing for mutational assessment and RNA sequencing for gene expression profiling. The m7-FLIPI is a clinicogenomic risk model incorporating the traditional FLIPI, patient performance status, and mutation statuses of 7 genes.41 Moreover, gene expression studies highlight associations between features of nonmalignant cells in FL and patient outcomes.42,43 However, these approaches are not integrated into patient care because of limitations regarding actionability and generalizability.7,44-46 To help overcome these dilemmas and expand our understanding of the relationships between patient outcomes and FL genetic alterations, we performed a large clinicogenomic study of FL, taking advantage of diverse presentations, ranging from initial diagnoses to relapsed/refractory (rel/ref) or transformed disease.
Methods
Study design
We assembled a cohort of patients with FL or t-FL and obtained tumor biopsies from Washington University School of Medicine (WUSM; St. Louis, MO), Cleveland Clinic (Cleveland, OH), and University of Miami (Miami, FL), or through the Alliance for Clinical Trials in Oncology (Cancer and Leukemia Group B [CALGB], now part of the Alliance, studies 50402/50701/50803; clinicaltrials.gov identifiers: NCT00117975 [CALGB 50402], NCT00553501 [CALGB 50701], and NCT01145495 [CALGB 50803]). All samples were collected within protocols and consenting processes approved by local regulatory offices; all patients from Alliance studies provided written informed consent for sample collection. Deidentified patient characteristics, treatment histories, and outcomes were compiled. Pathology review of tumor biopsies confirmed the diagnosis and estimated tumor cell distribution. Sections from frozen specimens or punches in tumor-containing areas from formalin-fixed, paraffin-embedded specimens were used for genomic DNA isolation. Data from 113 samples analyzed herein were generated previously.23
DNA sequencing and analysis
In addition to the WUSM-LPv1 reagent,23 a new custom capture reagent was designed: the 12-megabase WUSM-LPv2 reagent targets 2613 genes, including all genes from WUSM-LPv1 (N = 1716) plus 897 additional genes recurrently mutated in 14 previous lymphoma studies (supplemental Table 1). Genomic DNA was isolated using the QIAamp DNA Mini kit after paraffin removal using xylene or CitroSolv. Library preparation was performed using the KAPA HTP library prep kit for whole-exome sequencing (WES) on a SciClone next-generation sequencing instrument. Capture hybridization was performed using NimbleGen Custom Liquid Capture Reagent WUSM-LPv1, NimbleGen Custom Liquid Capture Reagent WUSM-LPv2, or NimbleGen SeqCap EZ Exome version 2.0 or IDT Exome kits. Sequencing was performed using a HiSeq4000 (WUSM-LPv2 or WES) or HiSeq2500 (WUSM-LPv1). Sequencing reads were processed using the Genome Modeling System.47 Data from 113 previously described samples were reprocessed in parallel with new samples. Paired-end reads were aligned to human reference GRCh37 using BWA-MEM and deduplicated using Picard. Pipeline details, variant calling, and manual review, plus significantly mutated gene (SMG) identification using MutSigCV,48 are described in the supplemental Materials. Selected data are accessible via dbGaP #phs001229 based on patients’ specific consent.
Clinical and statistical analyses
Data lock date was 10 November 2020. Median follow-up was 74.9 months. FLIPI and m7-FLIPI stratifications were performed as previously described.40,41 Watch-and-wait was defined as time from diagnosis to frontline treatment initiation of ≥12 months. Progression-free survival (PFS) was defined as time from treatment initiation (or watch-and-wait initiation if ≥12 months) to time of progressive lymphoma or death from any cause; patients were censored at last follow-up if alive without progression. Overall survival (OS) was defined as time from diagnosis to death from any cause; patients were censored at last follow-up if alive. POD24 was defined as progressive lymphoma or death within 24 months of frontline treatment initiation, irrespective of therapy.16,17,49,50 OS stratified by POD24 status was landmark-adjusted for this risk-defining event, measuring survival from time of progressive lymphoma for early progressors or from 24 months post-treatment initiation for the reference group, as previously described.16 The Kaplan-Meier method was applied to estimate survival probabilities and generate survival curves; comparisons were made using log-rank tests. Cox proportional hazard penalized splines were used to model PFS/OS vs mutation burden. Statistical testing included t tests to compare numerical data between 2 groups, one-way analysis of variance to compare numerical data between ≥3 groups, or Fisher exact tests to compare categorical data between 2 groups, with significance being defined as P < .05 without adjustment for multiplicity, unless stated. Benjamini-Hochberg (BH) correction for multiple comparisons was applied, as indicated, for assessments across multiple genes. Analyses used R, including packages survival and survminer.
Results
Clinical characteristics and outcomes
We performed genomic profiling on FL or t-FL samples from 370 patients (Table 1; supplemental Table 2; genomic sample set) using custom capture sequencing or WES (supplemental Figure 1). A unified 2595-gene set (supplemental Table 3) was applied to all samples for subsequent analyses. Baseline characteristics plus partial treatment histories and outcomes were available for all 370 patients. Complete treatment histories and long-term follow-up data were available for 343 patients (clinical sample set). No baseline characteristics, including age, sex, or stage, differed significantly between sample sets.
Patient information
| Characteristic . | Variable . | Genomic sample set (N = 370) . | Clinical sample set (N = 343) . | ||
|---|---|---|---|---|---|
| n . | % of evaluable patients . | n . | % of evaluable patients . | ||
| Sex | |||||
| Male | 202 | 56.1 | 192 | 56 | |
| Female | 158 | 43.9 | 151 | 44 | |
| Age, y | |||||
| Median | 60 | 60 | |||
| <40 | 25 | 7.2 | 25 | 7.6 | |
| 40–60 | 154 | 44.4 | 147 | 44.5 | |
| 60–80 | 154 | 44.4 | 144 | 43.6 | |
| ≥80 | 14 | 4 | 14 | 4.2 | |
| Lymphoma subtype | |||||
| FL | 323 | 87.3 | 296 | 86.3 | |
| t-FL | 47 | 12.7 | 47 | 13.7 | |
| FL grade | |||||
| Grade 1-2 | 247 | 82.6 | 235 | 83 | |
| Grade 3A | 52 | 17.4 | 48 | 17 | |
| Sequenced sample | |||||
| Diagnostic | 253 | 76.7 | 253 | 76.7 | |
| Relapse #1 | 36 | 10.9 | 36 | 10.9 | |
| Relapse #2+ | 41 | 12.4 | 41 | 12.4 | |
| FLIPI: nodal involvement | |||||
| 0-4 regions | 172 | 71.7 | 156 | 69.6 | |
| ≥5 regions | 68 | 28.3 | 68 | 30.4 | |
| FLIPI: LDH | |||||
| Normal | 231 | 83.1 | 218 | 82.9 | |
| Elevated | 47 | 16.9 | 45 | 17.1 | |
| FLIPI: age (y) | |||||
| <60 | 144 | 48 | 139 | 49.1 | |
| ≥60 | 156 | 52 | 144 | 50.9 | |
| FLIPI: stage | |||||
| Early (I-II) | 63 | 21.3 | 59 | 21.1 | |
| Advanced (III-IV) | 233 | 78.7 | 221 | 78.9 | |
| FLIPI: hemoglobin | |||||
| <12 g/dL | 32 | 13.5 | 29 | 13.2 | |
| ≥12 g/dL | 205 | 86.5 | 191 | 86.8 | |
| FLIPI: risk strata | |||||
| Low (0-1) | 101 | 33.9 | 97 | 34.4 | |
| Intermediate (2) | 106 | 35.6 | 100 | 35.5 | |
| High (3-5) | 91 | 30.5 | 85 | 30.1 | |
| Treatment history | |||||
| (any line of treatment) | Initial W&W | – | – | 48 | 14.5 |
| Anti-CD20 mAb | – | – | 282 | 85.2 | |
| CHOP/CVP | – | – | 168 | 50.8 | |
| Bendamustine | – | – | 56 | 16.9 | |
| Lenalidomide | – | – | 31 | 9.4 | |
| Autologous SCT | – | – | 25 | 7.6 | |
| Characteristic . | Variable . | Genomic sample set (N = 370) . | Clinical sample set (N = 343) . | ||
|---|---|---|---|---|---|
| n . | % of evaluable patients . | n . | % of evaluable patients . | ||
| Sex | |||||
| Male | 202 | 56.1 | 192 | 56 | |
| Female | 158 | 43.9 | 151 | 44 | |
| Age, y | |||||
| Median | 60 | 60 | |||
| <40 | 25 | 7.2 | 25 | 7.6 | |
| 40–60 | 154 | 44.4 | 147 | 44.5 | |
| 60–80 | 154 | 44.4 | 144 | 43.6 | |
| ≥80 | 14 | 4 | 14 | 4.2 | |
| Lymphoma subtype | |||||
| FL | 323 | 87.3 | 296 | 86.3 | |
| t-FL | 47 | 12.7 | 47 | 13.7 | |
| FL grade | |||||
| Grade 1-2 | 247 | 82.6 | 235 | 83 | |
| Grade 3A | 52 | 17.4 | 48 | 17 | |
| Sequenced sample | |||||
| Diagnostic | 253 | 76.7 | 253 | 76.7 | |
| Relapse #1 | 36 | 10.9 | 36 | 10.9 | |
| Relapse #2+ | 41 | 12.4 | 41 | 12.4 | |
| FLIPI: nodal involvement | |||||
| 0-4 regions | 172 | 71.7 | 156 | 69.6 | |
| ≥5 regions | 68 | 28.3 | 68 | 30.4 | |
| FLIPI: LDH | |||||
| Normal | 231 | 83.1 | 218 | 82.9 | |
| Elevated | 47 | 16.9 | 45 | 17.1 | |
| FLIPI: age (y) | |||||
| <60 | 144 | 48 | 139 | 49.1 | |
| ≥60 | 156 | 52 | 144 | 50.9 | |
| FLIPI: stage | |||||
| Early (I-II) | 63 | 21.3 | 59 | 21.1 | |
| Advanced (III-IV) | 233 | 78.7 | 221 | 78.9 | |
| FLIPI: hemoglobin | |||||
| <12 g/dL | 32 | 13.5 | 29 | 13.2 | |
| ≥12 g/dL | 205 | 86.5 | 191 | 86.8 | |
| FLIPI: risk strata | |||||
| Low (0-1) | 101 | 33.9 | 97 | 34.4 | |
| Intermediate (2) | 106 | 35.6 | 100 | 35.5 | |
| High (3-5) | 91 | 30.5 | 85 | 30.1 | |
| Treatment history | |||||
| (any line of treatment) | Initial W&W | – | – | 48 | 14.5 |
| Anti-CD20 mAb | – | – | 282 | 85.2 | |
| CHOP/CVP | – | – | 168 | 50.8 | |
| Bendamustine | – | – | 56 | 16.9 | |
| Lenalidomide | – | – | 31 | 9.4 | |
| Autologous SCT | – | – | 25 | 7.6 | |
CHOP, cyclophosphamide, hydroxydaunorubicin (doxorubicin), Oncovin (vincristine), and prednisone; CVP, cyclophosphamide, vincristine, and prednisone; LDH, lactate dehydrogenase; mAb, monoclonal antibody; SCT, stem cell transplantation; W&W, watch-and-wait.
PFS and OS of patients in our cohort aligned with expectations from real-world and trial populations, supporting the suitability of our cohort for the study of prognostic genomic biomarkers broadly generalizable in FL/t-FL. Specifically, OS was markedly different between patients with FL and t-FL: OS at 60 months was 94.8% vs 69.1%, respectively (median not reached [NR] for either cohort; hazard ratio [HR], 2.65; 95% confidence interval [CI], 1.53-4.60; P < .01; Figure 1A). In contrast, OS was similar between patients with FL undergoing initial watch-and-wait vs treatment (median NR for either cohort; HR, 0.88; 95% CI, 0.37-2.09; P = .78; Figure 1B), as expected.10,51 POD24 also conferred similar adverse risk to that observed in other series.17 Landmark-adjusted OS at 60 months was 96.0% vs 80.0% for patients without and with POD24, respectively (median NR for either cohort; HR, 4.27; 95% CI, 1.70-10.71; P < .01; Figure 1C). Also, PFS after frontline treatment was significantly longer than PFS after treatment in the relapsed setting (median 61.7 vs 19.7 months, respectively; HR, 1.23; 95% CI, 1.16-1.30; P < .01; Figure 1D),11 an important validity check, given the treatment diversity across our cohort. However, the traditional FLIPI demonstrated modest trends toward shorter frontline PFS and OS across strata (intermediate vs low PFS: HR, 1.42; 95% CI, 0.88-2.28; P = .15; high vs low PFS: HR, 1.67; 95% CI, 1.02-2.74; P = .04; Figure 1E-F), suggesting prognostication may be improved by incorporating biological insights from genomic profiling.
Patient outcomes. Kaplan-Meier curves displaying (A) OS in patients with FL vs t-FL, (B) OS in patients with FL undergoing initial watch-and-wait vs initial treatment, (C) landmark-adjusted OS in patients with FL experiencing POD24 or not experiencing POD24, (D) PFS in patients with FL by line of therapy (patients may be represented more than once), (E) PFS in patients with FL by traditional FLIPI stratification, and (F) OS in patients with FL by traditional FLIPI stratification.
Patient outcomes. Kaplan-Meier curves displaying (A) OS in patients with FL vs t-FL, (B) OS in patients with FL undergoing initial watch-and-wait vs initial treatment, (C) landmark-adjusted OS in patients with FL experiencing POD24 or not experiencing POD24, (D) PFS in patients with FL by line of therapy (patients may be represented more than once), (E) PFS in patients with FL by traditional FLIPI stratification, and (F) OS in patients with FL by traditional FLIPI stratification.
Mutational landscape of FL and t-FL
We identified the most frequently mutated genes in our FL (Figure 2) and t-FL (Figure 3) samples. The 2 most commonly mutated genes in FL were CREBBP (58.8%) and KMT2D (57.0%), with mutations in EZH2 (18.3%) and EP300 (13.3%) occurring less frequently. Overall, 40.2%, 8.7%, and 1.2% of samples possessed 2, 3, or 4 comutations, respectively, among these key histone modifiers. Notably, 19 FL cases possessed mutations in CREBBP and EP300 despite prior suggestion of mutual exclusivity of mutations in these histone acetyltransferases.52 Next, we identified SMGs, assessing observed enrichment of mutations above expected frequencies based on gene-specific properties (supplemental Table 3). Very large genes (eg, MUC16, OBSCN, and PCLO) with frequent nonsilent mutations failed to meet SMG criteria, suggesting that these may be irrelevant passenger alterations acquired over the long process of lymphomagenesis. In contrast, well-known FL drivers were identified as SMGs, as were several genes with less clear links to FL despite known roles in hematopoiesis, including EBF1. Several genes implicated in diffuse large B-cell lymphoma (DLBCL) were identified as SMGs despite being infrequently mutated in FL, including GNAI2. Although 25 genes were mutated in ≥10% of FL cases, we also identified a long tail of 526 genes mutated in 2% to 10% of FL cases; among these were several SMGs, such as B2M, with established roles in lymphoma pathogenesis.53 The diversity of these infrequently mutated yet pathogenically important genes suggests that FL clinical heterogeneity may not be driven only by common mutations but could also be influenced by combinations of infrequent events.
Mutational landscape of FL. Waterfall plot displaying patterns of mutations in 25 genes mutated in ≥10% of all FL samples. SMGs highlighted in bold on left y-axis.
Mutational landscape of FL. Waterfall plot displaying patterns of mutations in 25 genes mutated in ≥10% of all FL samples. SMGs highlighted in bold on left y-axis.
Mutational landscape of t-FL. Waterfall plot displaying patterns of mutations in 50 most frequently mutated genes in t-FL samples. SMGs highlighted in bold on left y-axis.
Mutational landscape of t-FL. Waterfall plot displaying patterns of mutations in 50 most frequently mutated genes in t-FL samples. SMGs highlighted in bold on left y-axis.
Treatment influences the m7-FLIPI
A prominent prior study integrating FL genomics into prognostication assessed 74 genes in a 151-patient training cohort.41 The resulting m7-FLIPI incorporates traditional FLIPI stratification, patient performance status, and mutation statuses of EZH2, ARID1A, MEF2B, EP300, FOXO1, CREBBP, and CARD11. We determined the m7-FLIPI risk group for the 231 patients in our cohort with newly diagnosed FL, applying a 10% variant allele fraction cutoff per the original publication. By this method, 15.2% (35/231) of patients were categorized as high-risk; this was unchanged using an alternative variant allele fraction cutoff of 1%, excluding methodological differences meaningfully influencing these findings. We compared how the m7-FLIPI and traditional FLIPI identify patients with favorable disease, using initial watch-and-wait as a proxy for indolent presentations. Of our 38 patients with newly diagnosed FL undergoing initial watch-and-wait with sequencing of their diagnostic sample, 33 (86.8%) were stratified as being at low-/intermediate-risk by the traditional FLIPI, whereas 94.7% (36 of 38) were stratified as low-risk by the m7-FLIPI. This supports the potential of the m7-FLIPI to identify patients with adverse clinical features (ie, high-risk FLIPI and frailty) who perform similarly to patients at low-risk in the context of favorable genomic alterations, such as mutated EZH2.54
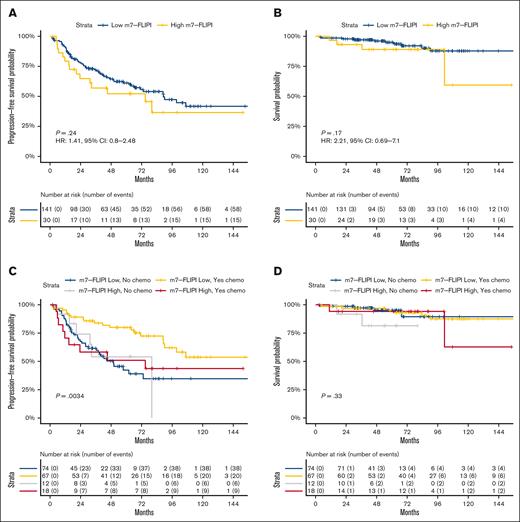
Next, we assessed the m7-FLIPI among 171 patients in our cohort with newly diagnosed FL receiving frontline therapy within 12 months of diagnosis and frontline PFS data. PFS was not significantly different between patients at low-risk vs high-risk by m7-FLIPI stratification (Figure 4A). PFS at 24 months was 77.7% vs 64.8%, and at 60 months was 61.2% vs 52.2% for patients at low-risk vs high-risk, respectively (median, 89.1 vs 74.9 months; HR, 1.41; 95% CI, 0.80-2.48; P = .24). Although the m7-FLIPI was developed to predict failure-free survival, it was also prognostic for OS in the initial study. In our cohort, OS was not significantly different between patients at low-risk vs high-risk by m7-FLIPI stratification (Figure 4B): OS at 60 months was 95.0% vs 89.1% for patients at low-risk vs high-risk, respectively (median, NR for either cohort; HR, 2.21; 95% CI, 0.69-7.10; P = .17).
Evaluation of the m7-FLIPI. Kaplan-Meier curves displaying (A) PFS in patients with FL by m7-FLIPI stratification, (B) OS in patients with FL by m7-FLIPI stratification, (C) PFS in patients with FL by m7-FLIPI stratification vs chemotherapy-treatment status, and (D) OS in patients with FL by m7-FLIPI stratification vs chemotherapy-treatment status.
Evaluation of the m7-FLIPI. Kaplan-Meier curves displaying (A) PFS in patients with FL by m7-FLIPI stratification, (B) OS in patients with FL by m7-FLIPI stratification, (C) PFS in patients with FL by m7-FLIPI stratification vs chemotherapy-treatment status, and (D) OS in patients with FL by m7-FLIPI stratification vs chemotherapy-treatment status.
Patients in our cohort received heterogeneous treatments compared with relatively homogeneous treatment received by patients in the original m7-FLIPI study. We hypothesized that the prognostic value of the m7-FLIPI may specifically apply to unique patient and treatment combinations. We assessed for an interaction between the m7-FLIPI prognostic value and patients’ frontline treatment, observing a strong relationship between receipt of cytotoxic chemotherapy (supplemental Table 5) and m7-FLIPI stratification for PFS (P < .01; Figure 4C), although not OS (Figure 4D). PFS at 48 months was 79.9% for patients with low-risk m7-FLIPI status receiving chemotherapy-based frontline treatment, vs 50% to 55% for all 3 other subsets (patients with low-risk m7-FLIPI status receiving chemotherapy-free treatment and patients with high-risk m7-FLIPI status irrespective of chemotherapy status). This provides further evidence that the prognostic value of the m7-FLIPI mutational pattern is influenced by patient treatment.55
Comparing the mutational landscapes of lymphoma subsets
Our cohort diversity permitted assessment of associations between the lymphoma genomic landscape and patient characteristics/outcomes. Conceptually integrating the model of multihit lymphomagenesis and the broad range of FL presentations, we hypothesized that adverse pretreatment features or clinical courses may reflect FL tumors further along a timeline of mutation acquisition. Thus, the abundance of genomic alterations at diagnosis could drive FL clinical heterogeneity. Comparing the number of genes with nonsilent mutations across multiple newly diagnosed FL subsets, we observed no significant differences between sample subsets by grade 1-2 vs 3A, early vs advanced stage, traditional FLIPI stratification, or initial treatment vs watch-and-wait (supplemental Figure 2). Given the increased frequency of focal SHM and phased variants in lymphoma,56,57 we also assessed the total nonsilent variant burden, a feature masked by condensing all mutations per gene (as used in the previous analysis) into a binary mutated/nonmutated status. FL subsets by grade, stage, FLIPI status, or initial treatment vs watch-and-wait did not significantly differ by total nonsilent variant burden (supplemental Figure 2), indicating that it is unlikely that features of FL at diagnosis are meaningfully driven by the extent of mutation accumulation from disease origination to presentation.
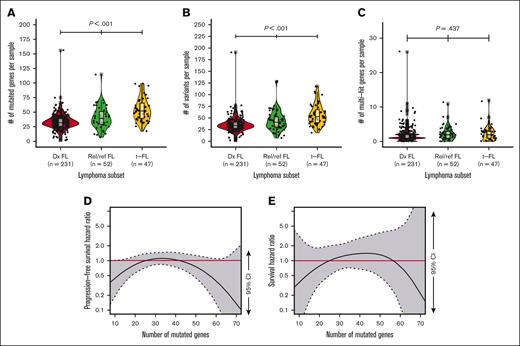
Having observed that higher mutation burden is not a feature of FL subsets by baseline characteristics, we asked whether comparisons along the natural history of FL would reveal key differences. Neither the number of nonsilent mutated genes nor total nonsilent variant burden significantly differed between newly diagnosed FL samples based on POD24 status (supplemental Figure 2). However, both the number of nonsilent mutated genes and total nonsilent variant burden were significantly higher (P < .01) in t-FL than rel/ref FL, and in rel/ref FL than in newly diagnosed FL (Figure 5A-B). The frequency of genes possessing ≥2 nonsilent mutations was not significantly different between newly diagnosed FL, rel/ref FL, and t-FL (Figure 5C), and largely similar trends were observed when focusing only on well-established SHM targets (supplemental Figure 2), despite SHM being described as a mechanism contributing to FL progression and transformation.58
Mutation burden increases across FL timeline but is not prognostic in newly diagnosed FL. (A) Box-and-whisker and violin plots of number of mutated genes per sample by newly diagnosed (Dx) FL, rel/ref FL, or t-FL; (B) box-and-whisker and violin plots of number of total nonsilent variants per sample by Dx FL, rel/ref FL, or t-FL; (C) box-and-whisker and violin plots of number genes with multiple mutations (multihit) per sample by Dx FL, rel/ref FL, or t-FL; (D-E) Cox proportional hazard penalized splines model of PFS or OS against number of mutated genes normalized to PFS or OS associated with median number of mutated genes; gray shading with dashed boundaries displays 95% CI; red horizontal line highlights null hypothesis HR of 1.0.
Mutation burden increases across FL timeline but is not prognostic in newly diagnosed FL. (A) Box-and-whisker and violin plots of number of mutated genes per sample by newly diagnosed (Dx) FL, rel/ref FL, or t-FL; (B) box-and-whisker and violin plots of number of total nonsilent variants per sample by Dx FL, rel/ref FL, or t-FL; (C) box-and-whisker and violin plots of number genes with multiple mutations (multihit) per sample by Dx FL, rel/ref FL, or t-FL; (D-E) Cox proportional hazard penalized splines model of PFS or OS against number of mutated genes normalized to PFS or OS associated with median number of mutated genes; gray shading with dashed boundaries displays 95% CI; red horizontal line highlights null hypothesis HR of 1.0.
Given the increasing mutation burden from FL diagnosis to rel/ref disease to transformed disease, we asked whether mutation burden at FL diagnosis conferred adverse clinical risk. Cox proportional hazard penalized splines modeling revealed no significant relationship between either frontline PFS or OS with either the number of nonsilent mutated genes or total nonsilent variant burden at diagnosis as continuous variables (Figure 5D-E; supplemental Figure 2). These findings indicate that adverse risk in FL is not simply the manifestation of FL presenting later along a timeline of mutation acquisition. Instead, specific mutated genes at diagnosis or acquired later may be drivers of unfavorable biology.
MAP predict poor outcomes in newly diagnosed FL
Exploring this, we evaluated for skewed frequencies of individual mutated genes in subsets of FL samples. We identified genes mutated in ≥10% of samples in either subset per comparison, then tested for statistical over-/underrepresentation plus SMG status, accounting for expected mutation frequency per gene. Surprisingly few mutated genes were over- or underrepresented in key FL subsets. GNAI2 was the only SMG in which mutations were significantly more common in patients undergoing initial watch-and-wait (BH false discovery rate [FDR] q < 0.1; Figure 6A). No SMG was more or less frequently mutated in patients with or without POD24 (BH FDR q < 0.1; Figure 6B). In contrast, comparisons between newly diagnosed FL, rel/ref FL, and t-FL were more striking. CREBBP was the only gene in which mutations were significantly more common in FL than in t-FL (BH FDR q < 0.1). Conversely, mutations in 20 genes were more common in rel/ref FL than in newly diagnosed FL, or t-FL than FL, including 6 SMGs: STAT6, TP53, IGLL5, B2M, SOCS1, and MYD88 (BH FDR q < 0.1; Figure 6C-D; supplemental Figure 3).
Enrichment of mutations in FL/t-FL subsets. (A-D) Scatter plots displaying frequency of mutations per gene in FL/t-FL subsets. Points are highlighted in red for genes over-/underenriched by Fisher exact testing (P < .05). Bar plots magnifying data for points highlighted in red on scatter plots, with genes also meeting SMG criteria for (A-B) dx FL or (C-D) rel/ref FL or t-FL being colored blue/yellow.
Enrichment of mutations in FL/t-FL subsets. (A-D) Scatter plots displaying frequency of mutations per gene in FL/t-FL subsets. Points are highlighted in red for genes over-/underenriched by Fisher exact testing (P < .05). Bar plots magnifying data for points highlighted in red on scatter plots, with genes also meeting SMG criteria for (A-B) dx FL or (C-D) rel/ref FL or t-FL being colored blue/yellow.
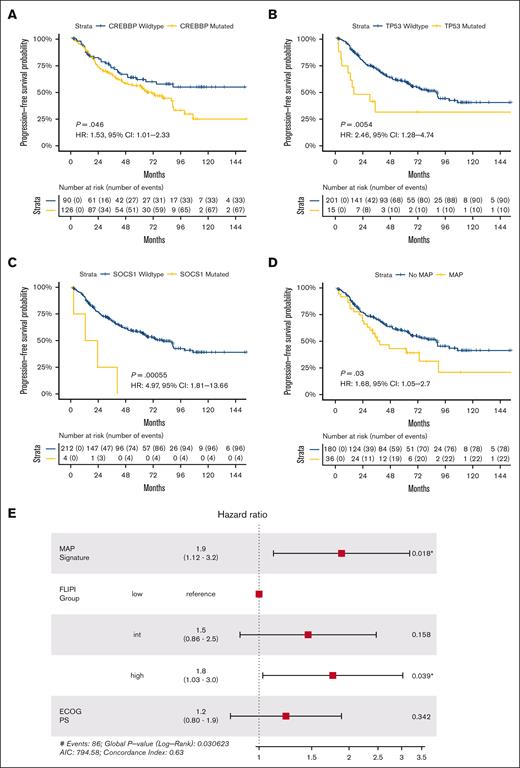
We examined the prognostic implications of these SMGs individually and in combination. Despite enrichment in FL over t-FL, CREBBP mutations were associated with shorter frontline PFS in patients with newly diagnosed FL (P < .05; Figure 7A), aligning with prior observations.41 We hypothesized that patients with newly diagnosed FL may suffer poor outcomes if their pretreatment tumors harbored mutations in genes more commonly mutated in rel/ref or transformed disease. Univariate analyses of the 6 SMGs enriched in rel/ref or transformed disease revealed TP53 and SOCS1 mutations to be associated with shorter frontline PFS in newly diagnosed FL (Figure 7B-C; supplemental Figure 4).
Mutations associated with progression confer adverse risk in newly diagnosed FL. Kaplan-Meier curves displaying univariate frontline PFS in patients with FL by (A) CREBBP, (B) TP53, (C) SOCS1, and (D) MAP signature status. (E) Forest plot of multivariate model for frontline PFS incorporating MAP signature status, traditional FLIPI group, and patient Eastern Cooperative Oncology Group performance status; significant P values are marked with asterisks.
Mutations associated with progression confer adverse risk in newly diagnosed FL. Kaplan-Meier curves displaying univariate frontline PFS in patients with FL by (A) CREBBP, (B) TP53, (C) SOCS1, and (D) MAP signature status. (E) Forest plot of multivariate model for frontline PFS incorporating MAP signature status, traditional FLIPI group, and patient Eastern Cooperative Oncology Group performance status; significant P values are marked with asterisks.
Taking an integrated approach, we defined a mutations associated with progression (MAP) signature as ≥2 mutations in these 7 genes (6 rel/ref/t-FL–enriched SMGs plus CREBBP). This was present in 16.7% (36/216) of newly diagnosed FL samples with frontline PFS data. By univariate analysis, patients with newly diagnosed FL possessing a MAP signature had significantly shorter frontline PFS; median PFS was 38.8 vs 88.9 months for patients with and without a MAP signature, respectively (HR, 1.68; 95% CI, 1.05-2.70; P = .03; Figure 7D). No difference in OS was observed by MAP signature status, but, similar to the m7-FLIPI, we observed a significant interaction between MAP signature status and receipt of cytotoxic chemotherapy (supplemental Figure 4). However, in contrast to the m7-FLIPI, patients at high-risk by the presence of a MAP signature had inferior 48-month PFS irrespective of frontline chemotherapy status.
To assess the generalizability of our MAP signature, we performed multivariate modeling including traditional FLIPI stratification and patient performance status, the 2 nongenomic components of the m7-FLIPI. Adjusting for these features, the adverse risk associated with our MAP signature remained significant (HR, 1.90; 95% CI, 1.12-3.20; P = .018; Figure 7E), revealing the potential for this approach to identify patients at high-risk irrespective of major baseline clinical variables. Nonetheless, high-risk traditional FLIPI stratification was also an independent adverse risk factor in the multivariate model, and there was not a statistically significant interaction between MAP signature status and low-/intermediate-risk vs high-risk FLIPI (supplemental Figure 4), highlighting the complementary roles of clinical and genomic prognostication.
We interrogated the robustness of our findings by performing internal crossvalidation with repeated trials of randomly sampling 50% of our cohort, followed by assessment of the impact of our MAP signature on 48-month PFS within each random cohort subset. Across 100 random subsets, the mean 48-month PFS difference by MAP signature status was −21.9% (95% CI, −23.9 to −19.8; supplemental Figure 4). Given the disproportionately high frequency of CREBBP mutations in newly diagnosed FL compared with the frequencies of mutations in the other 6 MAP genes, we also assessed the prognostic value of our MAP approach if CREBBP status was assessed independently. Although few newly diagnosed FL samples possessed ≥2 mutations in the 6 SMGs enriched in rel/ref FL or t-FL, these patients had notably poor outcomes independent of CREBBP status, with and without adjusting for traditional FLIPI and performance status (supplemental Figure 4). These observations highlight the adverse risk linked with mutations associated with progression when identified at FL diagnosis.
Discussion
We report a clinicogenomic analysis of FL/t-FL, revealing unique insights into pathogenesis and prognostication. Comparing newly diagnosed, rel/ref, and transformed FL, we disentangle the consequences of mutation burden from those of specific mutated genes. We found no strong links between mutational landscape and several important baseline characteristics including tumor grade and disease stage. Similarly, few alterations were associated with initial watch-and-wait or POD24, demonstrating that these trajectories are likely common consequences of varied mutational patterns. In contrast, we identified key pathways as implicated in rel/ref or transformed FL, and we assessed the impact of newly diagnosed FL possessing a mutational landscape similar to rel/ref or transformed disease. Although absolute mutational burden was not prognostic for FL frontline treatment, the specific mutated genes at diagnosis proved highly consequential. Based on our observations, a MAP signature was defined as ≥2 mutations in the 7-gene set of CREBBP, STAT6, TP53, IGLL5, B2M, SOCS1, and MYD88. Other than CREBBP, these mutations are less common at diagnosis individually, and demonstrate variable univariate prognostic impact. However, the presence of a MAP signature at FL diagnosis was a significant adverse risk factor independent of traditional FLIPI stratification and patient performance status.
Pathobiologically connecting MAP alterations with FL outcomes will be an important area of study, because it remains largely unknown whether cooccurring mutations are redundant, additive, or synergistic. Specifically, CREBBP and B2M mutations both promote immune evasion,53,59 whereas STAT6 is regulated by CREBBP60,61 and SOCS1.62CREBBP and STAT6 mutations frequently cooccur in t(14;18)− FL,63,64 and recent data suggest that specific CREBBP mutations create an evolutionary constraint on transformation.65 Considering our observed enrichment of STAT6 mutations exclusively in rel/ref FL but not transformed FL, this constraint may be reinforced by CREBBP and STAT6 comutation. Furthermore, TP53 mutations nearly universally confer adverse risk across oncology, including FL.66 We also observed hot spot and non–hot spot MYD88 mutations; interestingly, murine models of Bcl2 and Myd88 codysregulation develop DLBCL with plasmablast differentiation,67 similar to the unique activated B-cell cell-of-origin bias seen in MYD88-mutated t-FL.68 The role of IGLL5 in FL is the least well defined of our MAP genes. IGLL5 mutations were reported in chronic lymphocytic leukemia69 and DLBCL70-72 but were not assessed in other prominent FL cohorts,41,73 and there are limited prior studies of the surrogate light chain encoded by IGLL5.
Our study has limitations. Because of technical and practical considerations, we used multiple sequencing assays including custom panels with inherent biases due to target curation. Although mutations not covered by our assays may influence FL outcomes, our approach, to our knowledge, is among the broadest reported.22,35,41,73-75 We did not focus on copy number, structural, or noncoding alterations,65,76 which future work will explore. Furthermore, our patient cohort received nonrandomized treatments. Our observations may be influenced by complex interactions between FL genotype, patient characteristics, and therapy choice. However, our cohort diversity is also a strength, given the need for FL risk assessment to be broadly generalizable and not narrowly applicable to specific therapies. Future validation cohorts should account for evolution in FL treatments, including T-cell engagers and cellular immunotherapy.
Relatedly, we highlight nuances of the m7-FLIPI. Patients in the training cohort received rituximab (R) plus cyclophosphamide, doxorubicin, vincristine, and prednisone with interferon-α maintenance (GLSG2000 trial);77 and patients in the validation cohort received R with cyclophosphamide, vincristine, and prednisolone with R maintenance. Not only is R plus bendamustine increasingly favored today but because GLSG2000 included randomization to interferon-α maintenance vs high-dose chemotherapy/autologous stem cell transplantation, this biases the m7-FLIPI toward signals most applicable to young patients with high disease burden as enrolled to the trial. One previous study assessed the m7-FLIPI in patients receiving frontline R without chemotherapy,45 and the m7-FLIPI was not prognostic for time to treatment failure. Interestingly, the accuracies of our MAP signature and the m7-FLIPI to predict POD24 in our cohort were 50% and 51%, respectively, which contrasts with the 76% accuracy reported in the original m7-FLIPI study. Although the POD24-PI was later developed using the same data set for optimized POD24 sensitivity,78 a recent independent analysis of 252 patients with FL found that no established approach (eg, FLIPI, PRIMA-PI, m7-FLIPI, or POD24-PI) significantly predicted POD24.79 Notably, although, to our knowledge, we report one of the largest tests of the interaction between m7-FLIPI and treatment. Patients with low-risk m7-FLIPI status receiving chemotherapy experienced prolonged PFS. In contrast, patients with high-risk m7-FLIPI status receiving chemotherapy had suboptimal outcomes remarkably similar to patients receiving chemotherapy-free approaches irrespective of m7-FLIPI stratification. Whether patients with high-risk m7-FLIPI status receiving chemotherapy would have achieved similar outcomes with chemotherapy-free treatment is unknown. Nonetheless, low-risk m7-FLIPI (and/or the absence of a MAP signature) predicting long first remissions with standard R chemotherapy are important findings that may influence clinical trial design.
Overall, this study advances our understanding of FL/t-FL genomics, and it investigates the prognostic value of individual and combinations of mutations independent of clinical variables. The strongest adverse risk factor in the m7-FLIPI is a high traditional FLIPI score, which outweighs the model’s highest risk mutation combination. To evaluate FL risk without model tuning, we assessed the impact of mutations enriched in progressive or transformed FL when present at FL diagnosis. This MAP signature is an adverse risk factor independent of traditional FLIPI and performance status. Future studies are warranted to validate the poor prognosis associated with a MAP signature in newly diagnosed FL, and trials of novel agents may ideally be targeted to this high-risk subset of patients.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers U10CA180821, U10CA180882, and U24CA196171 (to the Alliance for Clinical Trials in Oncology); K22CA188163; P30CA91842; UG1CA189824; UG1CA233339; U01CA209936; U01CA231844; U01CA248235; and U24CA237719; the National Human Genome Research Institute of the National Institutes of Health under award numbers K99HG007940 and R00HG007940; the National Human Genome Research Institute of the National Institutes of Health under award number T32HL007088; the Lymphoma Research Foundation; the American Society of Clinical Oncology, Larry and Winnie Chiang Lymphoma Fellowship; and the Foundation for Barnes-Jewish Hospital (Steinback Fund). The research reported in this publication was also supported, in part, by funds from Celgene (CALGB 50803). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Samples and clinical data contributed by the Alliance for Clinical Trials in Oncology were collected via correlative study protocol A151303. Please refer to https://acknowledgments.alliancefound.org for support acknowledged by Alliance for Clinical Trials in Oncology and Alliance Foundation Trials programs.
Authorship
Contribution: D.A.R.-G., K.K., M.G., O.L.G., and T.A.F. designed the research; D.A.R.-G., K.K., C.R., M.M., M.P.W., F.G., Z.L.S., L.T., and E.D.H. performed the research and analyzed the data; D.A.R.-G., C.R., M.M., M.P.W., J.P.A., and S.L.O. collected and harmonized the clinical data; D.A.R.-G., K.K., F.G., and S.G. performed the statistical analyses; A.F.C., N.M.-S., B.S.K., N.L.B., I.S.L., E.D.H., P.M., J.P.L., and T.A.F. recruited cases, enrolled the patients, and managed their care during therapy; T.A.F. supervised the project; D.A.R.-G., K.K., J.P.L., M.G., O.L.G., and T.A.F. analyzed and interpreted the data; D.A.R.-G., K.K., and T.A.F. wrote the manuscript; and all authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: N.M.-S. has received institutional clinical trial funding from AstraZeneca, Bristol Myers Squibb, Celgene, C4 Therapeutics, Corvus Pharmaceuticals, Daiichi Sankyo, Genentech/Roche, Innate Pharmaceuticals, and Secura Bio/Verastem; and serves as a consultant for AstraZeneca, Genentech, Secura Bio/Verastem, Kyowa Hakko Kirin, and Janssen. B.S.K. receives research funding from AstraZeneca, Genentech/Roche, AbbVie, Celgene/Bristol Myers Squibb, BeiGene, and Hutchmed; and serves as a consultant/adviser for AstraZeneca, ADC Therapeutics, Genentech/Roche, AbbVie, MEI Pharma, AcertaPharma, Pharmacyclics, Celgene/Bristol Myers Squibb, Beigene, Kite/Gilead, Janssen, Incyte, Hutchmed, TG Therapeutics, Genmab, and Seattle Genetics. N.L.B. receives research funding from Autolus, Celgene/Bristol Myers Squibb, Forty-Seven, Janssen, Kite/Gilead, Merck, Millennium, and Pharmacyclics; and serves as a consultant/adviser for ADC Therapeutics, Genentech/Roche, and Seattle Genetics. I.S.L. receives honoraria from Adaptive. E.D.H. receives research funding from AbbVie, Eli Lilly, and Virtuoso; serves as a consultant/adviser for Astellas, Cytomx, Novartis, and Abcon; and holds stock options in Abcon. P.M. serves as a consultant/adviser for ADC Therapeutics. J.P.L. serves as a consultant/adviser for AbbVie, Astellas, AstraZeneca, Bayer, BeiGene, Celgene/Bristol Myers Squibb, Calithera, Constellation, Eisai, Epizyme, Genentech/Roche, Genmab, Kite/Gilead, Grail, Incyte, Janssen, Karyopharm, Lilly, MEI Pharma, Merck, Mustang Bio, Novartis, Pfizer, Seattle Genetics, Second Genome, Sutro, ADC Therapeutics, Regeneron, and Miltenyi. T.A.F. receives research funding from Affimed, HCW Biologics, and ImmunityBio; serves as a consultant/adviser for Wugen, Affimed, and Indapta; holds stock options in Wugen, Indapta, and Orca Bio; and is coinventor of patents licensed to Wugen by Washington University. The remaining authors declare no competing financial interests.
Correspondence: Todd A. Fehniger, Division of Oncology, Department of Medicine, Washington University School of Medicine, 660 S. Euclid Ave, Campus Box 8007, St. Louis, MO 63110; e-mail: tfehnige@wustl.edu.
References
Author notes
∗D.A.R.-G. and K.K. contributed equally to this study.
Selected data are accessible via dbGaP #phs001229 based on patients’ specific consent.
Data are available on request from the corresponding author, Todd A. Fehniger (tfehnige@wustl.edu).
The full-text version of this article contains a data supplement.