TO THE EDITOR:
Hematopoietic stem cell (HSC) gene therapy is a promising treatment option for a variety of genetic diseases affecting the hematopoietic system, for example, sickle cell disease, HIV/AIDS, and certain malignancies. However, current approaches target CD34+ hematopoietic stem and progenitor cells (HSPCs), a heterogeneous population that contains <1% true HSCs with long-term multilineage engraftment potential, which limits the efficiency and feasibility of such approaches. In order to increase the targeting efficiency of true HSCs, reduce the costs for expensive clinical grade reagents (eg, viral vectors and nucleases), and enhance the overall feasibility of HSC gene therapy, current efforts in the field focus on either purifying or directly targeting HSCs ex vivo1,2 as well as in vivo.3,4 Especially for strategies based on integrating viruses it remains unknown whether direct targeting of HSCs leads to changes in the integration site (IS) profile that may adversely affect the safety of these strategies in comparison with that of the currently used gold standard CD34-mediated approaches.
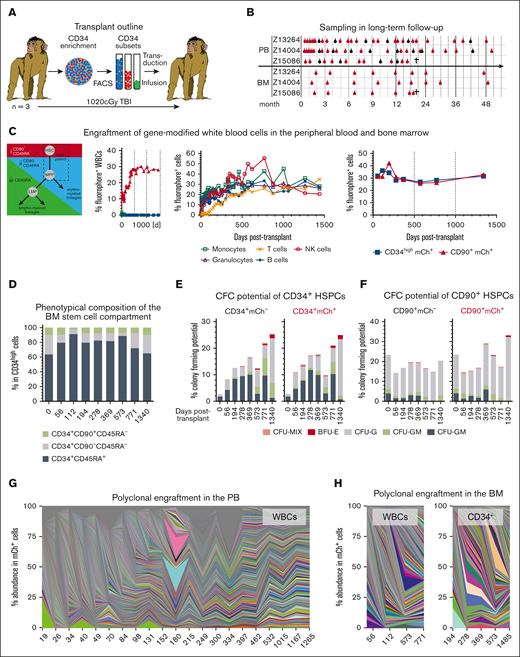
We have previously shown that the CD90+ subset of CD34+ HSPCs is highly enriched for HSCs and exclusively responsible for rapid recovery as well as robust long-term multilineage engraftment in a preclinical nonhuman primate stem cell transplantation and gene therapy model (Figure 1A).1,2 To confirm long-term stability of gene modification as well as safety of our HSC-targeted gene therapy approach, we closely followed up 3 animals that underwent transplantation with gene-modified CD34+CD45RA–CD90+ HSPCs for up to 5 years, analyzing their peripheral blood (PB) and bone marrow (BM) (Figure 1B). In the PB, CD34+CD45RA–CD90+ cells remained the exclusive source of all mature blood cells throughout the entire follow-up (Figure 1C; supplemental Figure 1A). Gene marking in peripheral white blood cells (WBCs) continuously increased within the first 9 to 12 months, reaching a plateau thereafter. Simultaneously, the WBC, lymphocyte, red blood cell, and platelet counts stabilized within the normal range and remained stable (supplemental Figure 1B). Nearly identical gene marking efficiency was found in T cells, B cells, natural killer cells, granulocytes, and monocytes starting at 9 months after transplantation, indicating successful gene modification and engraftment of long-term persisting multipotent HSCs without any evidence of lineage-bias or skewing at any time (Figure 1C; supplemental Figure 1C).
Stable and polyclonal gene modification in the PB and BM of Z13264. (A) CD34+ HSPCs from 3 pigtail macaques were enriched with immunomagnetic beads, CD34 subsets (CD90+CD45RA–, CD90-CD45RA–, and CD90–CD45RA+) FACS-purified, transduced, and cotransplanted after myeloablative conditioning with total body irradiation (TBI). (B) Animals were followed up for ∼50 months collecting PB and BM. Bulk WBCs were collected as indicated with red symbols, whereas blood lineages (T cells: CD3+; B cells: CD20+; natural killer cells: CD16+; monocytes: CD14+; and granulocytes: CD11b+CD14-) were FACS-purified, as indicated with black symbols. Z15086 was euthanized because of multiple re-occurring cytomegalovirus (CMV) infections. (C) Expression of fluorochromes in PB WBCs, PB lineages, and BM HSPCs longitudinally tracked via flow cytometry. (D) Longitudinal flow-cytometric quantification of HSPC subsets in the BM. (E-F) Colony-forming cell potential of unmodified (mCh– ) and gene-modified (mCh+) (E) CD34+ as well as (F) CD34+CD90+ HSPCs. Unmodified and gene-modified subsets were sort-purified into colony-forming cell assays, assays incubated for 10 to 14 days, and myeloid, erythroid, and as well as erythro-myeloid colonies quantified. (G-H) Polyclonal engraftment in the (G) PB and (H) BM. BFU-E, Burst forming unit–erythrocyte; CFU-G, granulocyte colony; CFU-GM, granulocyte-monocyte/macrophage colony; CFU-M, monocyte/macrophage colony; CFU-MIX, erythro-myeloid colony; GFP, Green fluorescent protein; mCer, monomeric Cerulean; mCh, monomeric Cherry.
Stable and polyclonal gene modification in the PB and BM of Z13264. (A) CD34+ HSPCs from 3 pigtail macaques were enriched with immunomagnetic beads, CD34 subsets (CD90+CD45RA–, CD90-CD45RA–, and CD90–CD45RA+) FACS-purified, transduced, and cotransplanted after myeloablative conditioning with total body irradiation (TBI). (B) Animals were followed up for ∼50 months collecting PB and BM. Bulk WBCs were collected as indicated with red symbols, whereas blood lineages (T cells: CD3+; B cells: CD20+; natural killer cells: CD16+; monocytes: CD14+; and granulocytes: CD11b+CD14-) were FACS-purified, as indicated with black symbols. Z15086 was euthanized because of multiple re-occurring cytomegalovirus (CMV) infections. (C) Expression of fluorochromes in PB WBCs, PB lineages, and BM HSPCs longitudinally tracked via flow cytometry. (D) Longitudinal flow-cytometric quantification of HSPC subsets in the BM. (E-F) Colony-forming cell potential of unmodified (mCh– ) and gene-modified (mCh+) (E) CD34+ as well as (F) CD34+CD90+ HSPCs. Unmodified and gene-modified subsets were sort-purified into colony-forming cell assays, assays incubated for 10 to 14 days, and myeloid, erythroid, and as well as erythro-myeloid colonies quantified. (G-H) Polyclonal engraftment in the (G) PB and (H) BM. BFU-E, Burst forming unit–erythrocyte; CFU-G, granulocyte colony; CFU-GM, granulocyte-monocyte/macrophage colony; CFU-M, monocyte/macrophage colony; CFU-MIX, erythro-myeloid colony; GFP, Green fluorescent protein; mCer, monomeric Cerulean; mCh, monomeric Cherry.
The BM stem cell compartment in all 3 animals fully replenished within 3 to 6 months, showing frequencies of phenotypic CD34 subsets similar to the pretransplantation baseline (Figure 1D; supplemental Figure 2A). Most importantly, gene marking of CD34+ HSPCs as well as CD34+CD45RA–CD90+ HSCs in the BM mirrored observed frequencies in the PB (Figure 1C; supplemental Figure 2B). Interestingly, high gene marking was observed significantly earlier in the BM stem cell compartment compared with that in PB WBCs, reaching stable levels of gene modification by week 6. Gene-modified CD34+ and CD34+CD45RA–CD90+ demonstrated almost identical erythro-myeloid colony-forming unit (CFU) potential in comparison with their unmarked counterparts, indicating no impact of fluorescence assisted cell sorting (FACS)-purification and ex vivo gene modification on their differentiation potential (Figure 1E; supplemental Figure 2C,D). CFU potential in bulk CD34+ cells was initially low and recovered to normal rates over time, whereas sustained colony formation was seen for CD34+CD45RA–CD90+ cells.
To evaluate the clonal diversity and confirm polyclonal engraftment, fluorochrome-expressing lineages from the PB, as well as CD34+ cells,. were FACS-purified DNA was extracted and integration site analysis (ISA) was performed (Figure 1B). ISA demonstrated highly polyclonal engraftment in PB WBCs (Figure 1G; supplemental Figure 3A), BM WBCs (Figure 1H; supplemental Figure 3B) as well as BM CD34+ cells (Figure 1I; supplemental Figure 3C). No evidence for the outgrowth of dominant clones was observed in any of the 3 animals or tissues longitudinally, confirming no increased risk of adverse viral integration in comparison with that of the current gold standard CD34-mediated strategies.
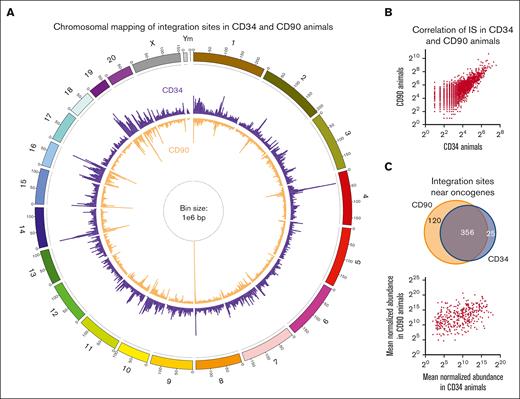
To further confirm the safety of our CD90-targeted gene therapy approach, we compared the ISA data of these 3 animals transplanted with CD90+ cells (CD90 animals) with that of 5 historical controls receiving CD34+ cells (CD34 animals) (supplemental Table 1). Chromosomal mapping of ISs demonstrated nearly identical patterns (R2=0.607) of transgene localization within genes in both groups (Figure 2A,B). Very high similarity of IS profiles between animals transplanted with genetically modified CD34- and those with CD90-enriched cells confirmed that historically described lentiviral IS patterns were not altered due to the direct gene-modification of CD34+CD45RA–CD90+ HSCs. Percentages of 4.8% and 4.2% in-gene ISs in 371 and 476 total genes were found to be associated with previously described oncogenes in CD34 and CD90 animals, respectively. The majority of ISs near oncogenes was shared, no correlation was observed in the abundance, and no clonal outgrowth was associated with these ISs in both groups (Figure 2C), showing no increased risk of insertional mutagenesis with our CD90-targeted gene therapy approach.
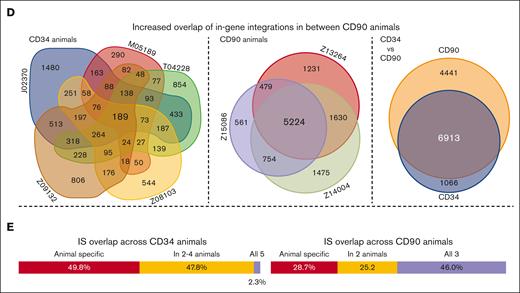
Comparison of IS profiles in CD34 and CD90 animals. (A) Chromosomal mapping of IS distribution in CD34 (purple outside ring) and CD90 (orange inside ring) animals. Bar height indicates the cumulative number of unique IS across all animals within a 1e6 base pair bin. (B) Correlation of the number of ISs within each bin across both groups. (C) comparison of shared and unique IS near oncogenes in between CD34 and CD90 animals (top). Correlation of the mean normalized abundance of shared IS near oncogenes between CD34 and CD90 animals (bottom). (D) Euler and Venn diagrams of in-gene IS overlap among animals within each group (left and middle) as well as across CD34 and CD90 animals (right). (E) Percentage of in-gene ISs that were animal specific (red), partly shared (orange), and shared in between all animals of 1 group (blue) for CD34 (left) and CD90 animals (right).
Comparison of IS profiles in CD34 and CD90 animals. (A) Chromosomal mapping of IS distribution in CD34 (purple outside ring) and CD90 (orange inside ring) animals. Bar height indicates the cumulative number of unique IS across all animals within a 1e6 base pair bin. (B) Correlation of the number of ISs within each bin across both groups. (C) comparison of shared and unique IS near oncogenes in between CD34 and CD90 animals (top). Correlation of the mean normalized abundance of shared IS near oncogenes between CD34 and CD90 animals (bottom). (D) Euler and Venn diagrams of in-gene IS overlap among animals within each group (left and middle) as well as across CD34 and CD90 animals (right). (E) Percentage of in-gene ISs that were animal specific (red), partly shared (orange), and shared in between all animals of 1 group (blue) for CD34 (left) and CD90 animals (right).
Lastly, we determined the impact of direct HSC modification and enhanced sampling on the reproducibility of IS patterns by comparing the data of individual animals within and across both groups (supplemental Figures 4 and 5). Correlations among CD34 animals were generally weak or very weak because of the vast majority of genes with IS not being shared. A weak to moderate correlation was reached when CD34 animals were compared with CD90 animals. A strong to very strong correlation was only observed for the comparison among CD90 animals, indicating that increased sampling leads to highly reproducible IS profiles (supplemental Figure 4). Overlap of in-gene IS across animals was further visualized via Euler/Venn diagrams (Figure 2D,E). Almost 50% of in-gene ISs were uniquely found in individual CD34 animals, with as little as 2.3% found in all 5, whereas 46% of all in-gene ISs were shared across CD90 animals, with 28.7% found only in individual animals. Despite lower sampling, the vast majority (86.6%) of ISs found in CD34 animals was detected in the CD90 cohort.
Here, we showed that direct lentiviral gene-modification of CD90+ HSCs resulted in stable long-term multilineage engraftment and high level of gene modification in the blood and BM without any increased risk of oncogenic events or the outgrowth of dominant clones. Furthermore, increased sampling, paired with direct modification of CD90+ cells enhanced the reproducibility of IS patterns in comparison with those of the current gold standard CD34-mediated protocols. Increased reproducibility and reliability of clonal tracking will be especially important, turning HSC gene therapy into a routine application, and will enhance the ability to monitor for rare adverse events in patients during recovery and in the long-term.
Our results are highly encouraging for all ongoing efforts in the field of ex vivo HSC gene therapy to enrich and directly target a more refined population in order to reduce the amount of expensive gene-modifying agents, increase the on-target efficiency, and enhance the overall feasibility of this promising approach. Beyond the application shown here and gene modification of HSCs ex vivo, our findings should be of high importance to HSC-targeted in vivo applications using either untargeted or targeted vectors. Our studies in the preclinical nonhuman primate model demonstrate that HSC-targeted gene therapy approaches are efficient and safe paving the way for the next generation of HSC gene therapy ex vivo as well as in vivo.
Acknowledgments: The authors thank Helen Crawford for her help in preparing this manuscript and the figures. This work was supported in part by grants from the National Institutes of Health (R01 AI135953-01 [NIAID] and R01 HL136135 [NHLBI]) (H.-P.K.). H.-P.K. also received support as a Markey molecular medicine investigator, as the inaugural recipient of the José Carreras/E. Donnall Thomas endowed Chair for Cancer research, and as the Stephanus family endowed chair for cell and gene therapy.
Contribution: S.R. and H.-P.K. designed the study; M.E. and D.P. performed ISA data analysis; S.R., D.P., and M.E. generated the figures; H.-P.K. funded the study; and all authors wrote, reviewed, and edited the final manuscript.
Conflict-of-interest disclosure: S.R. is consultant to Forty Seven Inc (Gilead Sciences) and Ensoma Inc. H.-P.K. reports consultancy to and ownership interests with Rocket Pharmaceuticals, Homology Medicines, Vor Biopharma, and Ensoma Inc, and has also been a consultant to CSL Behring and Magenta Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Stefan Radtke, Fred Hutchinson Cancer Center, 1100 Fairview Ave N, D1-100, Seattle, WA 98109; e-mail: sradtke@fredhutch.org; and Hans-Peter Kiem, Fred Hutchinson Cancer Center, 1100 Fairview Ave N, D1-100, Seattle, WA 98109; e-mail: hkiem@fredhutch.org.
References
Author notes
∗S.R. and D.P. contributed equally to this work.
Custom scripts as well as ISA data will be available upon publication or request from the corresponding authors, Stefan Radtke (sradtke@fredhutch.org) and Hans-Peter Kiem (hkiem@fredhutch.org).
The full-text version of this article contains a data supplement.