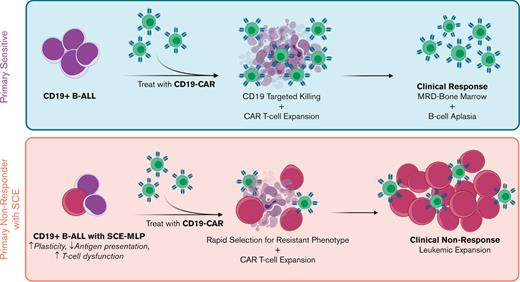
Key Points
Multiomic investigation of pretreatment bone marrow identifies a primary nonresponder signature to CD19-CAR in childhood B-ALL.
Primary nonresponders harbor myeloid and stem cell–like features while maintaining a pre–B-cell phenotype.
Abstract
CD19 chimeric antigen receptor T-cell therapy (CD19-CAR) has changed the treatment landscape and outcomes for patients with pre–B-cell acute lymphoblastic leukemia (B-ALL). Unfortunately, primary nonresponse (PNR), sustained CD19+ disease, and concurrent expansion of CD19-CAR occur in 20% of the patients and is associated with adverse outcomes. Although some failures may be attributable to CD19 loss, mechanisms of CD19-independent, leukemia-intrinsic resistance to CD19-CAR remain poorly understood. We hypothesize that PNR leukemias are distinct compared with primary sensitive (PS) leukemias and that these differences are present before treatment. We used a multiomic approach to investigate this in 14 patients (7 with PNR and 7 with PS) enrolled in the PLAT-02 trial at Seattle Children’s Hospital. Long-read PacBio sequencing helped identify 1 PNR in which 47% of CD19 transcripts had exon 2 skipping, but other samples lacked CD19 transcript abnormalities. Epigenetic profiling discovered DNA hypermethylation at genes targeted by polycomb repressive complex 2 (PRC2) in embryonic stem cells. Similarly, assays of transposase-accessible chromatin–sequencing revealed reduced accessibility at these PRC2 target genes, with a gain in accessibility of regions characteristic of hematopoietic stem cells and multilineage progenitors in PNR. Single-cell RNA sequencing and cytometry by time of flight analyses identified leukemic subpopulations expressing multilineage markers and decreased antigen presentation in PNR. We thus describe the association of a stem cell epigenome with primary resistance to CD19-CAR therapy. Future trials incorporating these biomarkers, with the addition of multispecific CAR T cells targeting against leukemic stem cell or myeloid antigens, and/or combined epigenetic therapy to disrupt this distinct stem cell epigenome may improve outcomes of patients with B-ALL.
Introduction
CD19 chimeric antigen receptor T cells (CD19-CAR) have improved treatment strategies and clinical outcomes for children with relapsed/refractory B-cell acute lymphoblastic leukemia (B-ALL), with ∼80% to 90% of patients initially responding to therapy.1-4 Unfortunately, primary nonresponse (PNR), defined as progressive CD19+ disease, occurs in 10% to 20% of treated patients despite the expansion of CD19-CAR. Significant efforts have been made to understand the mechanisms of relapse, which occur in ∼50% of the patients who initially respond.5,6 About 10% to 30% of the relapsed disease is CD19–, highlighting that target expression is a critical component of durable outcomes with CD19-CAR.2,7-11 Mechanisms of CD19 antigen loss in patients treated with CD19-CAR include preexisting or acquired mutations, aberrant splicing of the messenger RNA encoding the extracellular domain of CD19,12-16 downregulation of CD19 surface expression,8,17 and lineage switching to acute myeloid leukemia (AML).18-21 Alternatively, PNR, which is characterized by CD19+ progressive disease, has been associated with high T-cell exhaustion markers in the apheresis product used for cell manufacturing or a decreased rate of CAR T-cell expansion.22,23 Genome-wide loss-of-function screens in cell lines have also identified impaired death receptor signaling as another leukemia-intrinsic mechanism of PNR.24 However, this loss of expression of death receptor signaling pathways is not always correlated with PNR in patients, suggesting that these mechanisms of resistance are heterogenous and not completely understood.
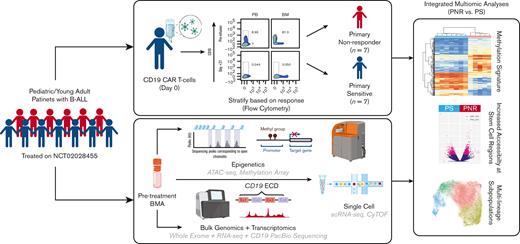
To address the gap in the understanding of PNR and identify pretreatment biomarkers, we performed a comprehensive molecular characterization of patient bone marrow aspirates (BMAs) before CD19-CAR treatment from a selected cohort of patients treated on the PLAT-02 trial (NCT02028455), excluding patients in whom the lack of response was due to the loss of CD19 expression or early loss of CD19-CAR engraftment.7,23 We used a multiomic approach consisting of a combination of genomic, transcriptomic, epigenetic, and single-cell methods to identify preexisting, leukemia-intrinsic mechanisms of CD19-CAR resistance (Figure 1). The identification of key markers of resistance in our study could positively influence clinical decision making by identifying patients likely to exhibit PNR to CD19-CAR and offer alternative, potentially curative, approaches.
Schema of multiomic platform to investigate mechanism of PNR vs PS to CD19-CAR in patients with pediatric B-ALL. Workflow using pretreatment, cryopreserved BMAs to investigate PNR. Clinical annotations were used in integrative multiomic analysis to detect novel mechanisms of PNR compared with that of PS.
Schema of multiomic platform to investigate mechanism of PNR vs PS to CD19-CAR in patients with pediatric B-ALL. Workflow using pretreatment, cryopreserved BMAs to investigate PNR. Clinical annotations were used in integrative multiomic analysis to detect novel mechanisms of PNR compared with that of PS.
Methods
Patient samples
Samples were cryopreserved prelymphodepletion BMAs from patients with CD19+ B-ALL enrolled in NCT02028455. Patients were stratified as primary sensitive (PS), defined as obtaining a durable minimal residual disease negative (MRD−) complete remission for more than 6 months, or PNR, defined as failing to achieve and maintain MRD– by 63 days despite the ongoing CD19-CAR engraftment and leukemic expression of CD19 via flow cytometry. Additional details on the clinical trial, patient inclusion, and sample processing are provided in the supplemental methods.
Bulk DNA and RNA sequencing and variant analysis
DNA and RNA were isolated and purified, and whole exome sequencing (WES) and RNA sequencing (RNA-seq) was performed using NextSeq 500 (Illumina, San Diego, CA); targeted, long-read sequencing of the CD19 locus was also performed (PacBio). Additional details are provided in the supplemental methods.
Illumina EPIC methylation array
DNA was extracted, and methylation data were generated per manufacturer protocols using the Infinium MethylationEPIC Bead Chip Kit (Illumina). Additional details, including bioinformatic analysis approaches, are described in the supplemental Methods.
ATAC-sequencing
Assays of transposase-accessible chromatin (ATAC) were performed as previously described.25 The resulting ATAC libraries were sequenced with NextSeq500 (Illumina) with paired-end reads. Additional details, including bioinformatic analysis approaches and identification of lineage-specific regions and motif enrichment analyses,26,27 are described in the supplemental methods.
Single-cell RNA-seq
Single-cell RNA-seq (scRNA-seq) experiments were done in collaboration with the Single-Cell Analysis Facility of the National Cancer Institute, Center for Cancer Research. Libraries were generated with 10x Genomics technology using the 5′ v1.1 single-cell gene expression assay and sequenced using NextSeq500 (Illumina). Additional details, including bioinformatic analysis approaches, are described in the supplemental methods.
Mass CyTOF
Results
Patient cohort demographics
We included 14 treated patients who were enrolled in NCT02028455. Available samples from PNR patients were selected and matched to an equal group of PS patients who had sufficient BMAs available. Seven were categorized as PNRs, and 7 as PS (Table 1; Figure 1). Patients were categorized as PNR if they failed to achieve and maintain MRD– complete responses (CRs), had concurrent CD19-CAR engraftment and expansion by day 63, and had retention of CD19 antigen on the leukemic cell surface. Our cohort were all CD19+, as detected via pretreatment flow cytometry. These leukemias were composed of a spectrum of cytogenetic and molecular subtypes, and these patients had varied numbers of relapses and prior hematopoietic cell transplants (HCTs) (Table 1). There were 3 patients who had previously received the CD19-directed targeted therapy, blinatumomab. Of these, 1 was a PNR, whereas the other 2 were PS. Among the PNRs, 2 patients had restaging marrows demonstrating MRD– CR, but within 3 weeks, they had reemergence of CD19+ ALL in the setting of ongoing engraftment of CD19-CARs, suggestive of CD19-independent resistance to CD19-CAR. The remaining PNR samples exhibited progressive, CD19+ disease. All PS leukemias obtained durable MRD– CRs. Although 2 patients, 1 with CD19– disease at 19 months and 1 with CD19+ disease at 11 months, ultimately became refactory, the other 5 patients continued in ongoing remission without subsequent therapies at least 5 years from treatment.
Cohort demographics, clinical characteristics, and response to CD19-CAR
| Patient . | Response categorization . | Relapse status at time of CD19-CAR . | Prior HCT . | Prior blinatumomab . | Pre-CAR leukemic burden (n %) . | Known cytogenetics . | Pre-CAR surface CD19 . | Post CD19-CAR surface CD19 . | Best response and duration . |
|---|---|---|---|---|---|---|---|---|---|
| S09 | PNR | Second relapse | Yes (1) | No | 92 | iAMP21 | Positive | Positive | PD |
| S27 | PNR | First relapse | No | No | 97 | iAMP21 | Positive | Positive | PD |
| S31 | PNR | second relapse | No | No | 80 | t(5;14) | Positive | Positive | Initial MRD– CR; CD19+ B-ALL present within 3 wk |
| S37 | PNR | Third relapse | Yes (1) | No | 59 | ETV6-RUNX1 | Positive | Positive | Initial MRD– CR; CD19+ B-ALL present within 3 wk |
| S46 | PNR | 2nd Relapse | Yes (1) | Yes | 94.5 | 46, XY, 22 PSTL+ | Positive | Positive | PD |
| S1023 | PNR | 3rd Relapse | No | No | 100 | 46, XY, t(9:12)(p24;p13)/46, idem, del(1)(q41)/47, idem, +8 IKZF deletion | Positive | Positive | PD |
| S1057 | PNR | 1st Relapse | No | No | 30 | 67-68 (3n), -X, i(X)(q10), +1, i(1)(q10), −3, −4, del(6)(q13q23), +6, −7, +8, −10, −12, −13, +14, −16, −17, −17, +20, +22, +22/46, XX(20) | Positive | Positive | PD |
| S01 | PS | 2nd Relapse | Yes (1) | No | 59 | Ph-like | Positive | MRD– | MRD– CR; ongoing >7 y |
| S07 | PS | 2nd Relapse | Yes (1) | Yes | 77 | KMT2A-rearrangement | Positive | MRD– | MRD– CR; ongoing >6 y |
| S19 | PS | 2nd Relapse | Yes (1) | No | 75.8 | 55, XX | Positive | MRD– | MRD– CR; CD19 negative recurrence at 19 mo |
| S36 | PS | 2nd Relapse | No | No | 85 | 46, XX with inv | Positive | MRD– | MRD– CR; ongoing >5 y |
| S43 | PS | 4th Relapse | Yes (2) | No | 23 | Ph+ | Positive | MRD– | MRD– CR; CD19+ recurrence at 11 mo |
| S53 | PS | 3rd Relapse | Yes (1) | Yes | 1.9 | Trisomy 1, 4, 9, 10, 12, 17, 21 | Positive | MRD– | MRD– CR; ongoing >5 y |
| S58 | PS | 1st Relapse | Yes (1) | No | 4.4 | Tetrasomy 21, Trisomy 8 | Positive | MRD– | MRD– CR; ongoing >5 y |
| Patient . | Response categorization . | Relapse status at time of CD19-CAR . | Prior HCT . | Prior blinatumomab . | Pre-CAR leukemic burden (n %) . | Known cytogenetics . | Pre-CAR surface CD19 . | Post CD19-CAR surface CD19 . | Best response and duration . |
|---|---|---|---|---|---|---|---|---|---|
| S09 | PNR | Second relapse | Yes (1) | No | 92 | iAMP21 | Positive | Positive | PD |
| S27 | PNR | First relapse | No | No | 97 | iAMP21 | Positive | Positive | PD |
| S31 | PNR | second relapse | No | No | 80 | t(5;14) | Positive | Positive | Initial MRD– CR; CD19+ B-ALL present within 3 wk |
| S37 | PNR | Third relapse | Yes (1) | No | 59 | ETV6-RUNX1 | Positive | Positive | Initial MRD– CR; CD19+ B-ALL present within 3 wk |
| S46 | PNR | 2nd Relapse | Yes (1) | Yes | 94.5 | 46, XY, 22 PSTL+ | Positive | Positive | PD |
| S1023 | PNR | 3rd Relapse | No | No | 100 | 46, XY, t(9:12)(p24;p13)/46, idem, del(1)(q41)/47, idem, +8 IKZF deletion | Positive | Positive | PD |
| S1057 | PNR | 1st Relapse | No | No | 30 | 67-68 (3n), -X, i(X)(q10), +1, i(1)(q10), −3, −4, del(6)(q13q23), +6, −7, +8, −10, −12, −13, +14, −16, −17, −17, +20, +22, +22/46, XX(20) | Positive | Positive | PD |
| S01 | PS | 2nd Relapse | Yes (1) | No | 59 | Ph-like | Positive | MRD– | MRD– CR; ongoing >7 y |
| S07 | PS | 2nd Relapse | Yes (1) | Yes | 77 | KMT2A-rearrangement | Positive | MRD– | MRD– CR; ongoing >6 y |
| S19 | PS | 2nd Relapse | Yes (1) | No | 75.8 | 55, XX | Positive | MRD– | MRD– CR; CD19 negative recurrence at 19 mo |
| S36 | PS | 2nd Relapse | No | No | 85 | 46, XX with inv | Positive | MRD– | MRD– CR; ongoing >5 y |
| S43 | PS | 4th Relapse | Yes (2) | No | 23 | Ph+ | Positive | MRD– | MRD– CR; CD19+ recurrence at 11 mo |
| S53 | PS | 3rd Relapse | Yes (1) | Yes | 1.9 | Trisomy 1, 4, 9, 10, 12, 17, 21 | Positive | MRD– | MRD– CR; ongoing >5 y |
| S58 | PS | 1st Relapse | Yes (1) | No | 4.4 | Tetrasomy 21, Trisomy 8 | Positive | MRD– | MRD– CR; ongoing >5 y |
AMP, amplification; inv, inversion; Ph, Philadelphia chromosome (BCR-ABL1); PD, progressive disease; PSTL, PSTL polymorphism; XX: duplication of X chromosome.
Available cryopreserved samples ranged in quantity from 3 × 105 to 50 × 106 cells per patient. A complete analysis required a minimum of 8 × 106 viable cells. For samples with fewer cells, we prioritized specific assays as per the data that were most likely to inform our analysis (supplemental Table 1). Samples with a leukemic burden of <5% in the marrow were excluded from bulk epigenetic and bulk differential expression assays to avoid confounding variables (Table 1; supplemental Table 1).
Undetected alterations in the CD19 extracellular domain exist in some, but not all, primary nonresponders before treatment
Associations between CD19-CAR resistance and CD19 dependent and independent genomic alterations have previously been reported.8,13,15,16 We hypothesized that definition of key resistance markers could be derived from genomic alterations in pretreatment leukemias. We therefore used WES and RNA-seq to interrogate for genomic alterations associated with PNR. Overall, we found no association between known leukemia specific alterations and responses (Figure 2A; supplemental Table 2-3). Consistent with previous findings, we identified genomic subtypes used for risk stratification or known to correlate with responses to conventional chemotherapy, but, interestingly, the risk associated with these subtypes was not associated with the therapeutic efficacy of CD19-CAR. For example, both patients with BCR-ABL1–positive or Philadelphia chromosome-like–positive ALL, leukemic subtypes known to be high-risk and correlate with poor response to therapy,30-32 were PS. Conversely, for patient S37's PNR leukemia harbored the ETV6-RUNX1 fusion, a marker for excellent prognosis.33 Remarkably, 1 patient (S07) with an MLL-rearranged (KMT2A-AFF1) leukemia was PS, despite this subtype’s known association with lineage switching to AML as an acquired resistance mechanism to CD19-CAR.19
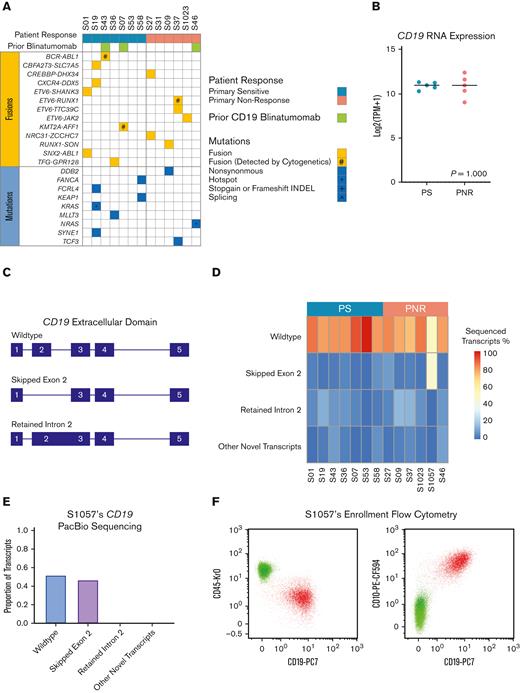
WES, Bulk RNA-seq, and targeted long-read PacBio sequencing of the CD19 of B-ALL from PNR and PS patients. (A) Pretreatment genomic landscape of pediatric patients with B-ALL within the study cohort stratified per the patient response. Prior CD19-targeting therapy with blinatumomab is designated in green. Fusions are in yellow, with alterations that were previously discovered by cytogenetics noted (#). Mutations are in blue with hotspot mutations noted (∗). No stop-gain, frameshift, or splicing variants were detected using WES. (B) Bulk RNA-seq showed no significant difference in CD19 expression between responders (PS) and nonresponders (PNRs), using Mann-Whitney two-tailed test (P = 1.000). Patient samples with <5% leukemic infiltration (S53 and S58) were excluded from the analysis. (C) Schematic depicting the extracellular domain of top transcripts of the CD19 locus identified by PacBio sequencing. (D) Heatmap showing the proportion as a percentage of detected CD19 transcripts via targeted PacBio long-read sequencing across the patient samples. Notably, patient S1057 had significant preexisting transcripts without CD19’s exon 2, the known binding domain of CD19-CAR. (E) Proportions of CD19 isoforms identified by PacBio sequencing in patient S1057. (F) Pre-CD19-CAR flow cytometry data for CD19 (clone J3-119) vs CD45 and CD10 from patient S1057 do not reveal a significant CD19– population.
WES, Bulk RNA-seq, and targeted long-read PacBio sequencing of the CD19 of B-ALL from PNR and PS patients. (A) Pretreatment genomic landscape of pediatric patients with B-ALL within the study cohort stratified per the patient response. Prior CD19-targeting therapy with blinatumomab is designated in green. Fusions are in yellow, with alterations that were previously discovered by cytogenetics noted (#). Mutations are in blue with hotspot mutations noted (∗). No stop-gain, frameshift, or splicing variants were detected using WES. (B) Bulk RNA-seq showed no significant difference in CD19 expression between responders (PS) and nonresponders (PNRs), using Mann-Whitney two-tailed test (P = 1.000). Patient samples with <5% leukemic infiltration (S53 and S58) were excluded from the analysis. (C) Schematic depicting the extracellular domain of top transcripts of the CD19 locus identified by PacBio sequencing. (D) Heatmap showing the proportion as a percentage of detected CD19 transcripts via targeted PacBio long-read sequencing across the patient samples. Notably, patient S1057 had significant preexisting transcripts without CD19’s exon 2, the known binding domain of CD19-CAR. (E) Proportions of CD19 isoforms identified by PacBio sequencing in patient S1057. (F) Pre-CD19-CAR flow cytometry data for CD19 (clone J3-119) vs CD45 and CD10 from patient S1057 do not reveal a significant CD19– population.
Per the eligibility requirements for the clinical trial, all leukemias were CD19+ by pretreatment flow cytometry.7 By RNA-seq, there were no statistical differences (P = 1.000) in CD19 gene expression between PNR and PS (Figure 2B). Because alternative splicing of the CD19 extracellular domain (ECD), particularly deletion of exon 2, is a well-established CD19-CAR resistance genotype, we used targeted PacBio long-read sequencing of complementary DNA generated from patient CD19 mRNA to determine if different pretreatment isoforms were associated with nonresponse. We identified multiple variant isoforms of CD19 in both PNR and PS, including the previously described deletion of exon 2 or retention of intron 2 (Figure 2C; supplemental Figure 1).8,13-17 Greater than 70% of all sequenced CD19 transcripts were the wild-type isoform in all leukemias except in 1 PNR, S1057. Notably, patient S1057 had a preexisting CD19 isoform with exon 2 alternatively spliced out in 47% of all detected transcripts (Figure 2D-E; supplemental Figure 2), and thus, the likely source of PNR in this case is antigen dependent.14 Clinical flow cytometry did not reveal this patient as having either insufficient expression of surface CD19 or subpopulations lacking CD19 surface expression (Figure 2F). Interestingly, this patient’s leukemia remained CD19+ after treatment with CD19-CAR. It is possible that this splicing event affects the efficacy of CD19-CAR specific killing without interfering with binding of the specific antibody used for flow cytometry. With our results indicating a paucity of recurrent driving genetic alterations associated with PNR, we hypothesized that epigenetic regulatory mechanisms could be a source of resistance to CD19-CAR.
Identification of a predictive methylation signature of PNR that is suggestive of a stem cell phenotype
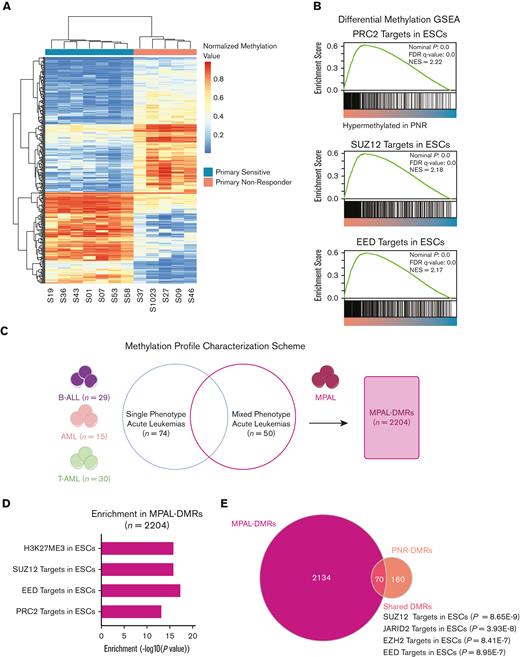
Because one established form of acquired resistance to CD19-CAR is lineage reprogramming from B-ALL to AML, we hypothesized that changes in the leukemic epigenome could confer plasticity and lead to alternative causes of PNR.19,21 Using array-based DNA methylation, we identified 238 differentially hyper- or hypomethylated regions (DMR) characteristic of pretreatment PNRs compared with that of PSs (P < .05; PNR-DMRs), (Figure 3A; supplemental Table 4). This analysis was performed on 11 samples (6 PNR and 5 PS), excluding S1057, S53, and S58 because of low sample availability or low tumor burden. Using gene set enrichment analysis (GSEA) and hypergeometric pathway enrichment of the hypermethylated PNR-DMRs, we identified an enrichment of promoter hypermethylation at genes known to be targets of PRC2 repression in embryonic stem cells (ESCs) (BENPORATH_PRC2_TARGETS; P = 8.15E-25 and BENPORATH_ES_WITH_H3K27ME3; P = 5.67E-29) (Figure 3B; supplemental Table 4).34 Interestingly, hypermethylation at these genes has previously been associated with tumor-intrinsic resistance of chronic lymphocytic leukemia to HCT, and therapy resistant cancer stem cells in other tumor types have been shown to share this repression pattern.34,35 Therefore, our results suggest that hypermethylation of these regions in PNR could predispose them to a less differentiated phenotype with inherent plasticity, rendering leukemia cells resistant to CD19-CAR–mediated killing.
Differential DNA methylation of PNR vs PS and MPAL. (A) Heatmap of methylation showing the top 500 differentially methylated loci in PNR vs PS. Scale depicts normalized methylation values at the probe loci. (B) GSEA of PNR vs PS leukemias shows increased methylation at targets of PRC2 in ESCs. (C) Differential methylation analysis strategy of a publicly available data set of MPAL vs single-phenotype acute leukemias36 (D) Pathways enriched in MPAL-DMRs (n = 2204) detected by a hypergeometric Fisher exact test. (E) Venn diagram showing that the 70 overlapping regions between MPAL-DMRs and PNR-DMRs are enriched for SUZ12, JARID2, EZH2, and EED targets based on ENRICHR analysis.
Differential DNA methylation of PNR vs PS and MPAL. (A) Heatmap of methylation showing the top 500 differentially methylated loci in PNR vs PS. Scale depicts normalized methylation values at the probe loci. (B) GSEA of PNR vs PS leukemias shows increased methylation at targets of PRC2 in ESCs. (C) Differential methylation analysis strategy of a publicly available data set of MPAL vs single-phenotype acute leukemias36 (D) Pathways enriched in MPAL-DMRs (n = 2204) detected by a hypergeometric Fisher exact test. (E) Venn diagram showing that the 70 overlapping regions between MPAL-DMRs and PNR-DMRs are enriched for SUZ12, JARID2, EZH2, and EED targets based on ENRICHR analysis.
DNA methylation has previously been shown to be a cause of the inherent plasticity seen in mixed phenotype acute leukemias (MPALs), independently of subpopulation lineage-specific genomic alterations.36 We hypothesized that our leukemias could harbor a similar DNA methylation pattern. To explore this, we analyzed a DNA methylation data set from a previously published, independent cohort of acute leukemias (n = 159) including MPALs.36 We compared the methylation profiles of the MPALs (n = 50) with the other single lineage acute leukemias, including AML (n = 15), B-ALL (n = 29), and T-ALL (n = 30) (Figure 3C). We identified 2204 uniquely differentially methylated regions characteristic of MPALs associated with lineage-independent, leukemic plasticity (MPAL-DMRs) and found that, of these, the hypermethylated genes are also enriched for ESC repressive signatures (BENPORATH_ES_WITH_H3K27ME3; P = 1.13E-16) and ESC PRC2 targets (BENPORATH_PRC2_TARGETS; P = 5.16E-14) (Figure 3D; supplemental Table 6-7). We then compared the MPAL-DMRs with our cohort’s PNR-DMRs and revealed an overlap of 70 hypermethylated genomic regions (Figure 3E; supplemental Table 8). Using ENRICHR, we found that genes associated with these specific regions are bound by multiple PRC2 components and cofactors in human ESCs (Figure 3E; supplemental Table 9).37,38
Thus, our results indicate that the relatively hypermethylated regions characteristic of PNR are also repressed in ESCs and cancer stem cells (CSCs) and associated with inherent leukemic plasticity, which may be important in resistance to CD19-CAR. Furthermore, these regions of hypermethylation are characteristic of PNR compared with those of PS and have the potential to be used as a pretreatment biomarker to identify these cases of nonresponse.
Maintenance of a B-ALL phenotype with acquisition of hematopoietic progenitor chromatin landscapes in PNR
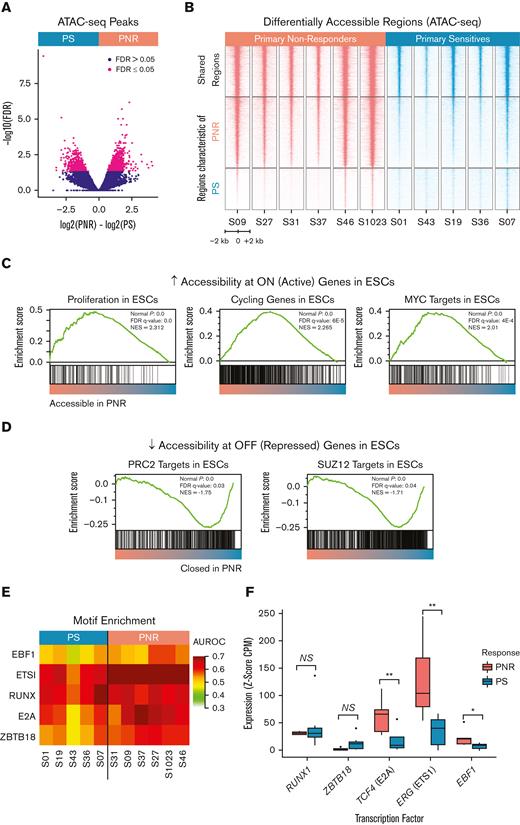
In order to further characterize the phenotype associated with our described methylation signature, we performed ATAC-seq. Comparison of PNR (n = 6) and PS (n = 5) leukemias revealed 2128 differentially accessible regions (FDR ≤ 0.05) and an overall increase in the accessible chromatin domains in PNR leukemias compared with that in PS (Figure 4A-B). GSEA and hypergeometric pathway enrichment analysis revealed stem cell and proliferation modules in the PNR defining accessible regions (Figure 4C).34 Increased expression of these genes has previously been associated with repression of the PRC2 target genes we found to be hypermethylated in PNR, further supporting a stem cell phenotype (Figure 4C; supplemental Table 10).34 Consistent with our discovered methylation signature, we observed a decreased accessibility at the same ESC PRC2 target genes found to be hypermethylated in our PNR leukemias (Figure 4D; supplemental Table 10).
Differential chromatin accessibility, motif, and gene expression analysis of PNR and PS leukemia. (A) Volcano plot comparing chromatin accessibility between PNR and PS leukemias. PNR and PS defining accessible regions (n = 2128) were determined by an FDR ≤ 0.5. (B) Heatmap depicting ATAC-seq peaks by sample at accessible regions shared and regions defining leukemias from PNR and PS. (C) GSEA shows increased accessibility at regions known to be active in embryonic and cancer stem cells in PNR. (D) GSEA shows decreased accessibility at regions known to be repressed in embryonic and cancer stem cells in PNR. (E) Heatmap showing the area under the receiver operating characteristic (AUROC) for key motif enrichment in regions whose accessibility was specific to leukemia cells, as determined by MEDEA.26 (F) Box plots showing increased RNA expression of the indicated TFs, measured for each leukemia sample as the Z-score of CPM (counts per million reads) values against 53 normal human tissues profiled by the GTeX consortium. Each of the presented TFs is both upregulated and known to bind to one of the motifs enriched in PNR leukemias. NS, not significant; ∗∗P ≤ .01; ∗P ≤ .05.
Differential chromatin accessibility, motif, and gene expression analysis of PNR and PS leukemia. (A) Volcano plot comparing chromatin accessibility between PNR and PS leukemias. PNR and PS defining accessible regions (n = 2128) were determined by an FDR ≤ 0.5. (B) Heatmap depicting ATAC-seq peaks by sample at accessible regions shared and regions defining leukemias from PNR and PS. (C) GSEA shows increased accessibility at regions known to be active in embryonic and cancer stem cells in PNR. (D) GSEA shows decreased accessibility at regions known to be repressed in embryonic and cancer stem cells in PNR. (E) Heatmap showing the area under the receiver operating characteristic (AUROC) for key motif enrichment in regions whose accessibility was specific to leukemia cells, as determined by MEDEA.26 (F) Box plots showing increased RNA expression of the indicated TFs, measured for each leukemia sample as the Z-score of CPM (counts per million reads) values against 53 normal human tissues profiled by the GTeX consortium. Each of the presented TFs is both upregulated and known to bind to one of the motifs enriched in PNR leukemias. NS, not significant; ∗∗P ≤ .01; ∗P ≤ .05.
Transcription factors (TFs) are known to determine cell identity through modulating chromatin accessibility patterns and driving transcription.39 In order to investigate which TFs were associated with the differential accessibility seen in our patient samples, we employed motif enrichment in differential elements of accessibility (MEDEA). Using a collection of 208 motifs representative of the known repertoire of human TF binding specificities, we found enrichment for 41 motifs as defined by AUROC ≥ 0.55 in at least 1 leukemia sample (Figure 4E; supplemental Figure 3).26 We then evaluated the RNA expression levels of the TFs known to recognize these enriched motifs and quantified their upregulation as the Z-score of counts per million reads relative to the 53 healthy human tissues profiled by the GTeX consortium. The RUNX motif was similarly enriched in PNR and PS, and both exhibited similar expression of RUNX1, a known master TF in B-ALL (Figure 4E-F). We additionally observed a pronounced enrichment for E2A, EBF1, ETS1, and ZBTB18 motifs, across all samples but higher in PNR than in PS (Figure 4E). We saw increased RNA expression of TCF4 (P = .005), ERG (P = .005), and EBF1 (P = .024), which are members of the E2A, ETS, and EBF families, respectively, in PNR relative to PS (Figure 4F). We validated this increased expression of TCF4 (P = .008) and ERG (P = .048) and the similar expression of RUNX1 (P = .867) using quantitative reverse transcription polymerase chain reaction (supplemental Figure 4). Interestingly, increased expression of ERG, an important lineage specifier for hematopoietic development, has been demonstrated to be leukemogenic and associated with poor clinical outcomes in both AML and ALL.40-43 Notably, patient S37 with PNR leukemia, harbors a ETV6-RUNX1 translocation, which could bind to the abundant RUNX and ETS motifs identified. Together, these results suggest that these TFs are associated with and could influence the chromatin landscapes observed in PNR.
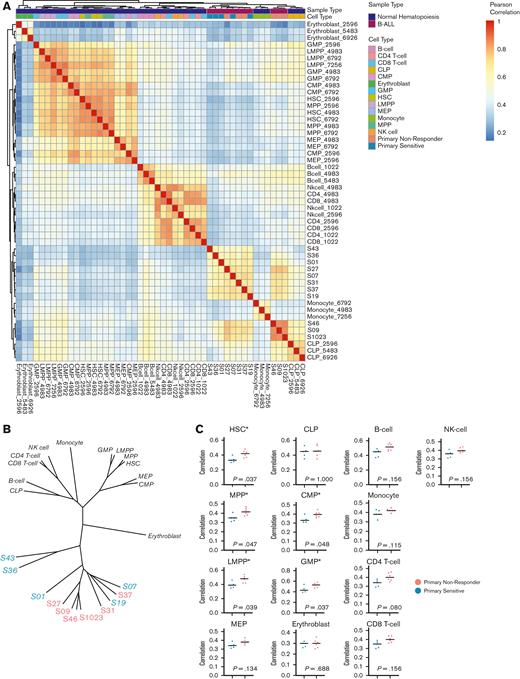
To examine if the differentially accessible regions, TF motif enrichments, and increased levels of specific TF expression were reflective of a loss of pre-B-cell identity, we compared our ATAC-seq data with those of a previously published data set of 13 healthy hematopoietic cell types from healthy human donors (n = 39).44 Overall, chromatin accessibility profiles from PNRs and PSs were clustered together relative to healthy hematopoietic cells (Figure 5A). Interestingly, PNR defining peaks were characteristic of multiple hematopoietic cell types, including both myeloid, lymphoid, and progenitor phenotypes. We further explored this relationship between the leukemias in our cohort and healthy hematopoietic cells by creating phylogenic trees based on distance as measured by Pearson correlation. We found that all the leukemia samples were closely clustered, suggesting a shared B-ALL accessible chromatin signature (Figure 5B). Interestingly, we observed no significant differences in regions defining CLPs or B-cells, confirming that PNR leukemias maintain their B-cell identity. However, the peaks in PNR leukemias were more highly correlated with more primitive cell types, including hematopoietic stem cells (P = .037), multipotent progenitors (P = .047), lymphoid-primed multipotent progenitors (P = .039), granulocyte-macrophage progenitors (P = .037), and common myeloid progenitors (P = .048) relative to those in PS leukemias (Figure 5C). By gaining accessibility at regions associated with hematopoietic progenitors, including those of both myeloid and lymphoid lineages, our data suggest that PNRs are less differentiated and may contain an increased plasticity when compared with PS. Furthermore, rather than losing a B-ALL phenotype, PNR leukemias gain characteristics of progenitor and myeloid lineages.
Comparisons of chromatin accessibility patterns from a data set of normal hematopoiesis with patients in this study. (A) Heatmap showing Pearson correlation values between pretreatment leukemias and 13 subtypes of cells from different stages of normal hematopoietic development. (B) Phylogenetic tree based on Pearson correlation between leukemias and subtypes of normal hematopoiesis from 3 separate healthy human donors. (C) Comparisons between PNR and PS stratified by similarity with healthy cells of hematopoietic development by Pearson correlation show that PNR (coral) are more similar to hematopoietic progenitors than PS (blue). There is no difference between the groups’ similarities to B cells or CLPs. Statistical significance was determined by Kolmogorov-Smirnov tests, and significance (P ≤ .05) is noted (∗). CMP, common myeloid progenitor; CLPs, common lymphoid progenitors; GMP, granulocyte-macrophage progenitor; HSC, hematopoietic stem cell; LMPP, lymphoid-primed multipotent progenitor; MPP, multipotent progenitor; MEP, megakaryocyte-erythroid progenitor; NK-cell, natural killer cell.
Comparisons of chromatin accessibility patterns from a data set of normal hematopoiesis with patients in this study. (A) Heatmap showing Pearson correlation values between pretreatment leukemias and 13 subtypes of cells from different stages of normal hematopoietic development. (B) Phylogenetic tree based on Pearson correlation between leukemias and subtypes of normal hematopoiesis from 3 separate healthy human donors. (C) Comparisons between PNR and PS stratified by similarity with healthy cells of hematopoietic development by Pearson correlation show that PNR (coral) are more similar to hematopoietic progenitors than PS (blue). There is no difference between the groups’ similarities to B cells or CLPs. Statistical significance was determined by Kolmogorov-Smirnov tests, and significance (P ≤ .05) is noted (∗). CMP, common myeloid progenitor; CLPs, common lymphoid progenitors; GMP, granulocyte-macrophage progenitor; HSC, hematopoietic stem cell; LMPP, lymphoid-primed multipotent progenitor; MPP, multipotent progenitor; MEP, megakaryocyte-erythroid progenitor; NK-cell, natural killer cell.
PNR samples have leukemic subpopulations coexpressing lymphoid, myeloid, and hematopoietic progenitor markers
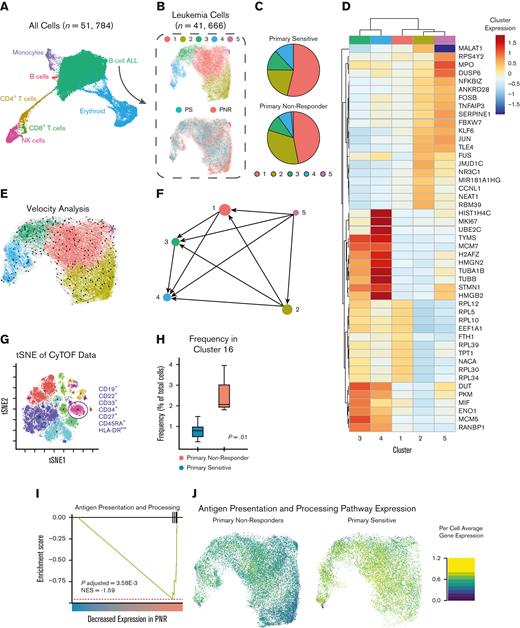
It is possible that the stem cell signatures we detected in our epigenetic analyses are due to subpopulations of leukemic stem cells. To further investigate our hypothesis of primitive and plastic subpopulations leading to PNR to CD19-CAR, we used single-cell transcriptomics and proteomics to define leukemic heterogeneity within PNR and PS leukemias not previously detected using flow cytometry. Using scRNA-seq, we captured a total of 51 784 cells, representing B-ALL as well as healthy cells along the hematopoietic spectrum (Figure 6A). We were able to identify the B-ALL cells and healthy cells of hematopoiesis using Uniform Manifold Approximation and Projections (UMAP) and cell type annotation with gene sets derived from previously published RNA-seq data of different stages along the hematopoietic spectrum.44 We then bioinformatically isolated the B-ALL cells (n = 41 666), clustered them, and focused our analysis only on leukemia-intrinsic differences (Figure 6A-B; supplemental Table 11).
scRNA-seq and CyTOF analyses of PNR and PS. (A) UMAP projection showing cells captured by scRNA-seq from pretreatment patient BMA. Cell type is classified and annotated using SingleR and referenced transcriptomic data sets.44 (B) UMAP of bioinformatically isolated B-ALL cells colored per the cluster (1-5; upper) and clinical response to CD19-CAR (PNR and PS; lower). (C) Pie chart depicting relative frequency of PNR or PS leukemic cells per cluster. (D) Heatmap of most significantly differentially expressed genes based on the cluster. Scale depicts normalized expression of each gene per cluster. (E) RNA velocity analysis of scRNA-seq data overlaid on UMAP colored by cluster. Arrows represent high-dimensional vectors predicting the future cell state of cells within the UMAP. (F) Partition-based graph abstraction (PAGA) summarizes the predicted cell trajectories between the clusters. Notably, Cluster 5 is the most primitive cluster, and cells in Cluster 2 are projected to progress to Clusters 1, 3, and 4. (G) t-distributed stochastic neighbor embedding (tSNE) plot of CyTOF analyses of PNR and PS leukemias. Cluster 16 was the only cluster occurring at a significantly different frequency. It is characterized as being CD19+, CD22+, CD10+, CD34+, CD33+, CD27+, and HLA-DRlow. (H) Plot of frequency (%) of cells being clustered into 16 per sample stratified by PNR and PS. Cluster 16 is differentially expressed in PNR leukemias (P = .01; two sample t test). (I) GSEA of cell count normalized scRNA-seq data revealed a significantly decreased expression of antigen presentation and processing pathway genes (NES, −1.59; P = .004). (J) UMAP showing per cell average expression of genes in the antigen presentation and processing pathway demonstrates that the observed decrease in the antigen presentation and processing pathway is independent of cell cluster based on single-cell transcriptomics.
scRNA-seq and CyTOF analyses of PNR and PS. (A) UMAP projection showing cells captured by scRNA-seq from pretreatment patient BMA. Cell type is classified and annotated using SingleR and referenced transcriptomic data sets.44 (B) UMAP of bioinformatically isolated B-ALL cells colored per the cluster (1-5; upper) and clinical response to CD19-CAR (PNR and PS; lower). (C) Pie chart depicting relative frequency of PNR or PS leukemic cells per cluster. (D) Heatmap of most significantly differentially expressed genes based on the cluster. Scale depicts normalized expression of each gene per cluster. (E) RNA velocity analysis of scRNA-seq data overlaid on UMAP colored by cluster. Arrows represent high-dimensional vectors predicting the future cell state of cells within the UMAP. (F) Partition-based graph abstraction (PAGA) summarizes the predicted cell trajectories between the clusters. Notably, Cluster 5 is the most primitive cluster, and cells in Cluster 2 are projected to progress to Clusters 1, 3, and 4. (G) t-distributed stochastic neighbor embedding (tSNE) plot of CyTOF analyses of PNR and PS leukemias. Cluster 16 was the only cluster occurring at a significantly different frequency. It is characterized as being CD19+, CD22+, CD10+, CD34+, CD33+, CD27+, and HLA-DRlow. (H) Plot of frequency (%) of cells being clustered into 16 per sample stratified by PNR and PS. Cluster 16 is differentially expressed in PNR leukemias (P = .01; two sample t test). (I) GSEA of cell count normalized scRNA-seq data revealed a significantly decreased expression of antigen presentation and processing pathway genes (NES, −1.59; P = .004). (J) UMAP showing per cell average expression of genes in the antigen presentation and processing pathway demonstrates that the observed decrease in the antigen presentation and processing pathway is independent of cell cluster based on single-cell transcriptomics.
Leukemic cells comprised 5 transcriptionally distinct clusters. Cluster frequency differed depending on patient response and no cluster was made up of only 1 sample (supplemental Table 12). Pretreatment PNR leukemias showed an increased number of cells in clusters 2 and 5 (Figure 6B-C; supplemental Figure 5A-C; supplemental Table 12-13). Cluster 2 is defined by relatively increased expression of genes known to interact with PRC2, cause cell proliferation, and prevent apoptosis (MALAT1); and those critical for maintaining leukemic stem cell phenotypes (JMJD1C) (supplemental Table 14).45-48 Notably, Cluster 5 had an increased expression of MPO (P = 2.91E-192), a myeloid lineage marker, with high expression in 91.4% of all cluster 5 cells (Figure 6D; supplemental Figure 5C; supplemental Table 14). In order to better understand the differentiation state of our identified clusters and elucidate if any of them are precursors to others, we employed RNA velocity analysis. Notably, Cluster 5 is predicted to be more primitive than all other clusters, and Cluster 2 is predicted to be a precursor to Clusters 1, 3, and 4 (Figure 6E-F). Taken together, this supports the existence of more primitive leukemic subpopulations that maintain a higher degree of multilineage potential in PNR.
To further evaluate this finding, we used our single-cell proteomic data, which was performed on the 6 samples available (2 PNR and 4 PS) (supplemental Table 1). CyTOF analyses based on our hematopoietic focused panel (supplemental Table 15) detected 17 distinct cell populations, including both healthy immune cells and multiple leukemic populations (Figure 6G-H; supplemental Figure 6A-B; supplemental Table 16). Only one cluster of leukemic cells (cluster 16) was found to significantly differ between PNR and PS leukemias (P = .01). This population expressed surface markers of myeloid (CD33) and lymphoid (CD19, CD22) lineage, as well as CD34, which is expressed on hematopoietic progenitors (Figure 6H; supplemental Figure 6A-B). These results further support the existence of leukemic populations with both stem cell and myeloid features that could confer resistance to CD19-CAR.
Decreased antigen presentation and a differential T-cell phenotypes exist in PNR prior to CD19-CAR
To determine if there were differences in gene expression between PNR and PS leukemias independent of cell clustering, we compared gene expression of PNR and PS B-ALL cells from our scRNA-seq data set. To avoid sample bias, up to 2000 randomly selected cells were used per sample. Interestingly, we found a decrease in apoptotic genes (BAX) and increased expression of genes known to inhibit apoptosis (MTRNR2L8) and maintain leukemic stem cell populations (CXCR4 and ID3) in PNR samples (supplemental Table 17).49-52 Concurrently, we found decreased expression in lineage-specific genes associated with B-cell development (JCHAIN and CD79B) and major histocompatibility complex class II genes, suggesting a less differentiated phenotype for PNR compared with PS samples (supplemental Table 17).53,54 Using GSEA of the same subset of leukemic cells, we then compared PNR with PS and found downregulation of discrete immune relevant and B-cell differentiation gene sets in PNR (supplemental Table 18). Notably, there was a significant decrease in antigen presentation and processing pathways in PNR (NES, −1.59; P = .004) (supplemental Table 18; Figure 6I). We then plotted the average expressions of the genes in this pathway in each cell, compared PNR and PS, and showed that cells from patients with PNR leukemia displayed a marked downregulation of antigen presentation and processing independent of cell cluster (Figure 6J).
In order to explore whether there were any preexisting phenotypic differences in the patients’ T cells, we then performed differential expression analysis of bioinformatically isolated T cells from our scRNA-seq data (supplemental Table 19). Comparing T cells from PNR with those from PS samples revealed further differences in pretreatment phenotypes (supplemental Figure 7, and Table 20). We found a significant downregulation of genes involved in T-cell cytotoxicity and peripheral homeostasis in PNR. These included the granzymes, GZMA (P = 1.67E-11) and GZMB (P = 3.73E-6), perforins (PRF1; P = 1.49E-12), and other key components of functional T-cell responses, such as EIF5A (P = 1.02E-75), PRDX1 (P = 5.75E-48), and RPS26 (P = 0), a gene previously shown to be important for T-cell homeostasis (supplemental Figure 7, and supplemental Table 20).55 This implies that not only is PNR leukemia less immunogenic, but it coexists with a differential phenotype of T cells prior to treatment with CD19-CAR.
Discussion
Factors contributing to leukemia-intrinsic PNR to CD19-CAR in pediatric ALL remain poorly understood, and current predictive markers are insufficient to reliably identify patient responses. To discover novel markers of PNR to CD19-CAR, specific to cases in which CD19 expression is maintained with ongoing CD19-CAR engraftment, we employed an integrated, multiomic approach, with focus on future clinical feasibility, to analyze pretreatment BMAs from patients known to respond or not respond to CD19-CAR.
Using CD19 targeted PacBio long-read sequencing, we detected exon 2 skipping, which has previously been associated with relapse after the initial response to CD19-CAR, in 1 case of PNR. This was not detected in the pretreatment clinical workup, indicating that the current evaluation approach using flow cytometry of the CAR directed antigen may not be able to predict both antigen dependent and independent PNR. Although this sequencing method is not directly quantitative, it suggests that using sequencing methods or careful matching of the flow cytometry antibody to the CAR binder could improve evaluation of antigen quality and quantity, and thus prediction of response.
Here, we uncovered a stem cell–like epigenome (SCE) as a novel candidate biomarker of PNR to CD19-CAR. Screening for a SCE phenotype with epigenetic analyses could be translated into clinical practice to prevent futile therapy in this high-risk patient population. We discovered that PNR leukemias harbor a distinct methylation profile, characterized by DNA hypermethylation at genes known to be targeted by PRC2 repression in ESCs and an associated SCE.34 Notably, we saw significant overlap between this profile and regions of DNA hypermethylation known to be associated with the plasticity seen in MPAL.36 PNR defining chromatin accessibility implicated a stem cell phenotype with characteristics of self-renewal and multilineage potential. Interestingly, we saw this epigenetic profile independently of leukemic subtype, and the 1 patient in our cohort who exhibited a KMT2A-AFF1 alteration, which has been previously associated with a lineage switch to AML under CD19-CAR immune pressure, was PS.19,21 Importantly, this patient did not harbor the SCE phenotype, including methylation signature or gained regions of chromatin accessibility characteristic of multipotent progenitors. This supports that SCE is a unique phenotype of CD19-CAR resistance rather than a precursor to lineage switching.
We further explored the leukemic phenotype associated with the epigenetic profile by bioinformatically isolating B-ALL cells from our scRNA-seq data and identified an increased expression of stem cell and myeloid lineage markers, including MPO, in clusters that were more frequent in PNR. These clusters also had increased expression of genes known to interact with PRC2. Overall, PNR leukemias had increased expressions of anti-apoptotic and leukemia stem cell associated genes, suggesting that these cells are likely more primitive and harbor increased plasticity. Further supporting this, we observed coexpression of myeloid, lymphoid, and hematopoietic progenitor surface protein markers in PNR subpopulations.
Although the change in the phenotype in B-ALL is often referred to as a lineage switch or a change to AML, our data, in which CD19 and other markers of lymphoblasts are retained, suggest that plasticity is an inherent property of the disease in a subset of patients and can confer resistance to immunotherapy. One could hypothesize that this discovered SCE, therefore, protects the leukemic cell from T-cell cytotoxicity, and the ongoing CD19 targeting pressure to downregulate antigen or lineage switch is therefore removed. The current understanding of blood cell development from long-lived progenitors is that lineage potential can be a retained property, even as certain malignant clones retain the phenotypic appearance of a committed lineage.56 Future studies to further characterize the leukemias in patients who undergo a lineage switch will be an important addition to this work.
Although our results provide several new lines of evidence for antigen-independent CD19-CAR resistance, this study had several limitations. Our cohort was small, with only 14 patients, and the leukemias were heterogenous, consisting of multiple subtypes. Additionally, our bulk analyses were performed on whole BMAs, rather than on isolated B-ALL cells. It is possible that some of our results arise from differences in the leukemic burden, although this was mitigated by our use of the single-cell analyses, which corroborated our findings. Previous work has described antigen-independent sources of CD19-CAR resistance, including the reduced expressions of death receptor genes in leukemia cells.24 Although we did not observe this in our cohort, this further highlights that genomic alterations of CD19 or downregulation of these death receptors are not the only factors contributing to nonresponse in these patients. It will be important for future studies to expand the number of patients screened to determine the sensitivity and specificity of SCEs to identify PNRs, and our described protocols would need to be refined to be suitable for use in the clinic. Furthermore, although we describe an association between SCE and PNR to CD19-CAR, we did not identify the exact mechanism causing the resistance to cytotoxic killing. Future mechanistic and in vivo studies using matched patient leukemias and CD19-CARs are required to uncover the underlying biology and improve the targeting of this PNR phenotype.
In summary, we developed a comprehensive multiomic approach to evaluate patient BMA and describe the association of SCE with PNR to CD19-CAR therapy, which is not readily identified by standard pretreatment flow cytometry. Our results support further refinement of the eligibility criteria for CD19-CAR, and future trials of combination therapy with epigenetic modifying agents. Continued investigation into the leukemic biology of PNR will provide informed clinical decision making, novel therapeutic strategies, and improved outcomes for patients with this devastating disease.
Acknowledgments
For this study, the authors used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH) (http://biowulf.nih.gov). This research was supported by the NIH National Cancer Institute’s intramural research program, the Seattle Children’s Research Institute, Stand Up to Cancer, Strong Against Cancer, Alex’s Lemonade Stand, a St. Baldrick’s Pediatric Dream Team Translational Research Grant, and the NIH/National Human Genome Research Institute (R01HG010501) (M.L.B.). K.E.M. was supported by the NIH Oxford-Cambridge Scholars Program, an NIH Cambridge Trust Scholarship, and the NIH Medical Research Scholars Program, a public-private partnership supported by the NIH and generous contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation (DDCF grant #2014194), Genetech, Elsevier, and other private donors. Biorender.com was used to generate figures.
Authorship
Contribution: R.J.O. and J.K. conceived the project and provided mentorship and supervision to K.E.M., who designed the study and experiments and coordinated the collaboration; R.A.G., A.L.W., and M.C.J. provided clinical samples and data; K.E.M., B.E.G., Y.K.S., B.Z.S., S.O.A., C.W., A.T-C.C., G.A-B., and M.C.K. performed experiments; K.E.M., H-C.C., A.A., B.E.G., V.G., L.M., S.O.A., X.W., Y.Z., and G.A-B. performed bioinformatic and statistical analyses; M.L.B. provided expertise, mentorship, and supervision; K.E.M., R.A.G., R.J.O., and J.K. wrote the manuscript; and all authors contributed to data interpretation and critical review and editing of the manuscript.
Conflict-of-interest disclosure: R.A.G. serves on a study steering committee for and is an inventor on a patent licensed to Juno Therapeutics, a Bristol Myers Squibb company, and has served on advisory boards for Novartis. M.C.J. has interests in Umoja Biopharma and Juno Therapeutics, a Bristol Myers Squibb company; is a seed investor and holds ownership equity in Umoja; serves as a member of the Umoja Joint Steering Committee; is a board observer of the Umoja Board of Directors; and holds patents, some of which are licensed to Umoja Biopharma and Juno Therapeutics. R.J.O. has received research support from Miltenyi Biotech and consults for Abound Bio (Pittsburgh, PA) and Umoja Biopharma (Seattle, WA).
Correspondence: Rimas J. Orentas, Seattle Children’s Research Institute, 1100 Olive Way, Suite 100, Seattle, WA 98101; e-mail: rimas.orentas@seattlechildrens.org; and Javed Khan, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 37 Convent Dr, Building 37, Room 2016, Bethesda, MD 20892; e-mail: khanjav@mail.nih.gov.
References
Author notes
Patient data from all experiments is in the process of being deposited in the database of Genotypes and Phenotypes (accession number phs001928).
Data are available on request from the corresponding author, Javed Khan (khanjav@mail.nih.gov).
The full-text version of this article contains a data supplement.