Key Points
Multiomic single-cell analysis of 3 different patient cohorts reveals few changes in T and NK cells following venetoclax monotherapy.
The data support the rationale for combining venetoclax with adjunct immune checkpoint inhibitors to achieve more durable responses.
Abstract
Venetoclax is an effective treatment for certain blood cancers, such as chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML). However, most patients relapse while on venetoclax and further treatment options are limited. Combining venetoclax with immunotherapies is an attractive approach; however, a detailed understanding of how venetoclax treatment impacts normal immune cells in patients is lacking. In this study, we performed deep profiling of peripheral blood (PB) cells from patients with CLL and AML before and after short-term treatment with venetoclax using mass cytometry (cytometry by time of flight) and found no impact on the concentrations of key T-cell subsets or their expression of checkpoint molecules. We also analyzed PB from patients with breast cancer receiving venetoclax long-term using a single-cell multiomics approach (cellular indexing of transcriptomes and epitopes by sequencing) and functional assays. We found significant depletion of B-cell populations with low expression of MCL-1 relative to other immune cells, attended by extensive transcriptomic changes. By contrast, there was less impact on circulating T cells and natural killer (NK) cells, with no changes in their subset composition, transcriptome, or function following venetoclax treatment. Our data indicate that venetoclax has minimal impact on circulating T or NK cells, supporting the rationale of combining this BH3 mimetic drug with cancer immunotherapies for more durable antitumor responses.
Introduction
Programmed cell death (apoptosis) is tightly regulated by the interaction between the proapoptotic and prosurvival members of the BCL-2 family. Among the most promising classes of emerging targeted therapies in hematological malignancies are the small molecule BH3 mimetics that selectively target the prosurvival proteins. Venetoclax1 was the first selective BCL-2 inhibitor to be introduced into clinical practice for the treatment of hematological malignancies, including chronic lymphocytic leukemia (CLL) and acute myeloid leukemia (AML). CLL leukemic cells express abnormally high levels of BCL-2, thus, allowing them to survive when they should die, and this leads to the accumulation of the cancerous cells. In CLL, venetoclax produced a 79% overall response rate among patients with relapsed and refractory disease2,3 when used as continuous monotherapy and produced higher rates when combined with anti-CD20 monoclonal antibodies in fixed duration regimens.4-6 In AML, venetoclax improved clinical responses (48% to 66% remission rates) when combined with hypomethylating agents or low-dose cytarabine as first-line therapy in older patients who could not tolerate intensive treatments.7,8 Unfortunately, the disease will ultimately recur in most patients. Rational combination therapies are needed to use venetoclax more effectively to achieve deeper and more enduring responses for better patient outcomes.9
T cells and the broader microenvironment play a crucial role in sustaining the growth and survival of cancer cells, including in leukemia (eg, in the lymph nodes or bone marrow). Therefore, targeting the T cells that can interact with leukemic cells is an attractive strategy to bolster the action of venetoclax and new immunotherapies are emerging to treat hematological cancers. Chimeric antigen receptor–based therapy is positioning itself as 1 of the most promising therapies for eradicating malignant cells. Moreover, multiple clinical trials currently focus on combination therapy of venetoclax with checkpoint inhibitors. In CLL and AML, clinical trials of venetoclax combined with inhibitors of PD-1 (#NCT04274907 and #NCT04284787), TIM-3 (#NCT04150029), OX-40 (#NCT03390296), or CD47 (#NCT04435691 and #NCT04778410) are underway. Such trials are predicated on there being sufficient T-cell function in patients treated with venetoclax to clear cancerous cells.
Studies in wild-type mice showed that venetoclax treatment decreased the number of CD4+ and CD8+ T cells.10-12 In a mouse model of colon cancer, venetoclax therapy elicited an increase of CD8+PD-1+ T effector/memory (EM) cells,13 whereas in a leukemia model, increased memory/naïve T-cell ratios and function were apparent following BCL-2 inhibition.14 Several studies of venetoclax combination therapies have been done to determine the impact of venetoclax on the immune state of human patients. These studies indicated few changes to immune subsets in treated subjects.15-17 However, the effect of venetoclax monotherapy directly on the immune system in patients has not been reported in detail.
In this study, we assessed the impact of venetoclax on immune states with deep profiling of cells from patients with 3 different cancers. Cytometry by time of flight (CyTOF) was performed on the circulating immune cells from patients with CLL and AML before and after venetoclax monotherapy. In addition, we performed single-cell cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq), flow cytometry, and functional assays on circulating immune cells from patients with breast cancer receiving long-term venetoclax therapy to determine the direct consequences of BCL-2 inhibition, independent of possible influences of blood cancer cells. We demonstrate that venetoclax has minimal impact on natural killer (NK) and T-cell populations, paving the way for future studies combining BCL-2 inhibitors with immune-targeting strategies.
Methods
Patient samples
Paired pre- and postvenetoclax treatment peripheral blood (PB) samples were obtained from cohorts of patients with CLL (n = 19; dose escalation to 400 mg daily; 1 month),18 AML (n = 16; 100-600 mg daily; 7 days),19 and breast cancer (n = 13; dose escalation to 200, 400, or 800 mg daily; median treatment duration, 88.6 weeks)20 (supplemental Tables 1-3). Studies were approved by the Human Research Ethics Committees at the Royal Melbourne Hospital (2005.008, 2012.244, 2014.226, 2016.305, and 2016.066), the Walter and Eliza Hall Institute (G15/05, G15/02, and 10/02), and The Alfred Health (575/19). Written informed consent was obtained from all patients. Total blood composition was analyzed by Advia; peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll gradient and cryopreserved for later analysis.
Mass cytometry
CyTOF data were acquired with a 39-antibody panel (supplemental Tables 4 and 5) on a Fluidigm Helios Mass Cytometer as previously described.21 Single-cell suspensions were thawed and stained for viability for 1 minute in 25 μM cisplatin (Sigma-Aldrich) at room temperature. Cells were then fixed with paraformaldehyde (PFA; Electron Microscopy Sciences) at a final concentration of 1.6% for 10 minutes at room temperature. Cells were pelleted washed once with cell-staining medium (CSM; phosphate-buffered saline [PBS] with 0.5% bovine serum albumin and 0.02% sodium azide) to remove residual PFA and stored at −80 °C.
Cells were barcoded using 20-plex palladium barcoding according to manufacturer instructions (DVS Sciences). After barcoding, cells were pelleted and washed once with CSM to remove residual PFA. Cells were stained with CD16/CD32 antibodies for 10 minutes and with surface antibodies (supplemental Tables 4 and 5) for 30 minutes at room temperature. Cells were permeabilized with 4°C methanol for 10 minutes. Cells were washed 3 times with CSM and stained with intracellular antibodies (supplemental Tables 4 and 5) for 30 minutes at room temperature. Cells were washed with CSM, then stained with 125 nM 191Ir/193Ir DNA intercalator (DVS Sciences) in PBS with 1.6% PFA at 4°C overnight. Cells were washed once with CSM, washed 3 times with double-distilled water, filtered to remove aggregates, and resuspended with EQ Four Element Four Element Callibration beads immediately before analysis using a Helios mass cytometer (DVS Sciences, maintained by Materials Characterisation and Fabrication Platform, University of Melbourne). Throughout the analysis, cells were maintained at 4°C and introduced at a constant rate of ∼300 cells per second. Antibodies conjugated in house were generated from carrier-free antibody solutions conjugated to custom isotypes (Trace Sciences and DVS Sciences) using the Maxpar X8 Antibody Labelling Kit (DVS Sciences).
Data concatenation, normalization, and debarcoding were done using Premessa. Single cells were gated using the Cytobank software (http://www.cytobank.org) based on event length and 191Ir/193Ir DNA contents to avoid debris and doublets. Data were then subjected to flow cytometry data analysis using self-organizing maps clustering or manual gating (FlowJo version 10.7.1). All flow and mass cytometry data have been made available from Flow Repository (ID: FR-FCM-Z57G, FR-FCM-Z57H, and FR-FCM-Z586).
Flow cytometry
Flow cytometry data were acquired with antibody panels (supplemental Table 6) on a Cytek Aurora Flow Cytometer. Analysis was performed using Premessa, Catalyst, R, and manual gating.
Cell concentration calculation
Concentrations of lymphocytes in PB were measured using the automated hematology analyzer (ADVIA2120i). Mass or flow cytometry were used to determine the percentage of each immune subset from total lymphocytes. The concentration of the immune subset was calculated using the following equation: percent of immune subset from total lymphocytes/100 from mass or flow cytometry × concentration of total lymphocytes calculated from ADVIA.
Single-cell RNA sequencing
Viable cells (propidium iodide–negative) were sort purified and samples from patients were processed on the same day and in 2 batches to minimize batch effects. Cells were stained with the TotalSeq-C Human Cocktail (Biolegend, #399905). Single cells were captured and barcoded using the Chromium X Instrument (10x Genomics) and libraries were prepared according to the manufacturer instructions. Analysis was conducted using scater version 1.20.022 and Seurat version 4.0.5.23 The data were integrated using harmony version 0.1.024 method before clustering and visualizing using uniform manifold approximation and projection (UMAP). A pseudobulk strategy and edgeR version 3.34.025 pipeline was used to find differentially expressed genes (DEGs) among different immune subsets. Additional details are provided in supplemental Methods.
Single-cell data analysis
CellRanger version 5.0.0 (GRCH38 human genome) was used to preprocess raw next-generation sequencing data. The fastq files were generated with the “cellranger mkfastq” function. Gene count and feature barcode matrix were made with “cellranger count.” We applied the isOutlier function of scater (v1.20.0) to identify low-quality cells which were >3 median absolute deviations from the median expression of the unique molecular identifier (UMI; both directions), detected genes (both directions), and mitochondrial gene expression (higher) in each sample. We conducted data normalization and highly variable gene selection using NormalizeData and FindVariableFeatures functions in Seurat version 4.0.5 with default parameters. We considered different patients as batches and corrected the gene expression matrix with harmony version 0.1.0. The protein matrix was normalized with DSB version 1.0.0, and binding levels of 4 rat/mouse antibodies were used as control. We then used the Seurat FindMultiModalNeighbors function to make UMAP and clusters, using gene expression profile (first 20 principal components) and protein expression of selected cell markers (CD3, CD4, CD8, CD45RA, CD45RO, CD25, KLRG1, CD56, CD62L, T-cell receptor γδ (TCR-αβ), TCR-γδ2, CD19, CD20, CD11c, and CD24). Doublets were identified by simultaneous expression of surface markers from multiple distinct hematological lineages (CD3, CD19, and CD56), higher UMIs, and higher detected gene numbers. The cluster with high mitochondrial gene expression and low detection of UMI genes was also removed. We used the cell-surface markers and selected gene expression to identify immune subsets (supplemental Figure 3B-C).
Clusters are annotated as C0-monocytes (protein: CD14+), C1-CD4 Tn (protein: CD4+CD45RA+CD62L+), C2-CD4 Temra (protein: CD4+CD45RA+CD62L−), C3-CD4 Tcm (protein: CD4+CD45RO+CD62L), C4-CD4 Tem (protein: CD4+CD45RO+CD62L−), C5-regulatory T cell (protein: CD4+CD25+, RNA: FOXP3+CTLA4+), C6-CD4 Tc (protein: CD4+, RNA: GZMB+PRF1+), C7-γδ T cells (protein: TCR−γδ2), C8-CD8 Tn (protein: CD8+CD45RA+CD62L+), C9-CD8 Tem (protein: CD8+CD45RO+), C10-CD8 Temra (CD8+CD45RA+), C11-CD8 CD16+ T cells (protein: CD3+CD8+CD16+), C12-naïve B cells (protein: CD20+CD27−, RNA: TCL1A+), C13-transitional B cells (protein: CD20+CD27−, RNA: TCL1A++), C14 and C15-memory B (protein: CD20+CD27+, RNA: TCL1A−), CD16−CD56high NK cells (protein: CD56++), and CD17−CD56dim NK cells (protein: CD56+).
We applied the pseudobulk strategy and edgeR (v3.34.0) pipeline to find DEGs in different immune subsets. Five patients were considered replicates in the design of DEG tests to identify consistent DEGs among all patients. In the DEG test for C6-Tc, 4 patients were used as replicates owing to the low cell counts in data from 1 patient (BC_01033). Raw data are available from European Genome-phenome Archive (accession number EGAS00001006241) and scripts are available at https://github.com/HongkePn/Circulating_T_NK_under_VEN. Results of DEG tests can be found in supplemental Table 7.
Ex vivo T-cell proliferation and activation assay
T cells were isolated from PBMCs using EasySep Human T Cell Isolation Kit (StemCell) following the manufacturer instructions. Isolated T cells (>98% purity) were then stained using CellTrace Violet Cell Proliferation Kit (Thermo Fisher Scientific) following manufacturer instructions. In a 96-well round bottom plate, 10 000 cells per well were plated in 200 μL of RPMI 1640 complete media (Gibco) supplemented with 2 mM l-glutamine (Gibco), 1% Minimum Essential Medium nonessential amino acids (Gibco), 1 mM sodium pyruvate (Gibco), 100 U/mL penicillin-streptomycin (Gibco), and 10% fetal bovine serum (Sigma). Recombinant human interleukin 2 protein (active; Abcam) (400 U/mL) and Dynabeads Human T-Activator anti-CD3/CD28 (Thermo Fisher Scientific) (at a 1:1 ratio with T cells) were added to appropriate wells and cultured for 48, 72, or 96 hours.
After 48 hours, the culture supernatant was collected and the cytokine and cytotoxic profiles examined using the LEGENDplex CD8/NK panel (BioLegend), following the manufacturer instructions. Samples were then run on a BD FACSCanto II cell analyzer, and data were analyzed using the LEGENDplex data analysis software.
For each time point, a known number of counting beads (CaliBRITE rainbow calibration particles, BD Biosciences) (10 000 per well) were added to samples before cell harvest for calculating cell numbers. Cells were stained with the following antibodies: fluorescein isothiocyanate antihuman CD3 (BioLegend; UCHT1), phycoerythrin antihuman CD25 (BD Biosciences; M-A251), APC/Fire 750 antihuman CD4 (BioLegend; SK3), and Brilliant Violet 785 antihuman CD8 (BioLegend; SK1) in staining buffer (PBS, 2mM EDTA, and 0.5% bovine serum albumin) for 20 minutes at 4°C, and finally propidium iodide (Sigma-Aldrich) (5 μg/mL) was included for dead cell exclusion before data acquisition on a BD LSRFortessa X-20 cell analyzer. Mean division number was calculated using a previously described method.26
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9 for macOS (GraphPad Software, San Diego, CA). For paired comparisons, paired t tests were used. For multiple comparisons with 2 independent variables, 2-way analysis of variance with Geisser-Greenhouse correction and Tukey multiple comparison test were used. A significant result is indicated where P < .05.
Results
Short-term venetoclax treatment in patients with CLL and AML does not alter T-cell composition or checkpoint phenotype
To assess whether venetoclax monotherapy has short-term impacts on circulating immune cells in vivo, we analyzed PBMCs from 19 patients with relapsed/refractory CLL in the VENICE trial (www.clinicaltrials.gov, #NCT02980731)18 before and after 1 month of single-agent venetoclax treatment. Patient demographics and clinical characteristics are summarized in supplemental Table 1. The median age of the overall cohort was 63 years (range, 52-77 years) and 74% were males. Seventeen patients (89%) had received prior therapies, of whom 8 (42%) had received ≥3 lines of therapy. Patients received oral venetoclax monotherapy daily, beginning at 20 mg on week 1 and increasing stepwise each week to achieve a target dose of 400 mg (20, 50, 100, and 200 mg). The 5-week ramp-up schedule was designed to gradually reduce tumor burden and minimize tumor lysis syndrome. PB samples were drawn at baseline and 1 week after patients received 200 mg venetoclax.
The paired patient samples were assayed using a tailor-made 39-parameter CyTOF panel composed of probes for key immune cell lineages, phenotypic states, transcription factors, signaling states, checkpoint molecules, and cell death regulators (Figure 1A; supplemental Table 4). We performed flow cytometry data analysis using self-organizing maps27 clustering to classify the main cell populations and visualized the data using UMAP (Figure 1B). Cells were partitioned into 11 clusters and the expression of lineage markers distinguished CLL cells and the major immune populations; CD4+ T cells, CD8+ T cells, NK cells, and myeloid cells (Figure 1B; supplemental Figure 1A). As expected, short-term venetoclax treatment induced a striking reduction in CLL cells (Figure 1C-D). For subsequent analysis, the CLL cells were excluded and standard 2-dimensional gating analysis was used to identify immune cell subsets, including CD4+ or CD8+ naïve, EM, central memory, and terminally differentiated EM cells reexpressing CD45RA (EMRA) populations (supplemental Figure 1B). We found no change in the concentrations of these T-cell subsets, basophils, monocyte subtypes, NK, or dendritic cells (Figure 1C-D; supplemental Figure 1C). The mean expression of the prosurvival proteins BCL-2, BCL-XL, and MCL-1 in CD4+ and CD8+ T cells was largely unchanged after venetoclax treatment, excepting increases in some patients (Figure 1E-F). Importantly, the expression of immune checkpoint proteins was unchanged by venetoclax treatment (Figure 1E-F).
Composition of T and NK cells immune cell types in patients with CLL remains unchanged after short-term venetoclax treatment. (A) Schematic representation of the CLL study. PB samples from 19 patients with CLL were collected before (pre) and after (post) 1 month of venetoclax treatment and analyzed by CyTOF. (B) UMAP projections of PB cells, colored by FlowSOM clusters with key immune cell types annotated. (C) UMAP of PB cells from patient CLL_028 pre- and postvenetoclax treatment. (D) Concentrations of CLL, NK, and T-cell subsets. (E) Mean expression of survival and checkpoint proteins expressed as a ratio of postvenetoclax:prevenetoclax treatment in CD4+ and CD8+ T cells. Each symbol represents an individual patient; paired t test; ∗P < .05, ∗∗P < .01. (F) Representative histograms of BCL-2, BCL-XL, MCL-1, CTLA-4, PD-1, TIM3, PD-L1, CD27, and OX40 protein pre- and postvenetoclax treatment in CD4 naïve (N) (CD45RAhighCD27high), EM CD4, CD8 naïve (CD45RAhighCD27high), and EM CD8 cells from patient CLL_025. CM, central memory.
Composition of T and NK cells immune cell types in patients with CLL remains unchanged after short-term venetoclax treatment. (A) Schematic representation of the CLL study. PB samples from 19 patients with CLL were collected before (pre) and after (post) 1 month of venetoclax treatment and analyzed by CyTOF. (B) UMAP projections of PB cells, colored by FlowSOM clusters with key immune cell types annotated. (C) UMAP of PB cells from patient CLL_028 pre- and postvenetoclax treatment. (D) Concentrations of CLL, NK, and T-cell subsets. (E) Mean expression of survival and checkpoint proteins expressed as a ratio of postvenetoclax:prevenetoclax treatment in CD4+ and CD8+ T cells. Each symbol represents an individual patient; paired t test; ∗P < .05, ∗∗P < .01. (F) Representative histograms of BCL-2, BCL-XL, MCL-1, CTLA-4, PD-1, TIM3, PD-L1, CD27, and OX40 protein pre- and postvenetoclax treatment in CD4 naïve (N) (CD45RAhighCD27high), EM CD4, CD8 naïve (CD45RAhighCD27high), and EM CD8 cells from patient CLL_025. CM, central memory.
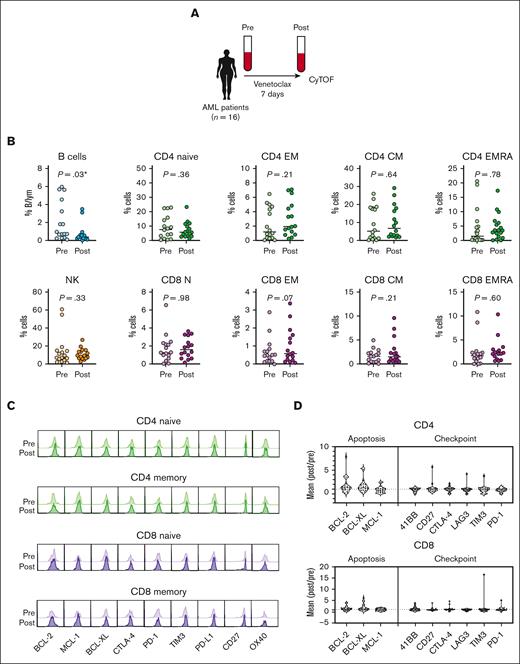
We also assessed immune cell changes in patients with AML treated for 7 days with venetoclax monotherapy from the CAVEAT trial (ANZ Clinical Trial Registry, #ACTRN12616000445471)19 (Figure 2A; patient characteristics are shown in supplemental Table 2). Patients with newly diagnosed de novo or secondary AML (median age, 71 years; range, 64-80 years; 69% male) received a 7-day venetoclax dose treatment (100-600 mg). Paired PB samples were analyzed before and after venetoclax treatment. Similar to the CLL cohort, we observed a significant decrease in B cells; however, circulating CD4+ and CD8+ T-cell subsets and NK cells remained unchanged in percentage (Figure 2B; supplemental Figure 2). Consistent with our observation in patients with CLL receiving venetoclax, the expression of prosurvival BCL-2 family proteins and immune checkpoint molecules were unchanged in key CD4+ and CD8+ T-cell populations (Figure 2C-D). Together, these results indicate that short-term venetoclax treatment does not alter the composition or checkpoint phenotype of circulating T and NK immune cells in patients with CLL or AML.
Composition of T and NK cells immune cell types in patients with AML remains unchanged after short-term venetoclax treatment. (A) Schematic representation of the AML study. PB samples from 16 patients with AML were collected before (pre) and after (post) 7 days of venetoclax treatment and analyzed by CyTOF. (B) Percentages of B cells, NK cells, and T cells from total lymphocytes measured by CyTOF. (C) Mean expression of survival and checkpoint proteins in CD4+ and CD8+ cells expressed as a ratio of postvenetoclax:prevenetoclax treatment. Each symbol represents an individual patient; paired t test; ∗P < .05, ∗∗P < .01. (D) Representative histograms and mean expression of BCL-2, MCL-1, BCL-XL, CTLA-4, PD-1, TIM-3, PD-L1, CD27, OX40, and T-BET protein expression pre- and postvenetoclax treatment in CD4 naïve (CD45RAhighCD27highCCR7high), CD4 EM (CD45ROhigh), CD8 naïve (CD45RAhighCD27highCCR7high), and CD8 EM (CD45ROhigh) cells from patient AML_027.
Composition of T and NK cells immune cell types in patients with AML remains unchanged after short-term venetoclax treatment. (A) Schematic representation of the AML study. PB samples from 16 patients with AML were collected before (pre) and after (post) 7 days of venetoclax treatment and analyzed by CyTOF. (B) Percentages of B cells, NK cells, and T cells from total lymphocytes measured by CyTOF. (C) Mean expression of survival and checkpoint proteins in CD4+ and CD8+ cells expressed as a ratio of postvenetoclax:prevenetoclax treatment. Each symbol represents an individual patient; paired t test; ∗P < .05, ∗∗P < .01. (D) Representative histograms and mean expression of BCL-2, MCL-1, BCL-XL, CTLA-4, PD-1, TIM-3, PD-L1, CD27, OX40, and T-BET protein expression pre- and postvenetoclax treatment in CD4 naïve (CD45RAhighCD27highCCR7high), CD4 EM (CD45ROhigh), CD8 naïve (CD45RAhighCD27highCCR7high), and CD8 EM (CD45ROhigh) cells from patient AML_027.
Long-term venetoclax treatment in patients with breast cancer diminishes B and T cells but does not alter T-cell composition
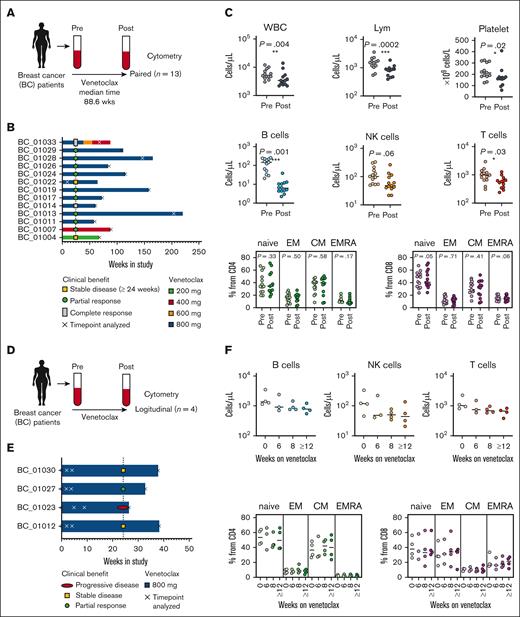
To assess the impact of long-term venetoclax therapy on circulating immune cells from patients without a hematological malignancy, we assayed paired PBMC samples from 13 patients with metastatic breast cancer (ANZ Clinical Trial Registry, #ACTRN12615000702516)20 before and after treatment with venetoclax for >50 weeks (Figure 3A-B; patient characteristics are shown in supplemental Table 3). All patients (median age, 64 years; range, 43-78 years) had ER- and BCL-2–positive tumors, had received prior therapies, and were treated with a combination of tamoxifen (20 mg) and venetoclax (200-800 mg daily). Samples from various time points were analyzed at the protein (flow cytometry), RNA (CITE-seq), and functional level (in vitro T-cell function assays). We found that long-term venetoclax treatment diminished circulating B cells by approximately an order of magnitude (Figure 3C). There was also a relatively modest (but significant) decrease in T cells over this period (Figure 3C; supplemental Figure 2). The reduction in T-cell concentration after long-term venetoclax treatment did not reflect the loss of a particular subset, as the proportions of naïve EM T-cell populations were unchanged (Figure 3C).
Flow cytometry profiling of circulating T cells in patients with breast cancer after short- and long-term venetoclax treatment reveals limited changes in T-cell subsets. (A) Schematic representation of immunophenotyping of the patients with breast cancer receiving long-term venetoclax treatment. Blood was collected from patients with breast cancer before (pre) and after (post) venetoclax treatment (median treatment, 88.6 weeks; range, 8.8-204.2 weeks). Pre- and posttreatment cells from each patient were analyzed by flow cytometry (n = 13 pairs). (B) Bar graphs show the study duration, early clinical benefit, dose of venetoclax, and analysis time point for each patient. (C) Concentration of lymphocytes, B cells, NK cells, and T cells. Percentages of CD45RA+CCR7+ naïve (N), CD45RA−CCR7+ CM, CD45RA−CCR7− EM, and CD45RA+CCR7− effector (EMRA) cells from CD4 (C) and CD8 (D) T cells measured by flow cytometry (bottom). (D) Schematic representation of the longitudinal immunophenotyping of the patients with breast cancer who received short-term venetoclax treatment. Blood was collected at 4 different time points and analyzed by flow cytometry (n = 4 sets). (E) Bar graphs show the time points analyzed, early clinical benefit, and dose of venetoclax for each patient in panel F. Measurements were made at pre- and post- (6, 8, and ≥12 weeks on venetoclax treatment). (F) Dot plots show concentrations of B cells, NK cells, and T cells. Percentages of CD45RA+CCR7+ naïve, CD45RA−CCR7+ CM, CD45RA−CCR7− EM, and CD45RA+CCR7− effector (EMRA) cells from CD4 (C) and CD8 (D) T cells were measured by flow cytometry. Lym, lymphocyte; WBC, white blood cell.
Flow cytometry profiling of circulating T cells in patients with breast cancer after short- and long-term venetoclax treatment reveals limited changes in T-cell subsets. (A) Schematic representation of immunophenotyping of the patients with breast cancer receiving long-term venetoclax treatment. Blood was collected from patients with breast cancer before (pre) and after (post) venetoclax treatment (median treatment, 88.6 weeks; range, 8.8-204.2 weeks). Pre- and posttreatment cells from each patient were analyzed by flow cytometry (n = 13 pairs). (B) Bar graphs show the study duration, early clinical benefit, dose of venetoclax, and analysis time point for each patient. (C) Concentration of lymphocytes, B cells, NK cells, and T cells. Percentages of CD45RA+CCR7+ naïve (N), CD45RA−CCR7+ CM, CD45RA−CCR7− EM, and CD45RA+CCR7− effector (EMRA) cells from CD4 (C) and CD8 (D) T cells measured by flow cytometry (bottom). (D) Schematic representation of the longitudinal immunophenotyping of the patients with breast cancer who received short-term venetoclax treatment. Blood was collected at 4 different time points and analyzed by flow cytometry (n = 4 sets). (E) Bar graphs show the time points analyzed, early clinical benefit, and dose of venetoclax for each patient in panel F. Measurements were made at pre- and post- (6, 8, and ≥12 weeks on venetoclax treatment). (F) Dot plots show concentrations of B cells, NK cells, and T cells. Percentages of CD45RA+CCR7+ naïve, CD45RA−CCR7+ CM, CD45RA−CCR7− EM, and CD45RA+CCR7− effector (EMRA) cells from CD4 (C) and CD8 (D) T cells were measured by flow cytometry. Lym, lymphocyte; WBC, white blood cell.
To assess the kinetics of these changes in lymphocyte concentration, we performed a longitudinal analysis of PBMCs from an independent batch of 4 patients with breast cancer at baseline and at 6, 8, and >12 weeks of 800 mg venetoclax treatment (Figure 3D-E; supplemental Figure 2A). Flow cytometry demonstrated that there was a steady, time-dependent decrease of B, NK, and T cells consistent with previous data following short-term venetoclax treatment20 (Figure 3F). Again, there were no subset-specific changes in T-cell subpopulations across this longitudinal analysis.
Limited impact of long-term venetoclax treatment on T-cell transcriptome and function
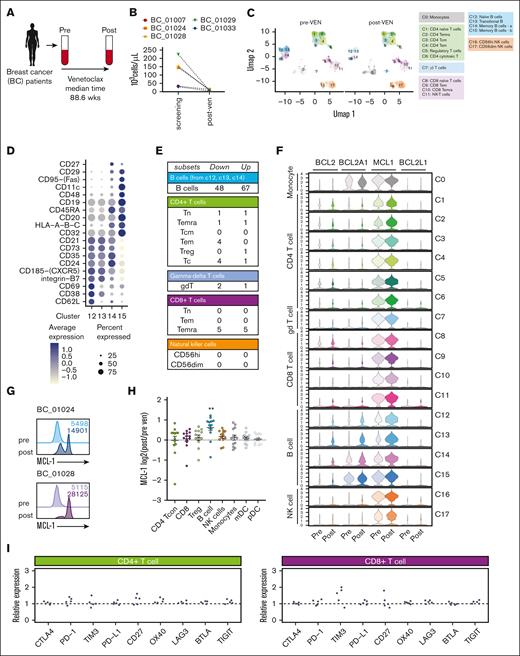
To assess the transcriptional impact of long-term venetoclax treatment on circulating lymphocytes, we employed single-cell CITE-seq with a 130-antibody panel in 5 paired samples from our breast cancer cohort (BC_01007 and BC_01033 receiving 400 mg; BC_01024, 01028, and 01029 receiving 800 mg venetoclax) (Figure 4A). As expected, B cells were reduced following long-term venetoclax treatment in this group (Figure 4B). We identified 17 clusters across 4 distinct lineages: B cells, T cells, NK cells, and monocytes (Figure 4C; supplemental Figure 3C). Marked reductions in most B-cell clusters were observed, with only 1 cluster of memory B cells remaining after long-term venetoclax treatment (Figure 4C; supplemental Figure 3C; cluster 15 shown in Figure 4D). Consistent with the flow cytometric analysis, there were no compositional changes in T-cell and NK cell subpopulations after venetoclax treatment (Figure 4C; supplemental Figure 4C).
Deep profiling of circulating lymphocytes cells in patients with breast cancer after long-term venetoclax treatment reveals limited changes in T-cell subsets. (A) Schematic representation of immunophenotyping of the patients with breast cancer who received long-term venetoclax (VEN) treatment. Blood was collected from patients with breast cancer before (pre) and after (post) venetoclax treatment (median treatment, 88.6 weeks; range, 8.8-204.2 weeks). Pre- and posttreatment cells from each patient were analyzed by CITE-seq (n = 5 pairs). (B) Concentrations of B cells from the 5 samples selected for CITE-seq. (C) UMAP and annotation of circulating immune cells from 5 patients with breast cancer on long-term venetoclax treatment analyzed by CITE-seq. (D) Dot plots showing a curated list of key cell-surface makers expressed in naïve (cluster 12), transitional (cluster 13), and memory (clusters 14 and 15) B-cell subsets. Color intensity reflects relative level of protein expression and the size of the dot indicates the fraction of cells expressing that protein in that cluster. (E) Table of the numbers of DEGs (false discovery rate < 0.05) found by pseudobulk DEG analysis (postvenetoclax:prevenetoclax). (F) Violin plots of BCL-2 survival gene transcription from pseudobulk analysis of all clusters. (G) Intracellular protein expression of MCL-1 measured by flow cytometry in B cells from patients BC_01024 and BC_01028. (H) Changes in MCL-1 protein expression measured in immune cell populations expressed as log2 changes in post- vs pretreatment measured by flow cytometry. (I) The expression ratio of checkpoint genes on CD4+ and CD8+ T cells, calculated by mean (postvenetoclax)/mean (prevenetoclax) measured by CITE-seq. In graphs in panel H, each symbol represents an individual patient, bar is at the mean, and error bars represent standard error of the mean. mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell; Tcon, concentration of T cell; Treg, regulatory T cell.
Deep profiling of circulating lymphocytes cells in patients with breast cancer after long-term venetoclax treatment reveals limited changes in T-cell subsets. (A) Schematic representation of immunophenotyping of the patients with breast cancer who received long-term venetoclax (VEN) treatment. Blood was collected from patients with breast cancer before (pre) and after (post) venetoclax treatment (median treatment, 88.6 weeks; range, 8.8-204.2 weeks). Pre- and posttreatment cells from each patient were analyzed by CITE-seq (n = 5 pairs). (B) Concentrations of B cells from the 5 samples selected for CITE-seq. (C) UMAP and annotation of circulating immune cells from 5 patients with breast cancer on long-term venetoclax treatment analyzed by CITE-seq. (D) Dot plots showing a curated list of key cell-surface makers expressed in naïve (cluster 12), transitional (cluster 13), and memory (clusters 14 and 15) B-cell subsets. Color intensity reflects relative level of protein expression and the size of the dot indicates the fraction of cells expressing that protein in that cluster. (E) Table of the numbers of DEGs (false discovery rate < 0.05) found by pseudobulk DEG analysis (postvenetoclax:prevenetoclax). (F) Violin plots of BCL-2 survival gene transcription from pseudobulk analysis of all clusters. (G) Intracellular protein expression of MCL-1 measured by flow cytometry in B cells from patients BC_01024 and BC_01028. (H) Changes in MCL-1 protein expression measured in immune cell populations expressed as log2 changes in post- vs pretreatment measured by flow cytometry. (I) The expression ratio of checkpoint genes on CD4+ and CD8+ T cells, calculated by mean (postvenetoclax)/mean (prevenetoclax) measured by CITE-seq. In graphs in panel H, each symbol represents an individual patient, bar is at the mean, and error bars represent standard error of the mean. mDC, myeloid dendritic cell; pDC, plasmacytoid dendritic cell; Tcon, concentration of T cell; Treg, regulatory T cell.
Many DEGs (false discovery rate < 0.05, log fold change > 0.5) were observed in B cells following long-term venetoclax treatment (Figure 4E; supplemental Figure 4). Concerted changes included heightened expression of TRAF1 and members of the S100 family of Ca2+ binding proteins in B-cell clusters sensitive to venetoclax (supplemental Figure 4B,D). Interestingly, these venetoclax-sensitive B-cell clusters had lower MCL-1 transcription (clusters 12-14) compared with the resistant memory B-cell (cluster 15) and other immune cell populations (Figure 4F-H). Higher expression of MCL1 and BCL2A1 was observed in those B cells from clusters 12 to 14 that persisted and the memory B cell (cluster 15) unaffected by venetoclax (Figure 4E). Increased MCL-1 expression in B cells after venetoclax was also evident at the protein level (Figure 4G-H).
By contrast, minimal transcriptional change was observed in T- and NK cell subsets in this single-cell analysis (Figure 4D; supplemental Figure 4). Moreover, consistent with the relatively normal T-cell proportions observed after long-term venetoclax treatment, cell-surface protein analysis of immune checkpoint molecules revealed few changes in CD4+ or CD8+ T-cell populations (Figure 4G; supplemental Figure 3D).
Finally, we examined CD4+ and CD8+ T-cell function in samples from 5 patients with breast cancer before and during venetoclax treatment (Figure 5A; T-cell gating strategy shown in Figure 5B). We observed no significant differences in T-cell activation kinetics measured by CD25 expression (Figure 5C) or proliferation (Figure 5D) in response to anti-CD3/CD28 stimulation. In addition, we examined the total T-cell cytokine and cytotoxic protein production after anti-CD3/CD28 stimulation in samples taken before and during venetoclax treatment. We found that there was no significant difference in T-cell production of proinflammatory cytokines and cytotoxic mediators across the venetoclax treatment time points (Figure 5E). Taken together, these results indicate that long-term venetoclax treatment does not impact T-cell proliferation or effector function, following TCR stimulation.
Venetoclax treatment does not alter T cell proliferation or effector function. (A) Schematic of the time points analyzed, early clinical benefit, and dose of venetoclax for each patient with breast cancer. Measurements were made at pre- (cycle 1) and post- (cycles 2 and/or 4-23); each cycle is 28 days. (B) Flow cytometry gating strategy to identify T-cell populations: lymphocytes, single cells, propidium iodide–negative live cells, CD3+ T cells, and either CD4+ or CD8+ cells. (C) Representative histograms showing expression of CD25 by CD4+ or CD8+ T cells from 1 patient during venetoclax treatment after incubation with anti-CD3/CD28 beads for 48, 72, or 96 hours. Graphs summarize CD25 expression by CD4+ or CD8+ T cells in patients during venetoclax treatment cycles. (D) Representative histograms showing cell trace violet (CTV) staining of CD4+ or CD8+ T cells from 1 patient during venetoclax treatment cycles after incubation with anti-CD3/CD28 beads for 48, 72, or 96 hours. Graphs show the percentage of proliferating cells (having undergone ≥1 division) in CD4+ or CD8+ T cells in patients at different venetoclax treatment cycles (n = 5). (E) Graphs showing the concentration of cytokines and cytotoxic molecules measured using the LEGENDplex CD8/NK panel produced by total T cells after 48 hours of culture with anti-CD3/CD28 beads for patients during venetoclax treatment cycles. In graphs in panels C-E, each symbol represents an individual patient, bar is at the mean, and error bars represent standard error of the mean. IFNγ, interferon gamma; IL-4, interleukin 4; NS, no stimulation; SSC, side scatter; TNF, tumor necrosis factor.
Venetoclax treatment does not alter T cell proliferation or effector function. (A) Schematic of the time points analyzed, early clinical benefit, and dose of venetoclax for each patient with breast cancer. Measurements were made at pre- (cycle 1) and post- (cycles 2 and/or 4-23); each cycle is 28 days. (B) Flow cytometry gating strategy to identify T-cell populations: lymphocytes, single cells, propidium iodide–negative live cells, CD3+ T cells, and either CD4+ or CD8+ cells. (C) Representative histograms showing expression of CD25 by CD4+ or CD8+ T cells from 1 patient during venetoclax treatment after incubation with anti-CD3/CD28 beads for 48, 72, or 96 hours. Graphs summarize CD25 expression by CD4+ or CD8+ T cells in patients during venetoclax treatment cycles. (D) Representative histograms showing cell trace violet (CTV) staining of CD4+ or CD8+ T cells from 1 patient during venetoclax treatment cycles after incubation with anti-CD3/CD28 beads for 48, 72, or 96 hours. Graphs show the percentage of proliferating cells (having undergone ≥1 division) in CD4+ or CD8+ T cells in patients at different venetoclax treatment cycles (n = 5). (E) Graphs showing the concentration of cytokines and cytotoxic molecules measured using the LEGENDplex CD8/NK panel produced by total T cells after 48 hours of culture with anti-CD3/CD28 beads for patients during venetoclax treatment cycles. In graphs in panels C-E, each symbol represents an individual patient, bar is at the mean, and error bars represent standard error of the mean. IFNγ, interferon gamma; IL-4, interleukin 4; NS, no stimulation; SSC, side scatter; TNF, tumor necrosis factor.
Discussion
Our analysis of 3 patient cohorts with different cancers (CLL, AML, and breast cancer) revealed that venetoclax has limited impact on circulating T and NK cells in vivo at the numerical, phenotypic, functional, and transcriptional levels. Despite preexisting T-cell defects in patients with leukemia,28 these findings provide reassurance that the cellular and molecular targets of immune checkpoint inhibitors are not perturbed by venetoclax therapy.
It is important to note that our study of patients with leukemia focused only on circulating leukocytes after venetoclax monotherapy. The different lymphocyte compositions and microenvironments of lymphoid or nonlymphoid tissues may engender different sensitivity to treatment, as observed by de Weerdt et al, after venetoclax and obinotuzumab treatment.16 In the context of this combination therapy, it should be noted that obinotuzumab treatment depletes CD4+ and NK cells,29 although this effect is thought to be indirect, caused by the decrease of tumor cells. Likewise, the Bruton tyrosine kinase inhibitor, ibrutinib, can revert T-cell defects in patients with CLL,30 and indeed, venetoclax and ibrutinib combination therapy leads to T-cell immune recovery, but not the recovery of B cells, in patients with mantle cell lymphoma.15 Our data showing the minimal impact of venetoclax monotherapy on T cells in patients with CLL or AML suggest that venetoclax will also be compatible with other more directed approaches to enhance T-cell activity.
We observed no impact of venetoclax treatment on activation-induced T-cell proliferation or effector function in our breast cancer cohort, consistent with recent preclinical studies.12,13 As noted above, prior studies of patients with CLL or mantle cell lymphoma found that combination therapies incorporating venetoclax could restore T-cell function.15,16,30,31 We suggest that the baseline T-cell fitness of the patients with breast cancer analyzed here may be less compromised than that of patients with leukemia. Moreover, those studies also involved additional therapies (eg, ibrutinib and obinotuzumab) that may impact T-cell phenotype or function. Future single-cell studies of immune subsets in patients with leukemia are warranted to pinpoint the changes induced by different treatments.
Previous studies of T-cell sensitivity to venetoclax in vitro found evidence of reactive oxygen species induction or selective ablation of naïve populations.13,32 Although we did not observe such phenotypic or transcriptional changes in T cells following long-term venetoclax treatment in the cohort of patients with breast cancer, this likely reflects differences between the in vivo vs in vitro settings. These observations suggest that homeostatic processes can compensate for potential venetoclax-induced loss. Whether venetoclax treatment might impact other aspects of T-cell homeostasis and/or the TCR repertoire are important questions.
Our work provides rationale to explore venetoclax in combination with checkpoint molecule inhibitors. Indeed, several clinical trials combining venetoclax with inhibitors of PD-1 (#NCT04274907 and #NCT04284787), TIM-3 (#NCT04150029), OX-40 (#NCT03390296), or CD47 (#NCT04435691 and #NCT04778410) are in progress, suggesting that this dual-targeting approach may be clinically feasible.
Acknowledgments
The authors thank Naomi Sprigg, Kirsten Hogg, Elliot Surgenor, and the Victorian Cancer Biobank for sample coordination and technical assistance; Andrew Mitchell and Tian Zheng (University of Melbourne) for assistance with mass cytometry maintenance/operation; Terence Speed for advice on data visualization; and Emma Carrington for critical review of the manuscript.
This work was supported by grants and fellowships from the Australian National Health and Medical Research Council (NHMRC) fellowships (1089072) (C.E.T.), (1090236 and 1158024) (D.H.D.G.), (1078730 and 1175960) (G.J.L.), (1079560) (A.W.R.), and (1043149 and 1156024) (D.C.S.H.); Investigator grants (1177718) (M.-A.A.), (1174902) (A.W.R.), and (2007884) (L.J.H.); a Fulbright Australia-America Postdoctoral Fellowship (C.E.T.); Victorian Cancer Agency Fellowships (MCRF20026) (C.E.T.) and (ECRF21014) (R.T.); Sir Clive McPherson Family Research Fellowship (V.L.B.); Cancer Council of Victoria Grants-in-Aid (1146518 and 1102104) (D.H.D.G.); Perpetual Impact Philanthropy funding (C.E.T.); Australian NHRMC grants (2002618) (C.E.T.), (1016647, 1113577, 1016701, 1113133, and 1079560) (A.W.R. and D.C.S.H.), (1113133 and 1153049) (G.J.L.), (2013478) (D.C.S.H. and R.T.), and the Metcalf Family (A.H.W.); the Leukemia and Lymphoma Society, US, Specialized Center of Research grant (7015-18) (A.H.W., A.W.R., and D.C.S.H.); a Medical Research Future Fund grant (1141460) (A.H.W.); National Breast Cancer Foundation Australia (IIRS-19-004); Breast Cancer Research Foundation (BCRF-20-182) (G.J.L.); Rae Foundation (V.L.B.); Jack Brockhoff Foundation Early Career Research grant (JBF4847-2021) (L.J.H.); University of Melbourne Melbourne Research Scholarship and Melbourne International Fee Remission Scholarship (H.P.); and Investigator Initiated Study support from AbbVie and Genentech (Roche) for the breast cancer study (#ACTRN12615000702516) (G.J.L).
This work was performed in part at the Materials Characterisation and Fabrication Platform at the University of Melbourne and the Victorian Node of the Australian National Fabrication Facility with support from the Victorian Comprehensive Cancer Centre. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institutes Infrastructure Support Scheme.
Authorship
Contribution: C.E.T., R.T., A.W.R., M.-A.A., D.H.D.G., and D.C.S H. conceived and designed the study; C.M., M.-A.A., A.W.R., A.H.W., and G.J.L. were responsible for patient care and recruited patients; C.E.T., M.L., M.T., T.T., C.C.C., F.B., H.P., L.J.H., V.L.B., M.E.R., and R.T. collected and analyzed data; C.E.T. and R.T. wrote the first version of the manuscript; and all authors reviewed the data and contributed to critical revision of the manuscript.
Conflict-of-interest disclosure: The Walter and Eliza Hall Institute of Medical Research receives milestone and royalty payments related to venetoclax. Employees are entitled to receive benefits related to these payments; C.E.T., H.P., M.L., T.T., M.T., C.M., F.B., M.E.R., L.J.H., V.L.B., A.H.W., A.W.R., M.-A.A., G.J.L., D.C.S.H., R.T., and D.H.D.G. report receiving these benefits. A.W.R. holds a patent related to venetoclax. G.J.L. has had a consulting/advisory role with AbbVie and Pfizer; has received research funding from Amgen and Servier; and institutional support for investigator initiated clinical trials from Amgen, AbbVie, Genentech (Roche), and Pfizer. D.H.D.G. has received research funding from Servier. A.H.W. has received research funding from Servier and AbbVie and serves as a medical adviser to both companies. C.C.C. declares no competing financial interest.
Correspondence: Daniel H. D. Gray, The Walter and Eliza Hall Institute Immunology, 1G Royal Parade, Parkville, Melbourne 3052, Australia; e-mail: dgray@wehi.edu.au.
References
Author notes
∗H.P. and M.L contributed equally to this study.
^R.T. and D.H.D.G. are joint senior authors.
The cellular indexing of transcriptomes and epitopes by sequencing data reported in this article have been deposited in the European Genome-phenome Archive (accession number EGAS00001006241).
A data transfer agreement is required, and limitations are in place on the disclosure of germ line variants. Additional details are provided in the supplemental Methods for data analysis.
Data are available on request from the corresponding author, Daniel H. D. Gray (dgray@wehi.edu.au).
The full-text version of this article contains a data supplement.