TO THE EDITOR:
Intravascular large B-cell lymphoma (IVLBCL) is an extremely rare extranodal B-cell lymphoma characterized by the selective growth of large lymphoma cells inside the lumens of small- and intermediate-sized blood vessels. IVLBCL has an age-adjusted incidence of 0.095 per 1 000 000 and has a propensity to involve the skin and central nervous system.1 Presenting symptoms in IVLBCL are highly heterogenous and most commonly include cutaneous lesions. However, approximately one-third of patients present with only transient neurologic changes, making IVLBCL challenging to diagnose,2 and in some patients, a diagnosis may only be confirmed at autopsy. The biology of IVLBCL is not well characterized, and the mechanism behind the unusual localization of lymphoma cells within the lumen of the blood vessels is poorly understood.
Given the rarity of the disease and difficulties obtaining sufficient diagnostic material for study, limited genomic data from IVLBCL is available. Shimada et al3 demonstrated genomic variants mainly using cell-free DNA obtained from patients with IVLBCL using whole-exome sequencing. Schrader et al4 and Suehara et al5 performed targeted sequencing in IVLBCL cases. The mutational spectrum of IVLBCL from these studies shows similarities to the MCD6,7 subtype of diffuse large B-cell lymphoma (DLBCL), characterized by MYD88L265P/CD79B mutation. MCD subtype is dominated by activated B-cell–type DLBCL and has a significantly worse prognosis.7MYD88L265P/CD79B mutation also typically coexists at higher frequency in extranodal DLBCL.6
In this study, we identified 6 cases of IVLBCL in autopsy patients seen at Washington University in St. Louis between 1996-2018. The diagnosis of IVLBCL was made antemortem in 1 case and postmortem in the remaining 5. Slides were rereviewed by a boarded hematopathologist (E.J.D.) and 2 neuropathologists (J.C. and P.J.C.) to confirm the diagnosis (Figure 1A-D; supplemental Figure 5). All autopsies were performed under unrestricted permits for research studies. Areas involved by IVLBCL were microdissected using 1 mm core punches of paraffin block with approximate tumor content of >90%. Paired normal tissue was used from 3 patients. Exome libraries were prepared using extracted DNA from involved areas in each case and sequenced on an Illumina NovaSeq 6000. Data were analyzed using the Illumina DRAGEN Bio-IT Platform v3.8 workflow for single-nucleotide variant and indel calling. Variants were annotated using Annovar.8 Variants were excluded if the variant allele frequency was <2%, total depth was <8×, the variant was not predicted to alter protein coding, or the variant was present in normal populations in >0.2% of individuals (Supplemental Materials). Copy number alterations were analyzed using CNVkit.9
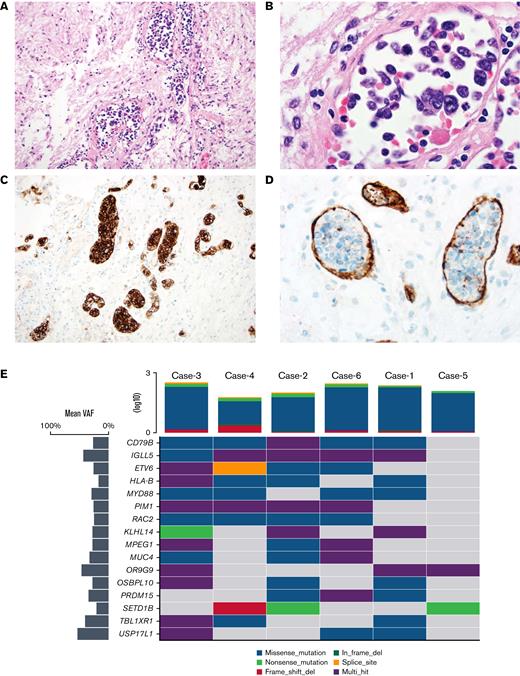
Histopathology and recurrent SNVs in IVLBCL. (A) H&E-stained section of the pituitary from case no. 4, which had a RAC2 switch 2 domain mutation p. R68Q (NM 002872.5) (×20 original magnification shows large, atypical lymphocytes within sized vessels). (B) High-power H&E image showing large, atypical lymphocytes within the vascular lumen (×100 original magnification). (C) A CD20 immunostain demonstrates large intravascular B cells (×20 original magnification). (D) A CD34 immunostain confirms that large cells are confined to the vascular lumen (×60 original magnification). (E) Oncoplot24 showing top cooccurring mutations detected in 6 patients with IVLBCL. Horizontal lines represent genes, and the vertical line indicates a single patient. The type of mutations is color-coded in the legend. Mean variant allele frequency for each gene is depicted in the bar plot on the left-hand side. The tumor mutation burden (TMB) for each patient is represented on the upper side of the plot. H&E, hematoxylin and eosin.
Histopathology and recurrent SNVs in IVLBCL. (A) H&E-stained section of the pituitary from case no. 4, which had a RAC2 switch 2 domain mutation p. R68Q (NM 002872.5) (×20 original magnification shows large, atypical lymphocytes within sized vessels). (B) High-power H&E image showing large, atypical lymphocytes within the vascular lumen (×100 original magnification). (C) A CD20 immunostain demonstrates large intravascular B cells (×20 original magnification). (D) A CD34 immunostain confirms that large cells are confined to the vascular lumen (×60 original magnification). (E) Oncoplot24 showing top cooccurring mutations detected in 6 patients with IVLBCL. Horizontal lines represent genes, and the vertical line indicates a single patient. The type of mutations is color-coded in the legend. Mean variant allele frequency for each gene is depicted in the bar plot on the left-hand side. The tumor mutation burden (TMB) for each patient is represented on the upper side of the plot. H&E, hematoxylin and eosin.
Mean exome coverage per patient was 144.9×. The median number of coding variants detected per patient was 195 (supplemental Data). C>T transitions were the most common alteration (supplemental Figures 1-2). All 6 IVLBCL cases had recurrent genetic aberrations that are part of the “MCD type” of DLBCL6 and genes commonly implicated in somatic hypermutations in lymphomas10,11 (Figures 1 and 2A; supplemental Figure 1; supplemental Table 1). Five of 6 cases had CD79B (NM_001039933.3) mutations (4 variants at p.Y197 in exon 5 and 1 at p.L200 in exon 6) and had high cooccurrence with MYD88 p.L265P (n = 4), RAC2 (n = 4), and genes that are common targets of somatic hypermutations like IGLL5, ETV6, HLA-B, and PIM13,11 (Figure 2A; supplemental Figure 3).
Distinct genetic features of IVLBCL. (A) Cooccurrence of top 10 mutations detected in IVLBCL cohort. (B) Lollipop plot showing SNVs detected in switch 2 domain of Rho GTPase family member RAC2 in 4 patients of IVLBCL. (C-D) Copy number loss of CDKN2A on chromosome 9p21 in 2 patients detected in our cohort. SNV, Single nucleotide variants.
Distinct genetic features of IVLBCL. (A) Cooccurrence of top 10 mutations detected in IVLBCL cohort. (B) Lollipop plot showing SNVs detected in switch 2 domain of Rho GTPase family member RAC2 in 4 patients of IVLBCL. (C-D) Copy number loss of CDKN2A on chromosome 9p21 in 2 patients detected in our cohort. SNV, Single nucleotide variants.
RAC2 mutations were detected in 4 patients (66%) in the highly conserved switch 2 domain12,13 (Figure 2B). Ras-related C3 botulinum toxin substrate 2 (RAC2) is a member of the ρ family GTPases involved in several cellular functions.14-16RAC2 is specifically expressed in hematopoietic lineage and regulates different cytoskeletal responses and homotypic adhesion in B cells.15,17 A similar pattern of mutations in the RAC2 gene was previously reported in cell-free DNA in only 5 cases (23.8%) within 21 patients with IVLBCL.3 Two large studies in primary DLBCL not otherwise specified (NOS) by Schmitz et al7 (n = 574) and Chapuy et al18 (n = 304) had only 10 cases with RAC2 mutations, of which, 4 cases had mutations in the switch 2 domain, which is between amino acid 56 to 71.19 Mutation in the switch 2 domain of RAC2 (p.E62K) results in sustained active guanosine triphosphate (GTP) bound RAC213,14 (supplemental Figure 4). RAC2 is activated on B-cell receptor (BCR) stimulation and acts as a link between BCR activation to cell adhesion via its physical association with B-cell linker protein.20RAC2 plays an important role in the adhesion of B cells to intracellular adhesion molecule-1 (ICAM-1), and constitutively active RAC2 creates more adherent B cells.16 In mice, RAC2 is one of the key genes responsible for lymphoma progression.21
Three cases in our cohort had a mutation in the tumor suppressor KLHL14, which has been previously reported in IVLBCL cases by Shimada et al.3KLHL14 inactivation in MCD cells maintains NF-κB signaling even in the presence of ibrutinib.22 Interestingly, a genome-wide CRISPR/Cas9 screen comparing effects of KLHL14 knockout on ibrutinib response in MCD-type DLBCL cell line TMD8 revealed RAC2 among the top 10 genes that sensitized to ibrutinib to a greater extent in wild-type KLHL14,22 and thus, the loss of KLHL14 might be used to predict ibrutinib treatment resistance in such cases. CDKN2A deletion at 9p21 is commonly seen in activated B-cell DLBCL and is associated with a poor prognosis.23 This deletion was present in ∼86% of the IVLBCL cases in a study by Shimada et al using cell-free DNA.3 In our cohort, we detected 2 out of 6 cases with CDKN2A deletion (Figure 2C-D)
Given the low incidence of IVLBCL and difficulty in establishing the diagnosis, we conducted a histologic review of sampled organs to determine the most frequent sites of involvement. Only 1 of the patients in our series had cutaneous disease documented before or at autopsy. Most frequently involved sites included the liver, heart, and kidney (5 of 6 cases). Central nervous system and pituitary were involved in 3 whereas spleen, lungs, and adrenals were involved in 2 of 6 cases. Organs with a low incidence of involvement included testes, prostate, and uterus (supplemental Data).
To our knowledge, this is the first study to evaluate histologically confirmed IVBCL tissue by exome sequencing as opposed to surrogate DNA sources such as cell-free DNA. Given the sparse nature of involvement in most IVBCL biopsies, the use of postmortem tissue allowed for wide sampling of involved areas that would be difficult to evaluate by antemortem biopsy. The study also demonstrates the continued utility of hospital-based autopsies in the genomic era. Using exome data obtained from 6 postmortem patients with IVLBCL, we demonstrate that the mutation spectrum of IVLBCL is most like the MCD type of DLBCL, characterized by gain-of-function mutations targeting CD79B BCR subunit and Toll-like receptor signaling adaptor MYD88. Recurrent mutations in the switch 2 domain of RAC2, which are rare in DLBCL not otherwise specified (NOS), were present in 4 of 6 patients in our cohort. The lower frequency of RAC2 mutations in IVLBCL cases reported by Shimada et al3 may be due to the use of surrogate circulating tumor DNA as opposed to actual tumor tissue used in this study. We hypothesize that activating RAC2 mutations in the background of chronic BCR signaling may lead to increased adhesion and sustained growth of B cells inside the lumen of the blood vessels. Targeted panels analyzing IVLBCL should include RAC2 gene in addition to the existing known recurrent mutations. Further studies are required to determine the role of RAC2 mutations in a chronic activated-BCR signaling state and the dynamics of interaction with the vascular lumen.
Acknowledgments: The authors would like to acknowledge Russell J.H. Ryan, University of Michigan, and David H. Spencer, Washington University, for their valuable insights.
Contribution: R.K. analyzed and interpreted the data and wrote the manuscript; J.C. and P.J.C. identified the autopsy cases, collected tumor samples for sequencing, and edited the manuscript; P.K. collected the clinical and histopathological details and edited the manuscript; J.R. extracted the DNA from the tumor tissue and organized the library preparation; A.D., L.C., C.F., and R.F. prepared and sequenced the NGS library; S.N.S. analyzed the data; and E.J.D. conceptualized the study, interpreted the data, and edited the manuscript
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric J. Duncavage, Department of Pathology and Immunology, Washington University in St. Louis, 660 Euclid Ave #8118, St. Louis, MO 63105; e-mail: duncavagee@wustl.edu.
References
Author notes
Data sharing requests should be sent to Eric J. Duncavage (duncavagee@wustl.edu).
The full-text version of this article contains a data supplement.