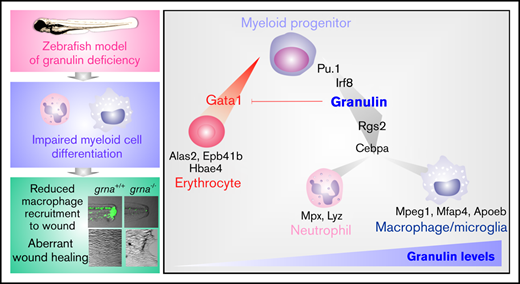
Key Points
Granulin a drives macrophage and neutrophil differentiation downstream of Pu.1 and Irf8 in normal and emergency myelopoiesis.
Macrophages fail to recruit to the wound in the absence of grna, resulting in abnormal wound healing.
Abstract
Granulin is a pleiotropic protein involved in inflammation, wound healing, neurodegenerative disease, and tumorigenesis. These roles in human health have prompted research efforts to use granulin to treat rheumatoid arthritis and frontotemporal dementia and to enhance wound healing. But how granulin contributes to each of these diverse biological functions remains largely unknown. Here, we have uncovered a new role for granulin during myeloid cell differentiation. We have taken advantage of the tissue-specific segregation of the zebrafish granulin paralogues to assess the functional role of granulin in hematopoiesis without perturbing other tissues. By using our zebrafish model of granulin deficiency, we revealed that during normal and emergency myelopoiesis, myeloid progenitors are unable to terminally differentiate into neutrophils and macrophages in the absence of granulin a (grna), failing to express the myeloid-specific genes cebpa, rgs2, lyz, mpx, mpeg1, mfap4, and apoeb. Functionally, macrophages fail to recruit to the wound, resulting in abnormal healing. Our CUT&RUN experiments identify Pu.1, which together with Irf8, positively regulates grna expression. In vivo imaging and RNA sequencing experiments show that grna inhibits the expression of gata1, leading to the repression of the erythroid program. Importantly, we demonstrated functional conservation between the mammalian granulin and the zebrafish ortholog grna. Our findings uncover a previously unrecognized role for granulin during myeloid cell differentiation, which opens a new field of study that can potentially have an impact on different aspects of human health and expand the therapeutic options for treating myeloid disorders such as neutropenia or myeloid leukemia.
Introduction
Neutrophils and macrophages differentiate from myeloid progenitors and are essential for clearing infections and promoting tissue repair. In addition, recent studies have elucidated a multitude of other functions beyond their classical roles in inflammation. For instance, we and others have demonstrated that neutrophils and macrophages are critical during hematopoietic stem cell specification.1-5 Numerous studies have shown that microglia, the tissue-resident macrophages of the brain, are involved in neurodegenerative disease.6,7 In addition, tumor-associated macrophages and tumor-associated neutrophils can be key in contributing to tumor metastasis. The tremendous implications that neutrophils and macrophages have in tissue homeostasis and how their disruption leads to human disease have put these cells in the spotlight of many recent investigations. The identification of new molecular regulators of myeloid cell differentiation thus can potentially have a broad impact on human health.
Granulin (GRN) is a protein with pleiotropic function that contains several cysteine-rich motifs unique to this molecule.8 It was first isolated from leukocytes,9 and it is known to regulate inflammation, wound healing, and tissue growth. It is also involved in neurodegenerative diseases, lipofuscinosis, and tumorigenesis.10 Granulin can bind to a wide variety of receptors, including tumor necrosis factor receptors, ephrin type-A receptor 2 (EphA2), Notch, Toll-like receptor 9 (TLR9), low-density lipoprotein receptor-related protein 1 (LRP1), and sortilin1.11 Consequently, granulin can modulate diverse signaling pathways such as NF-kB, WNT, MAPK/ERK, PI3K/Akt, and FAK.12-18
Many studies support a critical role for granulin during infection and inflammation. Although granulin can function as an anti-inflammatory factor, it can also exert a pro-inflammatory role. GRN knockout mice fail to clear Listeria monocytogenes, and their macrophages express high levels of pro-inflammatory cytokines. However, it is surprising that only a few macrophages were found in infected organs.19 Moreover, granulin functions as an antagonist of tumor necrosis factor signaling, thus playing a critical role in the pathogenesis of inflammatory diseases,20 and it has been reported as a promising therapeutic target for rheumatoid arthritis, psoriasis, and osteoarthritis.21-24
Several studies have demonstrated that granulin facilitates wound healing (for review, see Jian et al25 ). Administering GRN to fresh wounds increased the cell counts of neutrophils, macrophages, and fibroblasts, which collectively facilitated healing.26 In addition, it has been shown that macrophages produce granulin as a key regulatory factor in the processes of inflammation and wound healing.19
Mammals encode a single granulin gene (GRN) expressed in most tissues.27,28 However, human hematopoietic cells from the myeloid lineage contain the highest levels of granulin transcripts, being one of the most abundant transcripts in macrophages29 and monocyte-derived dendritic cells.30 Zebrafish (Danio rerio) possess many features that make them an ideal model of embryonic and adult hematopoiesis.31-35 A whole-genome duplication event in teleosts after evolutionary separation from the mammalian lineage resulted in 2 zebrafish granulin orthologs: granulin a (grna) and granulin b (grnb).36 Loss-of-function experiments, in combination with in vivo imaging and intracellular flow cytometry, demonstrate here that grna deficiency, but not grnb deficiency, leads to the loss of neutrophils and macrophages as a result of failure in differentiation from myeloid progenitors during embryonic development. In addition, examination of adult grna-mutant zebrafish shows failure in neutrophil differentiation in the adult hematopoietic system. Mechanistically, we have performed Cleavage Under Targets and Release Using Nuclease (CUT & RUN) and have shown that the master transcription factor of myeloid differentiation, Pu.1, directly binds granulin enhancers triggering its expression. Moreover, our studies demonstrate that Irf8, one of the main transcription factors that regulates macrophage development, also acts upstream of granulin. The regulation of mammalian GRN by PU.1 and IRF8 was confirmed by using empiric-based databases that highlight functional conservation between species in myeloid differentiation. Our functional studies in vivo demonstrate that loss of grna leads to defective recruitment of myeloid cells to wounds because of a lack of mature macrophages and neutrophils. This results in aberrant collagen deposition within the scar and therefore defective wound healing. Altogether, by using our zebrafish model of granulin deficiency, we have discovered that granulin is an essential regulator of myeloid cell differentiation. This study therefore opens a new field of investigation that will help shed light on the pleiotropic functions of this enigmatic protein and facilitate its use as a therapeutic target.
Materials and methods
Zebrafish husbandry
Wild-type AB* and transgenic zebrafish embryos and adults were mated, staged, raised, and processed as described37 in a circulating aquarium system at 28°C. See supplemental Data.
MO injection
Antisense targeting morpholinos (MOs) (Gene Tools) were resuspended in water at 0.2 to 2 mM and injected into 1-cell stage embryos. See supplemental Data.
qRT-PCR analysis
We performed quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. For more details, see supplemental Data.
Flow cytometry
Embryos were dechorionated with pronase, anesthetized in tricaine, and dissociated with liberase. The resulting suspension was filtered with a 30-μm cell strainer and flow cytometry was used for acquisition. Fluorescence-activated cell sorting (FACS) was performed on a FACS LSR-Fortessa flow cytometer. See supplemental Data.
In situ hybridization
Whole-mount in situ hybridization (WISH) was carried out as described.38 Probes for the grna, grnb, pu.1, mfap4, mpx, and apoeb transcripts were generated using the DIG RNA Labeling Kit (Roche Applied Science) from linearized plasmids. Fluorescence in situ hybridization (FISH) was performed as previously described.39 See supplemental Data.
WIHC and TUNEL
Whole-mount immunohistochemistry (WIHC) for immunofluorescence staining of P-H3Ser10 in 48-hours postfertilization (hpf) Tg(Mpx:eGFP) zebrafish embryos was performed as previously described.40 We also performed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays. See supplemental Data.
Fin amputation and enumeration of myeloid cells
See supplemental Data.
Cytology
See supplemental Data.
Morphological analyses and differential cell counts of kidney marrow hematopoietic cells
See supplemental Data.
CUT&RUN
See supplemental Data.
RNA sequencing (RNA-seq)
See supplemental Data.
Results
grna expression is restricted to myeloid cells in the zebrafish embryo
Despite mammalian granulin messenger RNA being among the most abundant transcripts in human macrophages and other myeloid cell lineages,41 functional studies on its potential roles in vivo have not been reported. This prompted us to identify an animal model of granulin deficiency amenable to determining the role of granulin during myelopoiesis.
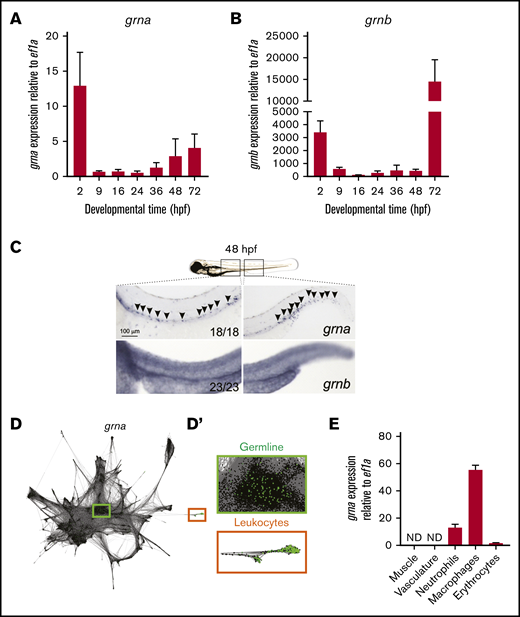
qPCR in zebrafish embryos showed that grna and grnb transcripts were maternally transferred because they were detected at 2 hpf. Zygotic grna and grnb transcripts (detected from 9 hpf) were detected throughout all embryonic stages evaluated (Figure 1A-B). grna and grnb were expressed by all cells in the early zygote as assessed by WISH (supplemental Figure 1). However, grna transcripts were restricted to the embryonic hematopoietic areas in the zebrafish at 48 hpf, including the dorsal aorta region and caudal hematopoietic tissue (CHT) (Figure 1C). In contrast, grnb was detected throughout the embryo (Figure 1C, lower panels). To confirm the cellular origin of grna expression, we used SPRING (a tool for interactive exploration of single-cell data).42 grna expression (green dots) was restricted to germline cells (green box) in the early zygote and leukocytes (orange box) at 24 hpf (Figure 1D-D'), further validating our WISH results. In contrast, grnb was expressed in all tissues (data not shown).
grna expression is restricted to the myeloid cell lineage during embryo development. Expression of grna (A) and grnb (B) during zebrafish embryonic and larval development. The messenger RNA (mRNA) levels were determined by real-time qPCR in 10 to 30 pooled larvae at the indicated times. The gene expression is normalized against ef1a; each bar represents the mean ± standard error of the mean (SEM) from 2 independent experiments. (C) Expression of grna (upper panel) and grnb (lower panel) by WISH at 48 hpf. Black arrowheads denote grna expression by distinct individual cells. Note that the grnb expression pattern is ubiquitous. Anterior is to the left, dorsal to the top. Numbers represent embryos with displayed phenotype. (D-D′) Single-cell RNA-seq graph showing grna expression (green dots) using the online tool SPRING by Wagner et al.42 Dots represent single cells from 4 hpf (center) to 24 hpf (periphery) zebrafish embryos. The cells that expressed grna (green dots) are magnified in panel D′. Notice that grna expression is restricted to germline cells (green box) and leukocytes (orange box). (E) Muscle (Myf5:eGFP+), vasculature (Flk:mcherry+; Gata1:DsRed–), neutrophils (Mpx:eGFP+), macrophages (Mpeg1:eGFP+), and erythrocytes (LCR:eGFP+) cells from dissected embryos were purified by FACS, and qPCR was performed for grna. Levels of grna transcripts along the x-axis are shown relative to the housekeeping gene ef1a. Bars represent mean ± SEM of 2 to 3 independent samples. ND, not detected.
grna expression is restricted to the myeloid cell lineage during embryo development. Expression of grna (A) and grnb (B) during zebrafish embryonic and larval development. The messenger RNA (mRNA) levels were determined by real-time qPCR in 10 to 30 pooled larvae at the indicated times. The gene expression is normalized against ef1a; each bar represents the mean ± standard error of the mean (SEM) from 2 independent experiments. (C) Expression of grna (upper panel) and grnb (lower panel) by WISH at 48 hpf. Black arrowheads denote grna expression by distinct individual cells. Note that the grnb expression pattern is ubiquitous. Anterior is to the left, dorsal to the top. Numbers represent embryos with displayed phenotype. (D-D′) Single-cell RNA-seq graph showing grna expression (green dots) using the online tool SPRING by Wagner et al.42 Dots represent single cells from 4 hpf (center) to 24 hpf (periphery) zebrafish embryos. The cells that expressed grna (green dots) are magnified in panel D′. Notice that grna expression is restricted to germline cells (green box) and leukocytes (orange box). (E) Muscle (Myf5:eGFP+), vasculature (Flk:mcherry+; Gata1:DsRed–), neutrophils (Mpx:eGFP+), macrophages (Mpeg1:eGFP+), and erythrocytes (LCR:eGFP+) cells from dissected embryos were purified by FACS, and qPCR was performed for grna. Levels of grna transcripts along the x-axis are shown relative to the housekeeping gene ef1a. Bars represent mean ± SEM of 2 to 3 independent samples. ND, not detected.
We next validated the restricted tissue specificity of grna in the late embryo (36-48 hpf) by qPCR of cells submitted to FACS. grna was absent in nonhematopoietic cells and highly expressed by myeloid cells (Figure 1E). Macrophages expressed 5 times more grna transcripts than neutrophils and 50 times more transcripts than developing erythrocytes (Figure 1E). Altogether, these results demonstrate that in zebrafish, grna expression is restricted to the myeloid cell lineage, whereas grnb is ubiquitously expressed throughout the embryo.
grna is required for proper myeloid development
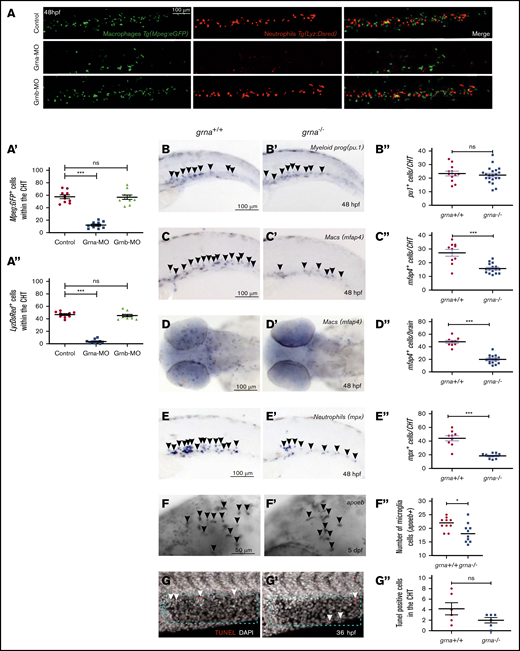
To test whether grna is essential for proper myeloid differentiation, we performed loss-of-function experiments for grna and grnb. In the zebrafish embryo, myeloid progenitors, macrophages, and neutrophils can be visualized by expression of pu.1, mfap4, and mpx/lyz, respectively. grna knockdown using previously validated MOs43 (supplemental Figure 2A) significantly reduced in vivo macrophage (mfap4) and neutrophil (mpx) numbers (Figure 2A-A"). An additional grna MO (grna-MO2) also showed decreased macrophage numbers by flow cytometry (supplemental Figure 2D-E). In contrast, Grnb knockdown (supplemental Figure 2B-C") led to normal numbers of neutrophils or macrophages (Figure 2A-A'). To verify these results, we made use of existing grna-null mutants.44 Although myeloid progenitor (pu.1+) numbers were similar (Figure 2B-B"), macrophages and neutrophils were significantly decreased in grna−/− compared with grna+/+ embryos (Figure 2C-E") as well as developing microglia (Figure 2F-F"). Intracellular flow for Mfap4 showed a fivefold decrease in grna−/− (supplemental Figure 2F-G). To ensure that the myeloid defects observed in the absence of grna were not the result of decreased proliferation, we performed WIHC for phospho-histoneH3 (p-H3) in Mpx:eGFP transgenic animals and found no colocalization in grna+/+ embryos, suggesting that neutrophils do not proliferate during embryonic development (supplemental Figure 2H). To validate this result, we performed a 5-ethynyl-2′-deoxyuridine incorporation assay in combination with WIHC using specific antibodies for macrophages (anti-Mfap4) and neutrophils (anti-Mpx). Confocal images of CHTs from grna−/− or grna+/+ embryos showed no proliferative myeloid cells (supplemental Figure 2I-J), demonstrating that grna does not have an impact on myeloid cell proliferation. In addition, a TUNEL assay and confocal analysis of the CHT of grna−/− and grna+/+ control embryos showed no increase in apoptotic cells in the absence of grna (Figure 2G-G”), indicating that grna leads to differentiation of myeloid cells rather than their survival. Altogether, these experiments demonstrate that grna is essential for the differentiation of myeloid progenitors into macrophages and neutrophils.
Absence of grna leads to decreased myeloid differentiation during embryo development. (A) Representative fluorescence images, and quantification by fluorescence microscopy (A′,A″) of the tails of 48 hpf Mpeg1:eGFP; Lyz:DsRed double transgenic embryos injected with Grna mismatch control, Grna, or Grnb MOs. (B,B′-F,F′) WISH for the myeloid progenitor (pu.1), macrophage (mfap4), neutrophilic (mpx), and microglia (apoeb) markers in grna−/− and grna+/+ control embryos at 48 hpf (B,B″-E,E″) or 5 days post fertilization (dpf) (F-F″). Black arrowheads depict cells expressing the indicated transcripts. (B″,C″,D″,E″,F″) Enumeration of apoeb, pu1+, mfap4+, and mpx+–expressing cells shown in (B-F and B′-F′). Each dot represents the number of positive cells in the photographed area in each embryo. Bars represent mean ± SEM. *P < .05; ***P < .001. (D) Magnification ×10. (G,G′) Maximum projections of the CHT (dotted blue region) of 36 hpf (G) grna+/+ or (G’) grna−/− embryos assayed for TUNEL (red) and 4′,6-diamidino-2-phenylindole (DAPI) (white nuclei). White arrowheads denote apoptotic nuclei. (G″) Enumeration of apoptotic cells in the CHT are quantified in panel G″. Horizontal lines indicate mean ± SEM. ***P < .001. ns, not significant.
Absence of grna leads to decreased myeloid differentiation during embryo development. (A) Representative fluorescence images, and quantification by fluorescence microscopy (A′,A″) of the tails of 48 hpf Mpeg1:eGFP; Lyz:DsRed double transgenic embryos injected with Grna mismatch control, Grna, or Grnb MOs. (B,B′-F,F′) WISH for the myeloid progenitor (pu.1), macrophage (mfap4), neutrophilic (mpx), and microglia (apoeb) markers in grna−/− and grna+/+ control embryos at 48 hpf (B,B″-E,E″) or 5 days post fertilization (dpf) (F-F″). Black arrowheads depict cells expressing the indicated transcripts. (B″,C″,D″,E″,F″) Enumeration of apoeb, pu1+, mfap4+, and mpx+–expressing cells shown in (B-F and B′-F′). Each dot represents the number of positive cells in the photographed area in each embryo. Bars represent mean ± SEM. *P < .05; ***P < .001. (D) Magnification ×10. (G,G′) Maximum projections of the CHT (dotted blue region) of 36 hpf (G) grna+/+ or (G’) grna−/− embryos assayed for TUNEL (red) and 4′,6-diamidino-2-phenylindole (DAPI) (white nuclei). White arrowheads denote apoptotic nuclei. (G″) Enumeration of apoptotic cells in the CHT are quantified in panel G″. Horizontal lines indicate mean ± SEM. ***P < .001. ns, not significant.
Absence of grna leads to long-lasting myelopoiesis defects
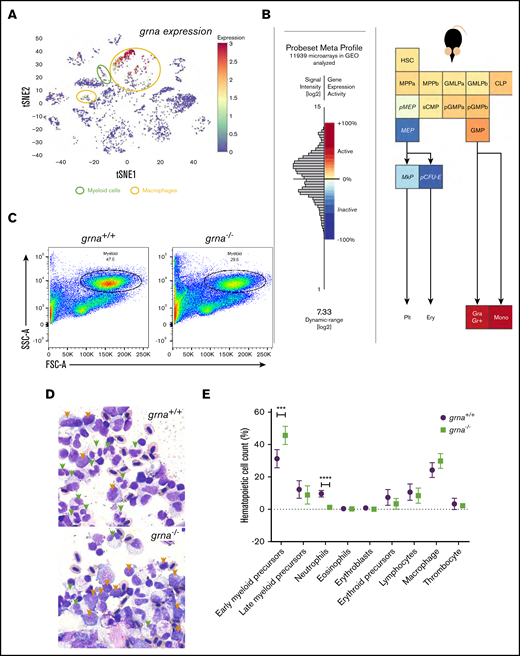
We next sought to investigate whether grna was also essential for adult myelopoiesis. We took advantage of the interactive single-cell RNA-seq (scRNA-seq) data published45 for adult wild-type zebrafish kidneys. grna expression was restricted to clusters of macrophages (yellow) and myeloid cells (green) (Figure 3A; supplemental Figure 3A). In contrast, grnb transcripts were present at low levels in hematopoietic cells (supplemental Figure 3A). We next wanted to determine whether mammalian granulin was also upregulated in myeloid cells. First, we used Gene Expression Commons46 to query the dynamic range of granulin within microarrays of the mouse (Mus musculus) hematopoietic system (https://gexc.riken.jp/models/3/genes/Grn?q=Grn). As shown in Figure 3B, Grn expression was active in hematopoietic stem cells, upregulated in granulocyte/macrophage progenitors, and reached its highest expression in granulocytes and monocytes. In contrast, cells of the megakaryocyte and erythrocyte lineages drastically downregulated Grn expression. Second, to confirm these results at the single-cell level, we used the hematopoietic single-cell interactive gene viewer from Olsson et al,47 which was based on mouse sorted hematopoietic cells, and found that murine Grn is highly expressed in myeloid cells, including monocytes, granulocytes, and myelocytes, mimicking the expression of PU.1 (supplemental Figure 3B, black squares). Both Grn and Pu.1 were downregulated in cells with erythroid lineage (supplemental Figure 3B, purple squares). Altogether, these data show conserved expression for the mammalian granulin and the zebrafish ortholog grna in hematopoietic lineages.
Granulin expression is upregulated in vertebrate myeloid cells and is essential for myeloid cell differentiation during adult hematopoiesis. (A) t-Distributed stochastic neighbor embedding (t-SNE) analysis showing grna expression levels (red, high; orange and yellow, medium; blue, absent) of single cells sequenced from wild-type zebrafish kidney marrow using the online visualizer Single Cell inDrops RNA-Seq Visualization of Adult Zebrafish Whole Kidney Marrow (https://molpath.shinyapps.io/zebrafishblood/#pltly).45 The main tSNE clusters identified expressing grna are denoted by open circles. Yellow open circles represent clusters defined as “macrophages.” Open green circles are grna-expressing clusters whose cells were identified as “myeloid cells.” (B) Mouse hematopoietic model showing the dynamic expression of Grn derived from microarray data (Affymetrix Mouse Genome 430 2.0 Array). Notice that lymphocyte differentiation beyond common lymphoid progenitor (CLP) is not shown here. (C) Representative flow cytometric light scatter profile showing the different hematopoietic populations present in grna+/+ (left) and grna−/− (right) kidney marrow. (D) Representative pictures from grna+/+ and grna−/− whole kidney marrows cytospins stained with Wright-Giemsa stain showing increased early myeloid precursors (orange arrowheads) and decreased mature neutrophils (green arrowheads) in the absence of grna (bottom panel) compared with grna+/+ control siblings (upper panel). Magnification ×100. (E) Manual quantification of kidney marrow hematopoietic cells in grna−/− (green squares, n = 5) compared with control grna+/+ (black dots, n = 5) from 2 independent experiments. Horizontal lines and error bars indicate mean ± SEM. ***P < .001; ****P < .0001. Ery, erythrocytes; FSC, forward scatter; GMLP, granulocyte-monocyte-lymphoid progenitor; GMP, granulocyte-macrophage progenitor; Gra Gr+, granulocyte; HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; MkP, megakaryocyte progenitor; Mono, monocyte; MPP, multipotential progenitor; pCFU-e, pre-colony-forming unit-erythroid; pGMP, pre-granulocyte-macrophage progenitor; Plt, platelet; pMEP, pre-megakaryocyte-erythroid progenitor; SSC, side scatter; sCMP, strict common myeloid progenitor.
Granulin expression is upregulated in vertebrate myeloid cells and is essential for myeloid cell differentiation during adult hematopoiesis. (A) t-Distributed stochastic neighbor embedding (t-SNE) analysis showing grna expression levels (red, high; orange and yellow, medium; blue, absent) of single cells sequenced from wild-type zebrafish kidney marrow using the online visualizer Single Cell inDrops RNA-Seq Visualization of Adult Zebrafish Whole Kidney Marrow (https://molpath.shinyapps.io/zebrafishblood/#pltly).45 The main tSNE clusters identified expressing grna are denoted by open circles. Yellow open circles represent clusters defined as “macrophages.” Open green circles are grna-expressing clusters whose cells were identified as “myeloid cells.” (B) Mouse hematopoietic model showing the dynamic expression of Grn derived from microarray data (Affymetrix Mouse Genome 430 2.0 Array). Notice that lymphocyte differentiation beyond common lymphoid progenitor (CLP) is not shown here. (C) Representative flow cytometric light scatter profile showing the different hematopoietic populations present in grna+/+ (left) and grna−/− (right) kidney marrow. (D) Representative pictures from grna+/+ and grna−/− whole kidney marrows cytospins stained with Wright-Giemsa stain showing increased early myeloid precursors (orange arrowheads) and decreased mature neutrophils (green arrowheads) in the absence of grna (bottom panel) compared with grna+/+ control siblings (upper panel). Magnification ×100. (E) Manual quantification of kidney marrow hematopoietic cells in grna−/− (green squares, n = 5) compared with control grna+/+ (black dots, n = 5) from 2 independent experiments. Horizontal lines and error bars indicate mean ± SEM. ***P < .001; ****P < .0001. Ery, erythrocytes; FSC, forward scatter; GMLP, granulocyte-monocyte-lymphoid progenitor; GMP, granulocyte-macrophage progenitor; Gra Gr+, granulocyte; HSC, hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; MkP, megakaryocyte progenitor; Mono, monocyte; MPP, multipotential progenitor; pCFU-e, pre-colony-forming unit-erythroid; pGMP, pre-granulocyte-macrophage progenitor; Plt, platelet; pMEP, pre-megakaryocyte-erythroid progenitor; SSC, side scatter; sCMP, strict common myeloid progenitor.
To assess the effect of grna loss on myeloid cell differentiation in the adult, we collected whole kidney marrow from grna−/− and grna+/+ siblings and performed flow cytometry.48 The percentage of myeloid cells within the grna−/− kidney marrow was significantly decreased compared with grna+/+ control kidneys (Figure 3C; supplemental Figure 3C). We next cytospun grna−/− and grna+/+ control sibling kidney cell suspensions and stained the cells with Wright-Giemsa stain. Examination by a pathologist revealed that grna−/− had increased early myeloid precursors (Figure 3D, orange arrowheads) with decreased differentiation into mature neutrophils (green arrowheads) (Figure 3D-E), indicating an inhibition in myeloid maturation in the absence of grna. No differences were seen in body size between grna+/+ and grna−/− sibling fish (supplemental Figure 3D). Taken together, these data indicate that grna is essential for driving myeloid cell differentiation from early myeloid precursors in adult zebrafish.
grna inhibits gata1 expression
Next, we performed bulk RNA-seq in kidney marrow from adult grna−/− and grna+/+ siblings to identify genes whose expression was dysregulated in the absence of grna. We found 154 genes significantly downregulated in the grna mutants (supplemental Table 2) and 116 genes significantly upregulated (Figure 4A-C; supplemental Table 3). As expected, we found important myelopoiesis-related genes downregulated in grna mutants, including cebpa, rgs2, and apoeb (Figure 4B-C; supplemental Figure 4A). Almost half (64 of 154) (supplemental Table 4) were restricted to myeloid cell subpopulations when assessed by scRNA-seq45 (supplemental Figure 4A-B). In addition, many genes downregulated in the grna mutants have been shown to participate in inflammation and immune response, including il4r.1, irf2a, nfkbie, tlr5b, cxcl12a, hbegfa, and mlx (Figure 4B-C; supplemental Figure 4A-B).
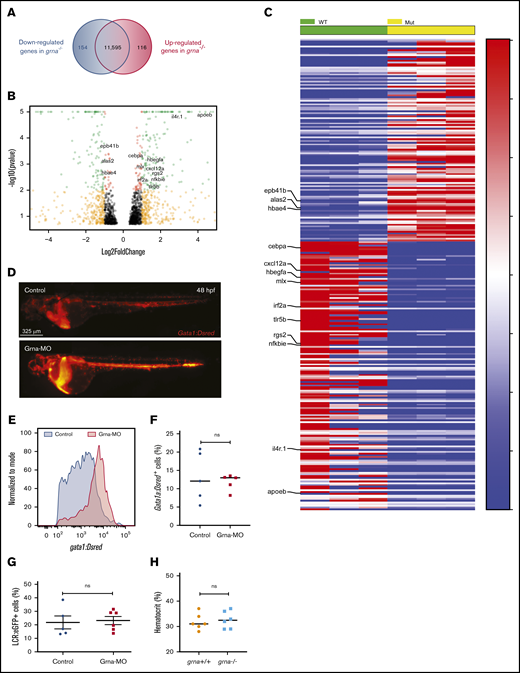
Grna inhibits gata1 expression. (A) RNA-seq analysis from grna−/− and grna+/+ adult zebrafish kidney marrows reveals 154 downregulated and 116 upregulated genes in grna−/− vs grna+/+ control. (B) Volcano plot obtained from DESeq2 analysis of grna−/− and grna+/+ kidney marrows. (C) Heat map of the enriched and depleted transcripts in kidney marrows from grna−/− vs grna+/+ adult fish. Color coding is based on rlog-transformed read count values. (D) Representative fluorescence images of 48 hpf Gata1:DsRed embryos injected with Grna-MO (bottom panel) and a 5-base Grna mismatch control (upper panel). (E) Quantification by flow cytometry showing the histogram of 3 pooled Gata1:DsRed embryos injected with Grna-MO (red) or mismatch Grna-MO control (gray). (F-G) Erythrocyte numbers quantified by flow cytometry of Gata1:DsRed (F) or LCR:eGFP (G) embryos injected with Grna-MO or Grna mismatch control MO. Dots represent independent biological replicates from 3 48-hpf Gata1:DsRed+ pooled embryos (F) or 3 48-hpf LCR:eGFP+ pooled embryos (G). (H) Hematocrit (%) in grna+/+ and grna−/− adult zebrafish. Horizontal lines indicate mean ± SEM. MUT, mutated; WT, wild-type.
Grna inhibits gata1 expression. (A) RNA-seq analysis from grna−/− and grna+/+ adult zebrafish kidney marrows reveals 154 downregulated and 116 upregulated genes in grna−/− vs grna+/+ control. (B) Volcano plot obtained from DESeq2 analysis of grna−/− and grna+/+ kidney marrows. (C) Heat map of the enriched and depleted transcripts in kidney marrows from grna−/− vs grna+/+ adult fish. Color coding is based on rlog-transformed read count values. (D) Representative fluorescence images of 48 hpf Gata1:DsRed embryos injected with Grna-MO (bottom panel) and a 5-base Grna mismatch control (upper panel). (E) Quantification by flow cytometry showing the histogram of 3 pooled Gata1:DsRed embryos injected with Grna-MO (red) or mismatch Grna-MO control (gray). (F-G) Erythrocyte numbers quantified by flow cytometry of Gata1:DsRed (F) or LCR:eGFP (G) embryos injected with Grna-MO or Grna mismatch control MO. Dots represent independent biological replicates from 3 48-hpf Gata1:DsRed+ pooled embryos (F) or 3 48-hpf LCR:eGFP+ pooled embryos (G). (H) Hematocrit (%) in grna+/+ and grna−/− adult zebrafish. Horizontal lines indicate mean ± SEM. MUT, mutated; WT, wild-type.
Among the upregulated genes in grna−/−, we found genes that drive red blood cell development (alas2), erythrocyte shape (epb41b), and hemoglobin transport (hbae4) (Figure 4B-C; supplemental Figure 4A). Thus, 27 of 116 upregulated genes were restricted to erythrocytes and, to a lesser extent, platelets (supplemental Figure 4; supplemental Table 5). We then hypothesized that grna might inhibit the erythroid program. To address this, we injected Grna-MO and Grna mismatch control MO into Tg(Gata1:DsRed) reporter zebrafish embryos. Gata1 is a master transcription factor that drives erythroid cell differentiation. We found a significant increase in DsRed expression in Grna morphants compared with control siblings (Figure 4D-E), whereas erythrocyte numbers were similar during embryo development (Figure 4F-G) as was hematocrit percentage in adult grna−/− and control grna+/+ (Figure 4H). Altogether, these results show that granulin reduces gata1 expression, but this is not sufficient to have an impact on the number of mature erythrocytes produced.
Pu.1 and Irf8 control grna expression in zebrafish, and this regulatory mechanism is conserved for mammalian GRN
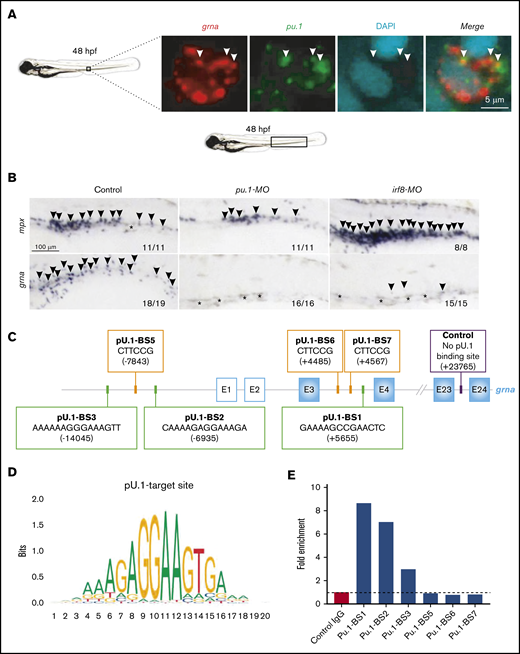
Pu.1 is a master transcription factor that leads to myeloid cell specification. We hypothesized that grna acted downstream of Pu.1. Double FISH analysis for grna and pu.1 showed colocalization at single-cell resolution (Figure 5A). To demonstrate that Pu.1 genetically acts upstream of grna, we used a specific MO against Pu.149 and found that grna expression was abolished (Figure 5B; supplemental Figure 5G) and expression of mpx and mpeg was decreased (Figure 5B; supplemental Figure 5A). To determine whether Pu.1 directly bound grna enhancers, we used Tomtom (http://meme-suite.org/tools/tomtom)50 and the PU.1 matrix ID: MA0080.5 (http://jaspar.genereg.net/matrix/MA0080.5/)51 (Figure 5D) and found several putative binding sites (BSs) (Figure 5C). We performed CUT&RUN for Pu.1 followed by qPCR to amplify each predicted Pu.1 BS. Putative Pu.1 BS5, BS6, and BS7 showed no enrichment compared with control isotype immunoglobulin G antibody, but Pu.1 BS1, BS2, and BS3 showed two- to eightfold enrichment (Figure 5E), demonstrating that Pu.1 directly binds grna regulatory sequences. To gain further insight into the grna regulatory gene network, we knocked down Irf8. It has been demonstrated that Pu.1 acts upstream of Irf8 and that both transcription factors cooperate to regulate granulocyte-macrophage fate decisions in myeloid progenitors and maturation of macrophage precursors.52-54 Irf8 depletion led to increased mpx expression (Figure 5B), loss of macrophages (supplemental Figure 5A) as previously reported,1 and decreased grna expression (Figure 5B; supplemental Figure 5G). By using Gene Expression Commons,46 we found that Irf8 is upregulated in myeloid precursors (supplemental Figure 5B). Together, these results indicate that Irf8 acts genetically upstream of grna.
TF network that controls granulin expression. (A) Double fluorescence in situ hybridization for grna (red) and pu.1 (green) shows colocalization of both transcripts. Nuclei are stained with DAPI (blue). Pictures were taken by confocal microscopy from the tail region of 48-hpf zebrafish embryos. Each image is a 1-µm z slice. (B) WISH for the neutrophilic marker mpx (upper panels, arrowheads) or grna (lower panels, arrowheads) after MO knockdown of Pu.1 (middle panels) or Irf8 (right panels) compared with standard MO control (left panels) at 48 hpf. Pu.1 or Irf8 knockdown abolished grna expression. Asterisks denote natural pigmentation occurring in the tail of zebrafish embryos. (C) Schematic representation of the grna gene and its 5′ enhancer locus denoting 6 putative Pu.1 BSs. Pu.1 BS1-3 (green squares) were found by searching the human PU.1 target site nucleotide matrix represented in panel D using the motif comparison tool Tomtom. Pu.1 BS5-7 (orange squares) were found searching the PU.1 target site nucleotide matrix available in ConSite (http://consite.genereg.net/cgi-bin/consite). BS positions are denoted by bracketed numbers. Positive numbering starts with +1 at the A of the grna ATG translation initiation (start) codon. Nucleotides upstream (5′) of the grna ATG translation initiation codon (start) are marked with a minus sign. E, exon. White squares with a blue line indicate grna exons from the untranslated region. Blue squares represent grna coding exons. Control primers to amplify 71 base pairs of a locus within the grna gene with no predicted Pu.1 BSs for CUT&RUN normalization are indicated with a purple square. (E) CUT&RUN experiment was performed in fresh zebrafish kidney marrows from adult AB* using a Pu.1 or control immunoglobulin G (IgG) antibody. Fold enrichment of Pu.1-associated DNA fragments was identified by qPCR using primers flanking the BSs denoted in panel C. To calculate the fold enrichment, qPCR results for each BS were normalized against spike-in DNA as described86 and control primers that amplify a locus of the grna gene lacking predicted Pu.1 BSs. This experiment was performed 3 times with similar enrichments. Panel E is a representative experiment from 3 independent biological replicates performed. The primers used are shown in supplemental Table 1.
TF network that controls granulin expression. (A) Double fluorescence in situ hybridization for grna (red) and pu.1 (green) shows colocalization of both transcripts. Nuclei are stained with DAPI (blue). Pictures were taken by confocal microscopy from the tail region of 48-hpf zebrafish embryos. Each image is a 1-µm z slice. (B) WISH for the neutrophilic marker mpx (upper panels, arrowheads) or grna (lower panels, arrowheads) after MO knockdown of Pu.1 (middle panels) or Irf8 (right panels) compared with standard MO control (left panels) at 48 hpf. Pu.1 or Irf8 knockdown abolished grna expression. Asterisks denote natural pigmentation occurring in the tail of zebrafish embryos. (C) Schematic representation of the grna gene and its 5′ enhancer locus denoting 6 putative Pu.1 BSs. Pu.1 BS1-3 (green squares) were found by searching the human PU.1 target site nucleotide matrix represented in panel D using the motif comparison tool Tomtom. Pu.1 BS5-7 (orange squares) were found searching the PU.1 target site nucleotide matrix available in ConSite (http://consite.genereg.net/cgi-bin/consite). BS positions are denoted by bracketed numbers. Positive numbering starts with +1 at the A of the grna ATG translation initiation (start) codon. Nucleotides upstream (5′) of the grna ATG translation initiation codon (start) are marked with a minus sign. E, exon. White squares with a blue line indicate grna exons from the untranslated region. Blue squares represent grna coding exons. Control primers to amplify 71 base pairs of a locus within the grna gene with no predicted Pu.1 BSs for CUT&RUN normalization are indicated with a purple square. (E) CUT&RUN experiment was performed in fresh zebrafish kidney marrows from adult AB* using a Pu.1 or control immunoglobulin G (IgG) antibody. Fold enrichment of Pu.1-associated DNA fragments was identified by qPCR using primers flanking the BSs denoted in panel C. To calculate the fold enrichment, qPCR results for each BS were normalized against spike-in DNA as described86 and control primers that amplify a locus of the grna gene lacking predicted Pu.1 BSs. This experiment was performed 3 times with similar enrichments. Panel E is a representative experiment from 3 independent biological replicates performed. The primers used are shown in supplemental Table 1.
We next wanted to investigate whether the transcriptional network that regulated grna in zebrafish myeloid cells was also conserved in mammals. We queried whether PU.1 bound the human GRN promoter by using the regulatory feature of ensembl.org55 and found that PU.1 bound to the first intron of GRN (supplemental Figure 5C). Chromatin immunoprecipitation enrichment analysis56 demonstrated that IRF8 and CEBPB bound the Grn promoter in mice (supplemental Figure 5D). In addition, the hematopoietic and inflammatory transcription factors (TFs) RUNX1, GF1, NFE2L2, and RELA also bound mammalian GRN (supplemental Figure 5D). Harmonizome57 in combination with chromatin immunoprecipitation enrichment analysis identified mammalian PU.1 and IRF8 to be co-expressed with GRN (supplemental Figure 5E) and myeloid-specific genes such as MPEG, CD68, and TREM2 with a Pearson correlation >0.6 (supplemental Figure 5F). Taken together, these data demonstrate that in zebrafish and mammals, Pu.1 and Irf8 activate upstream granulin for myeloid cell differentiation.
grna is required for emergency myelopoiesis and macrophage recruitment to the wound
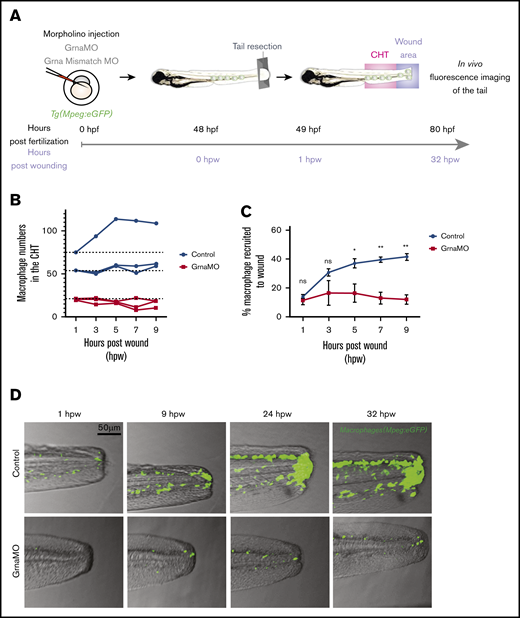
Emergency myelopoiesis is the proliferation and differentiation of hematopoietic progenitor cells toward the myeloid lineage as a result of increased demand following injury or infection.58 To investigate whether myeloid progenitors could respond to emergency myelopoiesis in the absence of Grna, Tg(Mpeg1:eGFP) embryos were injected with Grna or Grna control mismatch MOs, and caudal tails were resected (Figure 6A). Macrophage numbers increased in all control individuals at 9 hours post-wounding (hpw), but Grna morphants failed to generate more macrophages (Figure 6B), suggesting that Grna is required for emergency myelopoiesis. Although the absence of grna led to a remarkable decrease in macrophage number, a small percentage of macrophages were still present in grna mutants and morphants (Figure 2). Therefore, we determined whether these macrophages were able to respond and recruit to the injury site. As expected, the number of macrophages recruited to the wound were significantly decreased in the absence of Grna (Figure 6D). The percentage of macrophages that were recruited to the wound with respect to the total macrophage numbers was also significantly reduced in the absence of Grna (Figure 6C). These results indicate that Grna is essential for emergency myelopoiesis and that the macrophages produced in the absence of Grna have functional abnormalities.
Macrophages respond abnormally to injury in the absence of Grna. (A) Experimental workflow. Tg(Mpeg1:eGFP) 1-cell stage embryos were injected with either Grna MO or Grna mismatch MO. At 48 hpf, caudal tails were resected immediately after the end of the notochord. Fluorescence imaging of the tail (CHT, where the majority of neutrophils reside at this developmental time, and wound area) was taken every 2 hours from 1 hour post-wounding (hpw) to 32 hpw, and the number of neutrophils was quantified manually. (B) Neutrophil numbers in the CHT from individual Mpeg1:eGFP transgenic animals at 1, 3, 5, 7, and 9 hpw after depletion of Grna compared with Grna mismatch control morphants. (C) Percentage of Mpeg1:eGFP+ macrophages recruited to the wound region normalized to the total macrophage number in the tail (CHT and wound) in embryos injected with Grna MO (red line, n = 3) or Grna mismatch MO (blue line, n = 3) at indicated time points. Circle and square dots indicate means, and error bars indicate SEM. (D) Representative fluorescence images of tail fins from Mpeg1:eGFP Grna or Grna mismatch control morphant siblings at the indicated times. *P < .05; **P < .001.
Macrophages respond abnormally to injury in the absence of Grna. (A) Experimental workflow. Tg(Mpeg1:eGFP) 1-cell stage embryos were injected with either Grna MO or Grna mismatch MO. At 48 hpf, caudal tails were resected immediately after the end of the notochord. Fluorescence imaging of the tail (CHT, where the majority of neutrophils reside at this developmental time, and wound area) was taken every 2 hours from 1 hour post-wounding (hpw) to 32 hpw, and the number of neutrophils was quantified manually. (B) Neutrophil numbers in the CHT from individual Mpeg1:eGFP transgenic animals at 1, 3, 5, 7, and 9 hpw after depletion of Grna compared with Grna mismatch control morphants. (C) Percentage of Mpeg1:eGFP+ macrophages recruited to the wound region normalized to the total macrophage number in the tail (CHT and wound) in embryos injected with Grna MO (red line, n = 3) or Grna mismatch MO (blue line, n = 3) at indicated time points. Circle and square dots indicate means, and error bars indicate SEM. (D) Representative fluorescence images of tail fins from Mpeg1:eGFP Grna or Grna mismatch control morphant siblings at the indicated times. *P < .05; **P < .001.
Wound healing is impaired in grna mutants
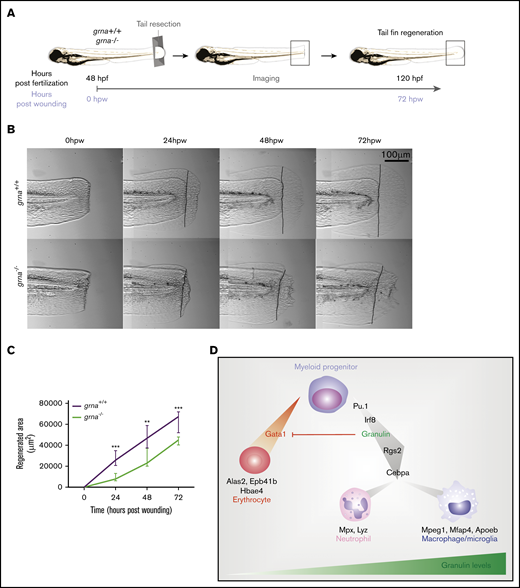
Macrophages are one of the main contributors to tissue repair in both mammals and zebrafish.59-64 Moreover, it is known that granulin facilitates wound healing by increasing macrophage numbers in the injured tissue.26 We hypothesized that wound healing would be impaired in grna−/− embryos because of the decreased and abnormal production of macrophages. To test our hypothesis, we performed tail fin resection in grna−/− and grna+/+ embryos and imaged regenerated tissue over 3 days (Figure 7A). Tissue regeneration was deeply impaired in grna−/− larvae (Figure 7B-C), and collagen organization was disrupted (supplemental Figure 6). Collagen fibers in grna+/+ were perpendicular to the wound edge, but aligned fibers were disarrayed in grna−/− (supplemental Figure 6). Altogether, these results link, for the first time, the role of granulin in wound healing with the myelopoietic defects described here and validate functional conservation between the mammalian granulin and the zebrafish ortholog grna. Overall, these data also reveal the TFs controlling grna expression and the proteins that act downstream to facilitate myeloid cell differentiation and inhibit the erythroid program (Figure 7D).
Grna mutants fail to regenerate the tail fin after resection. (A) Experimental workflow. grna+/+ or grna−/− 48 hpf embryos were subjected to caudal tail resection immediately after the end of the notochord. Bright field imaging of the wound was taken every 24 hours for 3 days (72 hpw, equivalent to 120 hpf). (B) Representative images of tail fins from grna+/+ (top panel) or grna−/− (bottom panel) larvae at the indicated times. Black lines indicate where the tail fins were resected. (C) Quantification of the regenerated tail fin area of grna+/+ (n = 5) and grna−/− (n = 5) larvae from panel B. (D) Schematic representation of signaling occurring during myeloid cell differentiation. Briefly, Pu.1 and Irf8 positively regulate granulin expression, which in turn controls the expression of rgs2 and cebpa for the differentiation of myeloid progenitors into neutrophils expressing mpx and lyz or macrophages (mpeg1 and mfap4). Granulin blocks gata1 expression, inhibiting erythroid development and the expression of the erythroid-related genes alas2, epb41b, and hbae4. Granulin expression levels are indicated in green. Error bars indicate SEM. **P < .001; ***P < .0001.
Grna mutants fail to regenerate the tail fin after resection. (A) Experimental workflow. grna+/+ or grna−/− 48 hpf embryos were subjected to caudal tail resection immediately after the end of the notochord. Bright field imaging of the wound was taken every 24 hours for 3 days (72 hpw, equivalent to 120 hpf). (B) Representative images of tail fins from grna+/+ (top panel) or grna−/− (bottom panel) larvae at the indicated times. Black lines indicate where the tail fins were resected. (C) Quantification of the regenerated tail fin area of grna+/+ (n = 5) and grna−/− (n = 5) larvae from panel B. (D) Schematic representation of signaling occurring during myeloid cell differentiation. Briefly, Pu.1 and Irf8 positively regulate granulin expression, which in turn controls the expression of rgs2 and cebpa for the differentiation of myeloid progenitors into neutrophils expressing mpx and lyz or macrophages (mpeg1 and mfap4). Granulin blocks gata1 expression, inhibiting erythroid development and the expression of the erythroid-related genes alas2, epb41b, and hbae4. Granulin expression levels are indicated in green. Error bars indicate SEM. **P < .001; ***P < .0001.
Discussion
Granulin has a profound impact on wound healing, autoimmune diseases, and tumorigenesis. In addition, loss-of-function mutations in the granulin gene are causative for frontotemporal lobar degeneration and ceroid lipofuscinosis.65,66 Despite its medical importance, why and how granulin can influence these diverse biological processes have remained unclear. Here, we have uncovered a new in vivo function for granulin in myeloid cell differentiation. We revealed that loss of granulin causes fewer macrophages and neutrophils to differentiate, which has consequences for wound healing and inflammation.
The tissue-specific segregation of the zebrafish granulin paralogues has allowed an unprecedented manner of assessing granulin function in hematopoiesis without perturbing other tissues. By using our zebrafish model of grna deficiency, we have demonstrated that grna is essential for neutrophil and macrophage differentiation from myeloid progenitors during normal and emergency myelopoiesis and therefore has an impact on inflammation and wound repair. Importantly, we show that the regulatory mechanisms of expression of grna and mammalian granulin by hematopoietic cells is highly conserved among zebrafish and mammals.
We consistently observed a profound defect in neutrophil differentiation in the absence of grna, but some macrophages still developed. Our data support a functional and transcriptional dysregulation in macrophages in the absence of grna, because macrophages failed to recruit to the wound, new macrophages failed to generate during emergency myelopoiesis, and many of the downregulated genes in the grna−/− vs grna+/+ kidneys are known anti-inflammatory genes, but some are completely restricted to macrophages (abca1b, il4r.1). This aberrant production of inflammatory genes in the absence of grna indicates a dysregulated inflammatory response, which is consistent with previous reports regarding mice that lack Grn19 and our previous data showing morphologic and transcriptional changes in microglial cells in zebrafish lacking grna and grnb, which is indicative of a pro-inflammatory phenotype.67 These observations also support functional conservation between the mammalian granulin and the zebrafish ortholog grna. We demonstrate here that terminal myeloid maturation in both adult and developmental hematopoiesis is compromised in the absence of grna, indicating a conserved role for grna during embryonic and adult hematopoiesis. However, although early myeloid precursors are increased in adult hematopoiesis, Pu.1+ embryonic myeloid progenitors remained unaltered. This difference could be a result of the distinct ontogeny of myeloid cells during adult and embryonic hematopoiesis34,68,69 or perhaps of an effort toward compensation activated only during adulthood. In addition, an outstanding question that derives from this work is which pathways granulin modulates in the context of myeloid differentiation. Because granulin can bind at least 6 different receptors70 and therefore modulate many signaling pathways, the exact molecular mechanism activated by granulin in this context still needs to be evaluated. These signaling pathways activated by granulin, which are also involved in myeloid cell differentiation, include NF-kB,71 β-catenin,72 MAPK/ERK,73 FAK,74 and STAT3.75
We demonstrated that wound healing is abnormal in individuals lacking grna, as previously reported in granulin knockout mice.26 However, the restricted grna expression within hematopoietic cells in our zebrafish model of grna deficiency allowed us to acknowledge that the defects in wound healing are caused, at least mostly, by aberrant myeloid cell differentiation. In accordance with our data in zebrafish, it has been reported that the majority of the granulin expressed in the wound derived from myeloid cells.26 Because macrophages greatly contribute to tissue repair, it is not surprising that impaired myeloid differentiation leads to aberrant healing of the wounded tissue. Altogether, we have been able to reproduce previously described phenotypes of mouse models of granulin deficiency in our grna zebrafish mutants and confirm that the cause of aberrant inflammation and wound healing is impaired myelopoiesis.
Our study also raises the question of whether the contributions described for granulin in tumorigenesis76 are also a result of a likely abnormal production of tumor-associated neutrophils, tumor-associated macrophages, and antitumor macrophages that constitute the tumor microenvironment, which has an impact on tumor progression.77 In addition, our discovery paves the way for understanding how granulin leads to frontotemporal dementia. Recently, multiple lines of evidence revealed the impact of microglia in neurologic disorders.7,78 The production of aberrant microglia in the absence of granulin has also recently been described in adult zebrafish67 and mice,79,80 being the cause of rather than the consequence of neurodegeneration. Therefore, a plausible hypothesis is that microglia-driven neuroinflammation in frontotemporal lobar degeneration is caused by the myeloid defects we described in myeloid development. Altogether, our results suggest genetic and functional differences in macrophages produced in the absence of grna, which most likely have an impact on the homeostasis of many tissues because of the ubiquitous presence of resident macrophages and the influences that macrophages and neutrophils have on tumor progression, autoinflammatory disease, and neurodegenerative disease. More experiments in each of these contexts will be needed to address the exact differences among microglia and tumor-associated myeloid cells in the absence of granulin, and our zebrafish model of granulin deficiency is an ideal model to use in performing these studies in vivo.
Among the complex regulatory network of TFs that regulate hematopoiesis, GATA1 and PU.1 are key in regulating the erythroid vs the myeloid program by antagonizing each other.81-84 These TFs are therefore main contributors to the pathogenesis of hematopoietic disorders.85 Despite decades of effort to address how PU.1 and GATA1 negatively regulate each other, little is known about this mechanism. In this study, we demonstrated that Pu.1 positively regulates grna expression, and that this regulatory mechanism is highly conserved in the mammalian granulin. Importantly, our results also indicate that granulin inhibits gata1 expression in vivo. Our findings identify granulin as a previously unrecognized regulator of Pu.1-Gata1 antagonism and extend previous results on the mechanism of gata1 inhibition by Pu.1. Further studies will be required to determine the precise molecular mechanism by which granulin inhibits gata1 expression.
In conclusion, with our discovery that granulin is essential for normal myelopoiesis, it is not surprising that its dysregulation leads to pleiotropic effects, because macrophages and neutrophils participate in inflammation, wound healing, tumorigenesis, and neurodegeneration. Our work therefore opens a new field of study that has the potential to impact different aspects of human health and fill a knowledge gap to advance the manipulation of GRN as a therapeutic target. Our results are expected to advance understanding of how this protein could be manipulated to treat hematopoietic disorders such as neutropenia or myeloid leukemia. Because granulin is a secreted factor, it may be plausible to use it as a therapeutic target to treat these hematologic disorders and expand treatment options for these patients.
Messenger RNA sequencing data are available at Gene Expression Omnibus (GEO) (GSE155258). For all other inquiries, please contact Raquel Espín-Palazón at espin@iastate.edu.
Acknowledgments
The authors thank Roy J. Carver Charitable Trust for the zebrafish research facility in the Advanced Teaching and Research Building at Iowa State University, Karen Ong for technical assistance, and Jesus Olvera and Cody Fine of the University of California–San Diego (UCSD) Human Embryonic Stem Cell Core Facility for technical assistance with flow cytometry experiments. The authors also thank Kristen Jepsen, Eugenia Ricciardelli, and Stephanie Hadimulia of the Institute of Genomics Medicine (IGM) at UCSD for technical assistance with RNA sequencing experiments. The authors are indebted to Jeffrey Essner and Maura McGrail for their support with fish husbandry.
This work was supported in part by a grant from California Institute for Regenerative Medicine Major Facilities (FA1-00607) to the Sanford Consortium for Regenerative Medicine. This article includes data generated at the UCSD IGM Genomics Center using an Illumina NovaSeq 6000 that was purchased with a Shared Instrumentation Grant from the Office of the Director, National Institutes of Health (S10 OD026929), by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (7K01DK115661 and R03DK125661), by Iowa State University startup funds, and by a postdoctoral fellowship from Fundacion Seneca, Agencia de Ciencia y Tecnologıa de la Region de Murcia and the American Heart Association (16POST30690005) (R.E.-P.).
Authorship
Contribution: R.E.-P., C.A.C., O.F., and D.T. designed the experiments; R.E.-P., C.A.C., O.F., E.S., X.C., L.L., A.M., B. Solchenberger, and M.M. performed the research; R.E.-P., C.A.C., X.C., O.F., E.S., L.L., A.M., B. Schmid, D.S., M.M., and D.T. analyzed the data; and R.E.-P., C.A.C., and D.T. wrote the paper with minor contributions from the remaining authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Raquel Espín-Palazón, Department of Genetics, Development and Cell Biology, Iowa State University, 2213 Pammel Dr, ATRB 3003, Ames, IA 50011; e-mail: espin@iastate.edu; and David Traver, University of California at San Diego, 9500 Gilman Dr, Natural Sciences Building 6107, La Jolla, CA 92093; e-mail: dtraver@ucsd.edu.
References
Author notes
The full-text version of this article contains a data supplement.