TO THE EDITOR:
KMT2A (MLL) abnormalities are common in leukemias of various lineages. Specifically, KMT2A chromosomal rearrangements are present in about 5% to 15% of all adolescents and young adults diagnosed with acute lymphoblastic leukemia (ALL) and in 3% of all adult patients with acute myeloid leukemia (AML).1,2 Thus far, more than 100 KMT2A partner genes have been described, AFF1 (AF4) and MLLT3 (AF9) being the most common ones, mainly associated with ALL and AML, respectively.3 Although the KMT2A recombinome has been largely studied in B-cell ALL (B-ALL), its characterization in T-cell ALL (T-ALL) is still limited, with KMT2A rearrangements accounting for about 2% to 5% of all T-ALL cases.4 In T-ALL, in turn, the most frequent partner genes involved in KMT2A rearrangements are MLLT1 (ENL) and AFDN (AF6).3,4
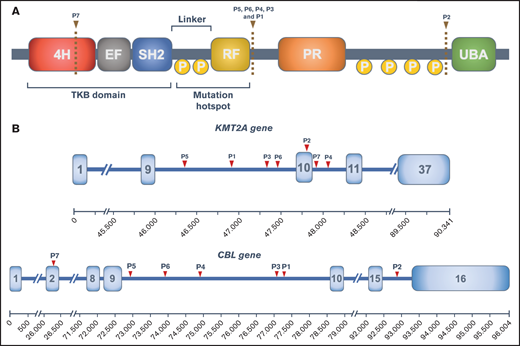
CBL (also known as c-CBL, casitas B-lineage leukemia) encodes for a ubiquitin-ligase (E3s) involved in the negative regulation of signaling mediated by multiple tyrosine kinase receptors. This protein is a member of the Cbl family of proto-oncogenes, which has 3 members (CBL, CBLB, and CBLC) and is ubiquitously expressed in the cytoplasm of many tissues.5 The protein domain structure of Cbl is presented in Figure 1A. At its N-terminal portion, there is a tyrosine kinase–binding (TKB) domain, an EF hand, and a Src homology 2 domain (SH2). Its main function is to bind multiple cytoplasmatic substrates, including tyrosine kinase proteins. The zinc-binding Really Interesting New Gene (RING) finger domain, which regulates the E3 ubiquitin ligase activity of c-Cbl, brings in an E2 ubiquitin-conjugating enzyme and therefore ubiquitylates the substrate bound to the TKB domain. Adjacent to the RING domain, there is a proline-rich region involved in protein/protein interactions via inducible association with SH2 and SH3. At the C-terminal portion, there is a ubiquitin associated/leucine zipper domain (UBA/LZ), crucial for Cbl homodimerization.
CBL structure and KMT2A-CBL rearrangements. (A) Structure of the CBL protein and its domains: 4-helix bundle (4H), EF-hand-calcium binding (EF), SRC homology 2 (SH2), RING finger (RF), proline-rich domain (PR), and ubiquitin-associated domain (UBA). Phosphorylation sites (P) and predicted protein breakpoints for each patient are also represented. (B) Schematic representation of KMT2A and CBL genes with the breakpoint for each patient.
CBL structure and KMT2A-CBL rearrangements. (A) Structure of the CBL protein and its domains: 4-helix bundle (4H), EF-hand-calcium binding (EF), SRC homology 2 (SH2), RING finger (RF), proline-rich domain (PR), and ubiquitin-associated domain (UBA). Phosphorylation sites (P) and predicted protein breakpoints for each patient are also represented. (B) Schematic representation of KMT2A and CBL genes with the breakpoint for each patient.
Several mutations in CBL have been described in various hematologic disorders, mostly in myeloid malignancies. The frequency of CBL mutations has been as high as 15% and 4% of adult myelodysplastic/myeloproliferative diseases and myeloproliferative neoplasms, respectively. In AML, its frequency is lower: approximately 1% to 2%.6 In juvenile myelomonocytic leukemia (JMML), CBL mutations are also common, with a frequency of 17%.7 In ALL, although rare, a few reports have described CBL mutations in both T- and B-cell lineages, especially in KMT2A-rearranged leukemias.8,9 CBL mutations consist mostly of point mutations clustering in exons 8 and 9, involving the linker and RING domain6 (Figure 1A). The proposed oncogenic mechanism of CBL mutations is based on intracellular signaling deregulation because of the loss of CBL inhibitory function.10 Interestingly, in CBL-mutated myeloid neoplasms, a loss of the germline CBL allele has been demonstrated, mostly because of uniparental disomy.11
To our knowledge, only 4 reports of hematologic neoplasms harboring a KMT2A-CBL gene fusion (3 of them under the diagnosis of AML) have been published.3,12,13 International collaborative efforts, such as the MLL recombinome database, are crucial to identify and collect these rare rearrangements. Along with the previously published cases, we have now identified 3 new cases with the same rearrangement but in distinct hematologic malignancies (Table 1). It has been described that KMT2A breakpoints mostly localize within exon 9 and intron 11, with an association of intron 9 breakpoints with AML or older patients and intron 11 with ALL and younger patients.3 In our cohort, all patients had their breakpoint within the previous described limits, 4 of them clustering in intron 9, although establishing associations between age or diagnosis with this finding seems far-fetched. CBL breakpoints also cluster in intron 9 (Figure 1). The KMT2A-CBL fusions of patients 2 and 7 are per se out of frame. However, the reverse transcriptase-polymerase chain reaction (RT-PCR) of case 2 revealed a cryptic exon composed of the truncated exon 10 of KMT2A and additional filler DNA that maintains the reading frame. This mechanism could also explain the KMT2A-CBL rearrangement of patient 7, but this hypothesis could not be verified because of the lack of material. This study has been approved by the Ethics Committee of Hospital Clínic de Barcelona. Code:HCB/2021/0010 and was conducted in accordance with the Declaration of Helsinki.
Patients with KMT2A-CBL rearrangement
| . | Patient 1 (Marseille) . | Patient 2 (Reims) . | Patient 3 (Barcelona) . | Patient 43 (Prague) . | Patient 512 (Taiwan) . | Patient 63 (Nantes) . | Patient 713 (London) . |
|---|---|---|---|---|---|---|---|
| Sex, age (y) | Male, 9 | Male, 11 | Male, 18 | Male, 3 | Female, 28 | Female, 59 | Female, 18 |
| Medical history | JMML - KRAS mut 6MP & CB-AlloSCT (Bu/Cy) | None | None | Development delay | None | Polycythemia vera JAK2+ Hydroxyurea and pipobroman | None |
| Hematologic diagnosis | T-ALL (EGIL T2) | T-ALL (EGIL T2) | T-ALL (EGIL T2) | B-ALL (EGIL B1) | AML (FAB M1) | sAML (FAB M5) | AML |
| White blood count (·109 /L) | 1.1 | 5.39 | 12.32 | 65.5 | 0.8 | 37.5 | 1.3 |
| Hemoglobin (g/L) | 90 | 97 | 83 | 107 | 44 | 107 | 74 |
| Platelets (·109 /L) | 109 | 69 | 24 | 71 | 15 | 45 | 55 |
| Blast count peripheral blood/bone marrow (%) | 0/97 | 33/99 | 85/99 | 80/93 | 47/96 | 65/83 | |
| Flow cytometry | cCD3+, sCD3−, TdT+, CD99+, CD7+, CD2+, CD5+ | cCD3+, sCD3−, CD7++, CD2++, CD5+/− (13%), CD34+dim, CD38++, CD13+ | cCD3+, sCD3+ dim, CD34−, TdT−, CD99+ dim, CD1a−, CD7++, CD38+ | CD34−, TdT+, CD19+, CD79a+, CD10+, CD7+ dim, CD38+ | CD34+, CD13+, CD33+ | CD34−, CD13+, CD33+, CD4+ | CD34+, cMPO+, CD33+, CD15+, CD38+, CD56+ |
| Cytogenetics | Pseudodiploid and monosomic with 3 clonal anomalies: del(2q),-16,r | 47,XY,?del(5)(q31),+8, del(9)(q13q34),del(11)(q21q23), del(12)(p11)[cp6]/46,XY[28] | 47, XY, der(7),+19[20] | NA | 50,XX,+22,+3 mar [5]/49,XX, −19, +22,+3mar [11]/46,XX,inv(9)[4] | 45∼46,XX,-5,add(6)(p21), -9,+i(9)(p10), add(11)(q23),-18,+mar[7], 47∼49,XX, +2,-5,-9,i(9q10),add(11)(q23), ?+13,-18,+19,+20,inc[9] | 46,XX[20] |
| KMT2A FISH | KMT2A 3' loss of signal | KMT2A 3' loss of signal | KMT2A 3' loss of signal | KMT2A 3' loss of signal | — | KMT2A 3' loss of signal | KMT2A 3' loss of signal |
| NGS | KRAS p.G13C (VAF 32%) ASXL1 p.G646Wfs12 (VAF 26%) STAT3 p.N481I (VAF 36%) | IL7R p.V253_A254insVA (VAF 13%) KMT2A p.N*2278fs (VAF 82%) NOTCH1 p.P2514fs (VAF 16%) NOTCH1 p.A1700D (VAF 10%) NOTCH1 p.L1678P (VAF 36%) ZFHX4 p.R2676Q (VAF 28%) | No mutations detected | KRAS p.A18D (VAF 44%) | CBL p.L380P (VAF 70%) | — | RUNX1 p.G372S (VAF 50%) |
| Method for KMT2A-CBL detection | RNAseq and targeted NGS | RNAseq and Targeted NGS | Targeted NGS | RNAseq and LDI-PCR | cDNA panhandle PCR Validated by RT-PCR + cloning and sequencing of the KMT2A fusion product. | RNAseq and LDI-PCR | Chromosomal microarray analysis |
| Breakpoint | KMT2A intron 9 CBL intron 9 | KMT2A exon 10 CBL intron 15 | KMT2A intron 9 CBL intron 9 | KMT2A intron 10 CBL intron 9 | KMT2A intron 9 CBL intron 9 | KMT2A intron 9 CBL intron 9 | KMT2A intron 10 CBL exon 2 |
| Clinical outcome | Induction Chemotherapy (CALL-T protocol) – CR MRD+ Consolidation + Intensification – CR MRD-CB-AlloSCT Alive, CR 1 y after diagnosis | Induction chemotherapy (CAALL-F01 protocol) NR Consolidation + Intensification CR MRD- RD-AlloSCT Alive, CR 3 y after diagnosis | Induction chemotherapy (PETHEMA 2019) – PR Re-induction (FLAG-IDA) – CR MRD- Consolidation – CR MRD- MUD-AlloSCT Alive, CR 6 mo after diagnosis | AEIOP/BFM 2000 ALL protocol CR MRD- Relapse after 8 y IntReALL SR 2010 protocol CR-MRD+ Blinatumomab × 2 MUD-AlloSCT Alive, CR 13 y after diagnosis | Induction chemotherapy CR Consolidation – CR Relapse 2 y after diagnosis Reinduction CR MUD-AlloSCT Deceased due to alloSCT complications | Induction chemotherapy CR Consolidation CR RD-AlloSCT Relapse 11 mo after diagnosis Azacytidine: No response Deceased because of disease progression | Induction chemotherapy CR Loss of follow-up |
| . | Patient 1 (Marseille) . | Patient 2 (Reims) . | Patient 3 (Barcelona) . | Patient 43 (Prague) . | Patient 512 (Taiwan) . | Patient 63 (Nantes) . | Patient 713 (London) . |
|---|---|---|---|---|---|---|---|
| Sex, age (y) | Male, 9 | Male, 11 | Male, 18 | Male, 3 | Female, 28 | Female, 59 | Female, 18 |
| Medical history | JMML - KRAS mut 6MP & CB-AlloSCT (Bu/Cy) | None | None | Development delay | None | Polycythemia vera JAK2+ Hydroxyurea and pipobroman | None |
| Hematologic diagnosis | T-ALL (EGIL T2) | T-ALL (EGIL T2) | T-ALL (EGIL T2) | B-ALL (EGIL B1) | AML (FAB M1) | sAML (FAB M5) | AML |
| White blood count (·109 /L) | 1.1 | 5.39 | 12.32 | 65.5 | 0.8 | 37.5 | 1.3 |
| Hemoglobin (g/L) | 90 | 97 | 83 | 107 | 44 | 107 | 74 |
| Platelets (·109 /L) | 109 | 69 | 24 | 71 | 15 | 45 | 55 |
| Blast count peripheral blood/bone marrow (%) | 0/97 | 33/99 | 85/99 | 80/93 | 47/96 | 65/83 | |
| Flow cytometry | cCD3+, sCD3−, TdT+, CD99+, CD7+, CD2+, CD5+ | cCD3+, sCD3−, CD7++, CD2++, CD5+/− (13%), CD34+dim, CD38++, CD13+ | cCD3+, sCD3+ dim, CD34−, TdT−, CD99+ dim, CD1a−, CD7++, CD38+ | CD34−, TdT+, CD19+, CD79a+, CD10+, CD7+ dim, CD38+ | CD34+, CD13+, CD33+ | CD34−, CD13+, CD33+, CD4+ | CD34+, cMPO+, CD33+, CD15+, CD38+, CD56+ |
| Cytogenetics | Pseudodiploid and monosomic with 3 clonal anomalies: del(2q),-16,r | 47,XY,?del(5)(q31),+8, del(9)(q13q34),del(11)(q21q23), del(12)(p11)[cp6]/46,XY[28] | 47, XY, der(7),+19[20] | NA | 50,XX,+22,+3 mar [5]/49,XX, −19, +22,+3mar [11]/46,XX,inv(9)[4] | 45∼46,XX,-5,add(6)(p21), -9,+i(9)(p10), add(11)(q23),-18,+mar[7], 47∼49,XX, +2,-5,-9,i(9q10),add(11)(q23), ?+13,-18,+19,+20,inc[9] | 46,XX[20] |
| KMT2A FISH | KMT2A 3' loss of signal | KMT2A 3' loss of signal | KMT2A 3' loss of signal | KMT2A 3' loss of signal | — | KMT2A 3' loss of signal | KMT2A 3' loss of signal |
| NGS | KRAS p.G13C (VAF 32%) ASXL1 p.G646Wfs12 (VAF 26%) STAT3 p.N481I (VAF 36%) | IL7R p.V253_A254insVA (VAF 13%) KMT2A p.N*2278fs (VAF 82%) NOTCH1 p.P2514fs (VAF 16%) NOTCH1 p.A1700D (VAF 10%) NOTCH1 p.L1678P (VAF 36%) ZFHX4 p.R2676Q (VAF 28%) | No mutations detected | KRAS p.A18D (VAF 44%) | CBL p.L380P (VAF 70%) | — | RUNX1 p.G372S (VAF 50%) |
| Method for KMT2A-CBL detection | RNAseq and targeted NGS | RNAseq and Targeted NGS | Targeted NGS | RNAseq and LDI-PCR | cDNA panhandle PCR Validated by RT-PCR + cloning and sequencing of the KMT2A fusion product. | RNAseq and LDI-PCR | Chromosomal microarray analysis |
| Breakpoint | KMT2A intron 9 CBL intron 9 | KMT2A exon 10 CBL intron 15 | KMT2A intron 9 CBL intron 9 | KMT2A intron 10 CBL intron 9 | KMT2A intron 9 CBL intron 9 | KMT2A intron 9 CBL intron 9 | KMT2A intron 10 CBL exon 2 |
| Clinical outcome | Induction Chemotherapy (CALL-T protocol) – CR MRD+ Consolidation + Intensification – CR MRD-CB-AlloSCT Alive, CR 1 y after diagnosis | Induction chemotherapy (CAALL-F01 protocol) NR Consolidation + Intensification CR MRD- RD-AlloSCT Alive, CR 3 y after diagnosis | Induction chemotherapy (PETHEMA 2019) – PR Re-induction (FLAG-IDA) – CR MRD- Consolidation – CR MRD- MUD-AlloSCT Alive, CR 6 mo after diagnosis | AEIOP/BFM 2000 ALL protocol CR MRD- Relapse after 8 y IntReALL SR 2010 protocol CR-MRD+ Blinatumomab × 2 MUD-AlloSCT Alive, CR 13 y after diagnosis | Induction chemotherapy CR Consolidation – CR Relapse 2 y after diagnosis Reinduction CR MUD-AlloSCT Deceased due to alloSCT complications | Induction chemotherapy CR Consolidation CR RD-AlloSCT Relapse 11 mo after diagnosis Azacytidine: No response Deceased because of disease progression | Induction chemotherapy CR Loss of follow-up |
In cytogenetic studies, number of metaphases studied is represented in brackets. 6MP, mercaptopurine; AlloSCT, allogeneic stem cell transplantation; CB, cord blood; CR: complete response; EGIL, European Group for the Immunological Classification of Leukemia; FAB, French-American-British classification; LDI-PCR, long-distance-inverse PCR; MRD, minimal residual disease; MUD, matched unrelated donor; NR, no response; PR, partial response; RD, related donor; VAF, variant allele frequency.
Despite the limited number of patients, some biological insights about this rare rearrangement can be extracted from our findings. We report for the first time that KMT2A-CBL rearrangements are present not only in AML and B-ALL but also in T-ALL. Moreover, 2 of these cases were secondary neoplasms (JMML and polycythemia vera; both received prior chemotherapeutic agents), which is also common in KMT2A-rearranged leukemias. CBL is located in chromosome 11q23, at a distance of 680 kb from KMT2A, suggesting that KMT2A-CBL rearrangements are necessarily caused either by an interstitial deletion or by a translocation event occurring between both homologous chromosomes 11. Split-signal FISH analysis can reveal these types of rearrangements, whereas conventional cytogenetics usually fails to detect them. Subsequently, different diagnostic techniques can be used to identify CBL as the KMT2A fusion partner, although their implementation in the clinical routine might be difficult. Targeted next-generation sequencing (NGS) is a feasible alternative to detect this and other rare rearrangements included in the panel.
In terms of the leukemogenic effect of KMT2A-CBL rearrangements, to date, there are no specific models that can clearly explain its mechanism. CBL is a cytoplasmatic protein so its effect in the leukemic cell might be different from that of nuclear factors like AFF1 or MLLT3.14 In fact, it has been hypothesized that rearrangements of KMT2A with cytoplasmatic proteins could trigger their oncogenic effects through dimerization of the chimeric KMT2A protein, enhancing a specific gene expression program.15 This is supported by the retention of the UBA domain responsible for the homodimerization of CBL in all the KMT2A-CBL cases reported in the present study. Thus, dimerization of KMT2A-CBL through the UBA domains could be an important leukemogenic mechanism to be explored. Another alternative explanation is that the translocated protein domains might exert novel functions in the wrong cellular compartments. In addition, the resulting truncated CBL protein most likely represents a genetic “loss-of-function” situation, as already observed in hematological neoplasms with CBL inactivating mutations.10
To conclude, KMT2A-CBL rearrangements are rare events identified in leukemias of various lineages. Conventional cytogenetics may miss such rearrangements but the increasingly widespread use of targeted sequencing techniques in the diagnostic work-up will probably increase the number of cases identified. International collaborative efforts are necessary to improve the characterization of the KMT2A-CBL rearrangement and investigate its prognostic impact. Ultimately, functional experiments of KMT2A-CBL gain-of-function models and CD34+ hematopoietic progenitor cells may provide fundamental knowledge about the leukemogenic potential and the cell-of-origin of KMT2A-CBL leukemias.
Acknowledgments: The authors thank Centres de Recerca de Catalunya/Generalitat de Catalunya and Fundació Josep Carreras‐Obra Social la Caixa for institutional support.
Contribution: A.B., F.G., M.S., A.S., O.M., P.C., M.Z., J.Z., P.M., L.M., N.G., P.C.-L., J.-F.F., L.-Y.S., T.B., E.N., S.B., and M.L.-G. provided clinical and biological information of all cases; A.C.-E., C.B., P.M., P.L., C.M., and R.M. provided genetic data; and A.B. and F.G. collected the data and wrote the manuscript along with C.B., P.M., J.E., P.L., C.M., and R.M. All authors reviewed the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alex Bataller, Department of Hematology, Hospital Clínic de Barcelona, Carrer Villarroel 170, 08036 Barcelona, Spain; email: abataller@clinic.cat and Francesca Guijarro, Hematopathology Section, Hospital Clínic de Barcelona, Carrer Villarroel 170, 08036 Barcelona, Spain; email: fguijarro@clinic.cat.
References
Author notes
A.B. and F.G. contributed equally to this study.
Requests for data sharing may be submitted to Alex Bataller (abataller@clinic.cat).