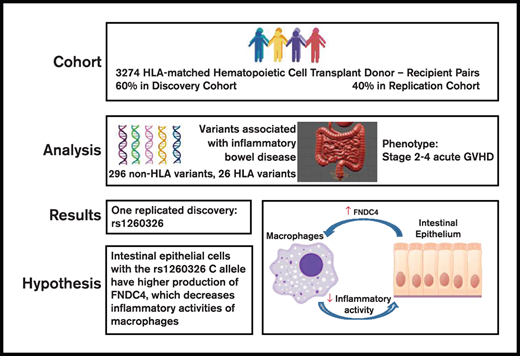
Key Points
An IBD-associated locus tagged by rs1260326 is associated with gut GVHD after allogeneic HCT.
Genetic variation in anti-inflammatory activity mediated by FNDC4 within this locus could account for the association with gut GVHD.
Abstract
Previous studies have identified genetic variants associated with inflammatory bowel disease (IBD). We tested the hypothesis that some of these variants are also associated with the risk of moderate to severe gut graft-versus-host disease (GVHD) after allogeneic hematopoietic cell transplantation (HCT). Associations were evaluated initially in a discovery cohort of 1980 HCT recipients of European ancestry with HLA-matched related or unrelated donors. Associations discovered in this cohort were tested for replication in a separate cohort of 1294 HCT recipients. Among the 296 single-nucleotide polymorphisms and 26 HLA alleles tested, we found that the recipient rs1260326 homozygous T allele in GCKR was associated with a higher risk of stage 2 to 4 gut GVHD. No other candidate variants were associated with stage 2 to 4 gut GVHD. The rs1260326 variant resides in an IBD-associated locus containing FNDC4, a gene that encodes a secreted anti-inflammatory factor that dampens macrophage activity and improves colitis in mice. Our results suggest that targeting inflammatory macrophages with recombinant FNDC4 offers an attractive avenue of clinical investigation for management of IBD and gut GVHD.
Introduction
Steroid-refractory acute graft-versus-host disease (GVHD) of the intestinal tract and inflammatory bowel disease (IBD) have similarities in clinical manifestations, pathogenic mechanisms, and genetic risk factors.1 Pathogenic mechanisms in both diseases involve inflammatory cytokines, epithelial barrier dysfunction, and dysbiosis, and genetic polymorphisms of TLR4, NOD2, and ATG16L1 have been associated with the risk of both GVHD and IBD.2-11TLR4 and NOD2 represent microbial-sensing mechanisms of innate immunity, whereas ATG16L1 prevents necroptosis in the intestinal epithelium.
The International IBD Genetics Consortium (IIBDGC) has compiled a comprehensive list of genetic loci containing variants having statistically significant associations with IBD.12-14 The extent to which these variants are also associated with the risk of acute intestinal graft-versus-host disease (GVHD) after allogeneic hematopoietic cell transplantation (HCT) has not been previously determined. We therefore tested these variants for association with moderate to severe gut GVHD in a cohort of 1980 HCT recipients of European ancestry with related or unrelated donors at our center. Associations discovered in this cohort were tested for replication in a separate cohort of 1294 HCT recipients. The IIBDGC also identified 26 HLA alleles that had independent associations with IBD.15 We therefore used the same approach to test these alleles for association with stage 2 to 4 gut GVHD.
Methods
Study population
All recipient and donor blood samples were collected before HCT according to research protocols approved by the Fred Hutchinson Cancer Research Center Institutional Review Board. Project-specific Institutional Review Board approval was obtained for the use of clinical data and research biospecimens. The study was conducted in accordance with the Declaration of Helsinki.
The study cohort included 3274 recipients of European ancestry who had a first allogeneic HCT at the Fred Hutchinson Cancer Research Center and Seattle Cancer Care Alliance from 1990 through 2011. Syngeneic donors and cord blood donors were excluded. A single prior autologous HCT was allowed. This study was limited to recipients of European ancestry because the number of non-European recipients in our cohort was too small to account for population stratification in the analysis. European ancestry was defined by using the minimum covariant determinant method as implemented by Conomos et al.16 Donors and recipients were HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 allele matched at the level of 4 digits. Indications for HCT included hematologic malignancy or myelodysplasia. Conditioning regimens were categorized as myeloablative or nonmyeloablative according to the intensity of chemotherapy and total body irradiation. Grafts used for HCT included bone marrow cells or growth factor–mobilized blood cells.
Selection of variants
The candidate analysis focused on the lead variants identified by the IIBDGC as summarized in supplemental materials from reports by de Lange et al12 and Huang et al.13 We also considered candidate HLA alleles associated with IBD identified by the IIBDGC as summarized in supplemental materials from the report by Goyette et al.15 Other variants of TLR4, NOD2, and ATG16L1 have been associated with gut GVHD in previous studies.1 These were not considered candidates but were analyzed in the combined discovery and replication cohorts.
Sample preparation, genotyping, quality assurance and quality control, and imputation
Details regarding preparation of genomic DNA samples from donors and recipients have been described previously.17 Details of DNA amplification, genotyping platforms, hybridization and genotyping, and imputation algorithms are presented elsewhere.17 Quality assurance and quality control followed standard methods as described previously.17
Statistical analysis
The outcome tested in this study was stage 2 to 4 acute gut GVHD. Evaluation of the association of genotype with stage 2 to 4 gut GVHD was based on cause-specific hazard ratio (HR) analysis using Cox regression, with death and relapse treated as competing risks. All candidate variants in the recipient and donor genomes were evaluated for allelic and genotypic (recessive and dominant) association with stage 2 to 4 gut GVHD.
All analyses of candidate variants were conducted in 2 phases. The study cohort was randomized in a 3:2 split into discovery and replication cohorts. Candidate variants with P values ≤.005 for associations with stage 2 to 4 acute GVHD in the discovery cohort were tested in the replication cohort with Bonferroni adjustment for multiple comparisons. Bonferroni adjustment was applied separately for the donor and recipient genomes but did not include different genetic models applied to the same variant. Post hoc power estimates for discovery were calculated for an HR ≥1.5 or ≤0.67, based on the estimated standard error of the log HR and a two-sided 0.005 significance level. Post hoc power estimates for replication were calculated based on the discovery log HR point estimate and the estimated standard error of the replication log HR, with a Bonferroni-corrected significance level reflecting the number of discovery findings.
Biological interpretation of associations
We used results from the Genotype-Tissue Expression (GTEx) Project and the Mouse Genome Database18 to explore biological mechanisms that could account for associations identified in our results.
Results
Association of candidate variants with stage 2 to 4 gut GVHD
Table 1 summarizes demographic, clinical, and transplant characteristics of patients in the study cohort. Donor samples were not available for 156 (5%) pairs, and recipient samples were not available for 283 (9%) pairs. Supplemental Table 1 summarizes the quality control assessment of the 296 candidate variants successfully genotyped or imputed on at least 1 of the 3 platforms used for our study. Supplemental Table 2 summarizes 13 variants that could not be analyzed because they did not pass quality control, because they were not included in our data set, or because results were not validated between de Lange et al12 and Huang et al.13
Characteristics of the study cohort (N = 3274 recipients)
| Characteristic . | Value . |
|---|---|
| Recipients genotyped | 2991 (91%) |
| Donors genotyped | 3118 (95%) |
| Patient age at transplantation, y | |
| Median | 45 |
| Range | 0-78 |
| Donor age, y | |
| Median | 39 |
| Range | 0-83 |
| Diagnosis | |
| Acute leukemia | 1300 (40%) |
| Chronic myeloid leukemia | 720 (22%) |
| Myelodysplastic syndrome or myeloproliferative neoplasm | 595 (18%) |
| Chronic lymphocytic leukemia | 104 (3%) |
| Malignant lymphoma or multiple myeloma | 555 (17%) |
| Disease risk* | |
| Low | 699 (21%) |
| Intermediate | 925 (28%) |
| High | 1438 (44%) |
| Not classified | 212 (6%) |
| Donor–recipient sex | |
| Male to male | 1123 (34%) |
| Male to female | 734 (22%) |
| Female to male | 790 (24%) |
| Female to female | 626 (19%) |
| Donor type | |
| 10/10 HLA–matched sibling | 1827 (56%) |
| 10/10 HLA–matched unrelated | 1447 (44%) |
| Graft source | |
| Bone marrow | 1606 (49%) |
| Mobilized blood cells | 1668 (51%) |
| Conditioning | |
| Myeloablative <900 cGy total body irradiation | 1296 (40%) |
| Myeloablative ≥900 cGy total body irradiation | 1338 (41%) |
| Nonmyeloablative | 640 (20%) |
| Initial posttransplant immunosuppression | |
| Cyclosporine and methotrexate | 1643 (50%) |
| Cyclosporine and mycophenolate mofetil | 589 (18%) |
| Tacrolimus and methotrexate | 486 (15%) |
| Tacrolimus and mycophenolate mofetil | 160 (5%) |
| Cyclosporine or tacrolimus alone | 206 (6%) |
| Methotrexate or mycophenolate mofetil alone | 73 (2%) |
| Other (cyclophosphamide or anti-thymocyte globulin) | 117 (4%) |
| Characteristic . | Value . |
|---|---|
| Recipients genotyped | 2991 (91%) |
| Donors genotyped | 3118 (95%) |
| Patient age at transplantation, y | |
| Median | 45 |
| Range | 0-78 |
| Donor age, y | |
| Median | 39 |
| Range | 0-83 |
| Diagnosis | |
| Acute leukemia | 1300 (40%) |
| Chronic myeloid leukemia | 720 (22%) |
| Myelodysplastic syndrome or myeloproliferative neoplasm | 595 (18%) |
| Chronic lymphocytic leukemia | 104 (3%) |
| Malignant lymphoma or multiple myeloma | 555 (17%) |
| Disease risk* | |
| Low | 699 (21%) |
| Intermediate | 925 (28%) |
| High | 1438 (44%) |
| Not classified | 212 (6%) |
| Donor–recipient sex | |
| Male to male | 1123 (34%) |
| Male to female | 734 (22%) |
| Female to male | 790 (24%) |
| Female to female | 626 (19%) |
| Donor type | |
| 10/10 HLA–matched sibling | 1827 (56%) |
| 10/10 HLA–matched unrelated | 1447 (44%) |
| Graft source | |
| Bone marrow | 1606 (49%) |
| Mobilized blood cells | 1668 (51%) |
| Conditioning | |
| Myeloablative <900 cGy total body irradiation | 1296 (40%) |
| Myeloablative ≥900 cGy total body irradiation | 1338 (41%) |
| Nonmyeloablative | 640 (20%) |
| Initial posttransplant immunosuppression | |
| Cyclosporine and methotrexate | 1643 (50%) |
| Cyclosporine and mycophenolate mofetil | 589 (18%) |
| Tacrolimus and methotrexate | 486 (15%) |
| Tacrolimus and mycophenolate mofetil | 160 (5%) |
| Cyclosporine or tacrolimus alone | 206 (6%) |
| Methotrexate or mycophenolate mofetil alone | 73 (2%) |
| Other (cyclophosphamide or anti-thymocyte globulin) | 117 (4%) |
Low risk is chronic myeloid leukemia in chronic phase or myelodysplastic syndrome-refractory anemia; intermediate risk, acute leukemia, chronic lymphocytic leukemia, or non-Hodgkin lymphoma in remission; high risk, all others.
Supplemental Tables 3 and 4, respectively, summarize associations of 296 candidate single-nucleotide polymorphisms (SNPs) and 26 candidate HLA alleles with stage 2 to 4 gut GVHD in the discovery cohort. Among the 296 SNP variants, 136 were associated with IBD defined as the combination of Crohn’s disease and ulcerative colitis (UC), 107 were associated with Crohn’s disease, 6 were associated with Crohn’s disease and IBD, 1 was associated with both Crohn’s disease and UC, 3 were associated with IBD and UC, and 43 were associated with UC. Further details are provided in supplemental Table 2 in DeLange et al12 and supplemental Table 1 in Huang et al.13 Five donor variants in the discovery cohort met criteria for replication testing (Table 2). For rs12722504, the replication results met criteria for statistical significance, but the HR point estimates were on opposite sides of 1.0. For the other 4 variants, the replication results were not statistically significant. Five recipient variants in the discovery cohort met criteria for replication testing. For HLA-A*03:01 and rs12722504, the replication results approached statistical significance, but the HR point estimates were on opposite sides of 1.0. For rs9319943 and rs1736137, the replication results were not statistically significant.
Association of candidate variants with stage 2 to 4 gut GVHD disease
| Chr . | Gene . | SNP . | Alleles* . | Genome . | Model . | MAF† . | Discovery results . | Replication results . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P . | HR . | LB . | UB . | Bf-C . | P . | HR . | LB . | UB . | Power‡ . | |||||||
| 1 | None | rs7517810 | C/T | Donor | Recessive | 0.25 | .004 | 2.02 | 1.3 | 3.1 | 0.01 | .47 | 1.27 | 0.7 | 2.4 | 36 |
| 6 | CCND3 | rs67289879 | C/T | Donor | Recessive | 0.21 | .001 | 4.25 | 2.0 | 9.0 | 0.01 | .50 | 0.54 | 0.1 | 3.9 | 13 |
| 10 | ZMIZ1 | rs1250563 | G/C | Donor | Allelic | 0.31 | .0008 | 1.42 | 1.2 | 1.7 | 0.01 | .82 | 1.03 | 0.8 | 1.3 | 52 |
| 10 | ZMIZ1 | rs1250563 | G/C | Donor | Dominant | 0.31 | .0002 | 1.70 | 1.3 | 2.3 | 0.01 | .34 | 1.19 | 0.8 | 1.7 | 62 |
| 10 | IL2RA | rs12722504 | GATAA/G | Donor | Recessive | 0.26 | .001 | 2.15 | 1.4 | 3.3 | 0.01 | .002 | 0.12 | 0.02 | 0.9 | 3 |
| 20 | TNFRSF6B | rs6062496 | A/G | Donor | Dominant | 0.44 | .002 | 0.53 | 0.4 | 0.8 | 0.01 | .39 | 1.28 | 0.7 | 2.3 | 32 |
| 2 | GCKR | rs1260326 | C/T | Recipient | Allelic | 0.42 | .0002 | 1.53 | 1.2 | 1.9 | 0.01 | .04 | 1.38 | 1.0 | 1.9 | 56 |
| 2 | GCKR | rs1260326 | C/T | Recipient | Recessive | 0.42 | .0004 | 1.96 | 1.4 | 2.8 | 0.01 | .02 | 1.87 | 1.2 | 3.0 | 56 |
| 6 | HLA-A | NA | Other/HLA-A*0301 | Recipient | Dominant | 0.15 | .004 | 0.62 | 0.4 | 0.9 | 0.01 | .05 | 1.43 | 1.0 | 2.0 | 55 |
| 10 | IL2RA | rs12722504 | GATAA/G | Recipient | Allelic | 0.27 | .003 | 1.40 | 1.1 | 1.7 | 0.01 | .01 | 0.67 | 0.5 | 0.9 | 29 |
| 18 | None | rs9319943 | T/C | Recipient | Allelic | 0.18 | .003 | 1.43 | 1.1 | 1.8 | 0.01 | .51 | 1.12 | 0.8 | 1.6 | 32 |
| 18 | None | rs9319943 | T/C | Recipient | Dominant | 0.18 | .005 | 1.50 | 1.1 | 2.0 | 0.01 | .56 | 1.12 | 0.8 | 1.7 | 31 |
| 21 | LOC101927745 | rs1736137 | A/G | Recipient | Allelic | 0.43 | .0004 | 1.42 | 1.2 | 1.7 | 0.01 | .42 | 1.11 | 0.9 | 1.5 | 53 |
| 21 | LOC101927745 | rs1736137 | A/G | Recipient | Recessive | 0.43 | .001 | 1.72 | 1.3 | 2.4 | 0.01 | .76 | 1.08 | 0.7 | 1.7 | 40 |
| Chr . | Gene . | SNP . | Alleles* . | Genome . | Model . | MAF† . | Discovery results . | Replication results . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P . | HR . | LB . | UB . | Bf-C . | P . | HR . | LB . | UB . | Power‡ . | |||||||
| 1 | None | rs7517810 | C/T | Donor | Recessive | 0.25 | .004 | 2.02 | 1.3 | 3.1 | 0.01 | .47 | 1.27 | 0.7 | 2.4 | 36 |
| 6 | CCND3 | rs67289879 | C/T | Donor | Recessive | 0.21 | .001 | 4.25 | 2.0 | 9.0 | 0.01 | .50 | 0.54 | 0.1 | 3.9 | 13 |
| 10 | ZMIZ1 | rs1250563 | G/C | Donor | Allelic | 0.31 | .0008 | 1.42 | 1.2 | 1.7 | 0.01 | .82 | 1.03 | 0.8 | 1.3 | 52 |
| 10 | ZMIZ1 | rs1250563 | G/C | Donor | Dominant | 0.31 | .0002 | 1.70 | 1.3 | 2.3 | 0.01 | .34 | 1.19 | 0.8 | 1.7 | 62 |
| 10 | IL2RA | rs12722504 | GATAA/G | Donor | Recessive | 0.26 | .001 | 2.15 | 1.4 | 3.3 | 0.01 | .002 | 0.12 | 0.02 | 0.9 | 3 |
| 20 | TNFRSF6B | rs6062496 | A/G | Donor | Dominant | 0.44 | .002 | 0.53 | 0.4 | 0.8 | 0.01 | .39 | 1.28 | 0.7 | 2.3 | 32 |
| 2 | GCKR | rs1260326 | C/T | Recipient | Allelic | 0.42 | .0002 | 1.53 | 1.2 | 1.9 | 0.01 | .04 | 1.38 | 1.0 | 1.9 | 56 |
| 2 | GCKR | rs1260326 | C/T | Recipient | Recessive | 0.42 | .0004 | 1.96 | 1.4 | 2.8 | 0.01 | .02 | 1.87 | 1.2 | 3.0 | 56 |
| 6 | HLA-A | NA | Other/HLA-A*0301 | Recipient | Dominant | 0.15 | .004 | 0.62 | 0.4 | 0.9 | 0.01 | .05 | 1.43 | 1.0 | 2.0 | 55 |
| 10 | IL2RA | rs12722504 | GATAA/G | Recipient | Allelic | 0.27 | .003 | 1.40 | 1.1 | 1.7 | 0.01 | .01 | 0.67 | 0.5 | 0.9 | 29 |
| 18 | None | rs9319943 | T/C | Recipient | Allelic | 0.18 | .003 | 1.43 | 1.1 | 1.8 | 0.01 | .51 | 1.12 | 0.8 | 1.6 | 32 |
| 18 | None | rs9319943 | T/C | Recipient | Dominant | 0.18 | .005 | 1.50 | 1.1 | 2.0 | 0.01 | .56 | 1.12 | 0.8 | 1.7 | 31 |
| 21 | LOC101927745 | rs1736137 | A/G | Recipient | Allelic | 0.43 | .0004 | 1.42 | 1.2 | 1.7 | 0.01 | .42 | 1.11 | 0.9 | 1.5 | 53 |
| 21 | LOC101927745 | rs1736137 | A/G | Recipient | Recessive | 0.43 | .001 | 1.72 | 1.3 | 2.4 | 0.01 | .76 | 1.08 | 0.7 | 1.7 | 40 |
Bf-C indicates the threshold P value after Bonferroni correction for the number of SNPs in each genome.
Chr, chromosome; LB, lower boundary of the 95% CI; MAF, minor allele frequency; NA, not applicable; UB, upper boundary of the 95% CI.
Plus strand major/minor alleles.
MAF in the combined discovery and replication samples used for the evaluation.
Estimates represent the power to detect the discovery effect size at the two-sided Bonferroni-corrected significance level using the estimated replication standard error.
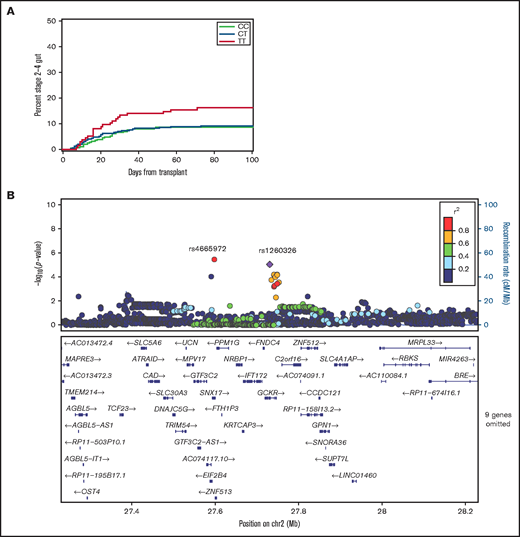
Results for rs1260326 (GCKR) approached the Bonferroni-adjusted 0.01 threshold of statistical significance in the replication cohort (Table 1; Figure 1A). The respective HR point estimates comparing the recipient homozygous T/T genotype with the collective recipient T/C and C/C genotypes in the discovery and replication cohorts were 1.96 (95% confidence interval [CI], 1.4-2.8) and 1.87 (95% CI, 1.2-3.0). Multivariate analysis with adjustments for donor type, donor–recipient sex combination, patient and donor age, marrow or growth factor–mobilized blood cell graft, intensity of the condition regimen and use of total body irradiation, and initial GVHD prophylaxis regimens did not change the HR in the replication cohort (HR, 1.93; 95% CI, 1.2-3.2). For 10/10 HLA–matched sibling and unrelated donor–recipient pairs in the combined discovery and replication cohorts, the respective HR point estimates were 2.31 (95% CI, 1.2-4.5) and 1.49 (95% CI, 0.7-3.0) (P = .37 for statistical interaction), indicating that the association did not differ significantly between the 2 types of donor.
The recipient rs1260326 T allele is associated with an increased risk of stage 2 to 4 intestinal acute GVHD. (A) Cumulative incidence plots of stage 2 to 4 gut GVHD according to the recipient rs1260326 genotype in the combined discovery and replication cohorts. (B) Locus-zoom plot shows –log10(P values) for association with stage 2 to 4 gut GVHD in the combined discovery and replication cohorts as a function of position in a 0.5 Mb region on either side of rs1260326 on chromosome 2 (chr2). Correlation coefficient r2 values for linkage disequilibrium with rs1260326 are coded according to the inset. Genes within this region are displayed in the lower panel.
The recipient rs1260326 T allele is associated with an increased risk of stage 2 to 4 intestinal acute GVHD. (A) Cumulative incidence plots of stage 2 to 4 gut GVHD according to the recipient rs1260326 genotype in the combined discovery and replication cohorts. (B) Locus-zoom plot shows –log10(P values) for association with stage 2 to 4 gut GVHD in the combined discovery and replication cohorts as a function of position in a 0.5 Mb region on either side of rs1260326 on chromosome 2 (chr2). Correlation coefficient r2 values for linkage disequilibrium with rs1260326 are coded according to the inset. Genes within this region are displayed in the lower panel.
Figure 1B shows associations with stage 2 to 4 gut GVHD in the combined discovery and replication cohorts for variants across a region within 0.5 Mb on either side of rs1260326. The results show that rs1260326 has strong linkage disequilibrium with rs4665972 despite its 155 000 bp distance from rs1260326 (r2 = 0.97). The HR point estimate for stage 2 to 4 gut GVHD with the homozygous recipient rs4665972 T/T genotype compared with the collective T/C and C/C genotypes in the combined discovery and replication cohorts was 1.86 (95% CI, 1.1-3.0), comparable to results with rs1260326.
Detailed analysis of TLR4, NOD2, and ATG16L1 variants
Our study did not confirm associations of TLR4, NOD2, or ATG16L1 polymorphisms with stage 2 to 4 gut GVHD in the combined discovery and replication cohorts (supplemental Tables 5 and 6).
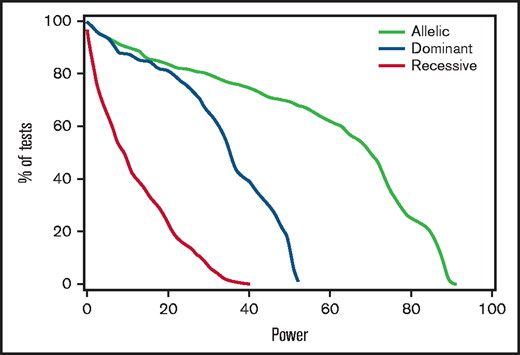
Power constraints
Post hoc analysis of results in the discovery cohort showed that power to detect HRs ≤ 0.67 or ≥ 1.5 was 80% or better in only 162 of the 1776 tested associations derived from the 296 candidate SNPs, 2 genomes, and 3 genetic models (supplemental Table 3). Figure 2 shows the percentages of tests that met any given level of power for the allelic, dominant, and recessive models. For example, 28% of tests with the allelic model had at least 80% power for this effect size, whereas none of the tests with the recessive model had more than 40% power. To support meta-analysis with results from other cohorts, supplemental Table 7 summarizes the overall discovery plus replication cohort-wide associations of donor and recipient variants with stage 2 to 4 gut GVHD.
Power to detect associations with stage 2 to 4 gut GVHD was limited. For each genetic model, the figure shows the percentage of tested recipient variants with post hoc power at or exceeding the level on the x-axis, using an assumed clinically significant effect size with an HR of 1.5 or 0.67, based on the observed discovery standard error and a 0.005 threshold of statistical significance. Results for donor variants are essentially the same.
Power to detect associations with stage 2 to 4 gut GVHD was limited. For each genetic model, the figure shows the percentage of tested recipient variants with post hoc power at or exceeding the level on the x-axis, using an assumed clinically significant effect size with an HR of 1.5 or 0.67, based on the observed discovery standard error and a 0.005 threshold of statistical significance. Results for donor variants are essentially the same.
Discussion
Our study had limited success in showing similarities in genomic risk factors between IBD and stage 2 to 4 acute GVHD. In many respects, this limited success is not surprising. The IIBDGC cohort included ∼38 000 European cases,14 whereas our cohort contained only 3274 European cases, not all of which could be genotyped for any given variant. The large size of the cohorts used for the IIBDGC studies provides at least 80% power for odds ratios >1.3 in loci with minor allele frequencies >1% in persons of European ancestry and for much lower odds ratios in loci with higher minor allele frequencies19 ; the much smaller size of our cohort had negligible power to detect HRs within the range of 0.67 to 1.5 and limited power outside this range. With these limitations, it is likely that our analysis missed IBD-associated loci that are relevant to stage 2 to 4 gut GVHD. To overcome this limitation, the cohort-wide results in supplemental Tables 6 and 7 could be used for meta-analysis with results in other cohorts.
A further limitation of the current study is that the analysis was restricted to European ancestry samples, as they comprised the predominant ancestral group (86%) in our study. Including a relatively small number of samples from multiple other non-European or admixed populations could lead to false-positive or false-negative results due to non-genetic associations of ancestry with gut GVHD. Analysis data representing non-European populations combined from multiple centers could overcome this limitation. To that end, deidentified individual donor and recipient genomic and disease data from our study are available from the National Center for Biotechnology Information database of Genotypes and Phenotypes (dbGAP)(www.ncbi.nlm.nih.gov; accession number phs001918).
Our study did not replicate previously reported associations of TLR4, NOD2, or ATG16L1 polymorphisms with stage 2 to 4 gut GVHD. Toll -like receptor-4 (TLR4) functions as a sensor for lipopolysaccharide. Because lipopolysaccharide has an important role in the pathogenesis of acute GVHD, it was postulated that the presence of hypomorphic variants in either the donor or recipient could decrease the risk of acute GVHD.2 Our results did not show any statistically significant associations of TLR4 variants with the risk of stage 2 to 4 gut GVHD, although the HR point estimates were <1.0. Power was low, and our results do not exclude a protective effect of hypomorphic TLR4 variants.
NOD2 functions as a bacterial sensor. Some studies reported associations of NOD2 variants with overall grade II to IV or grade III to IV GVHD or other outcomes but did not analyze stage 2 to 4 GVHD.4,7,10 In at least 2 studies, however, the presence of NOD2 variants in either the donor or recipient was specifically associated with an increased risk of stage 2 to 4 gut GVHD.3,5 The HR point estimate for the association of stage 2 to 4 gut GVHD with the presence of any donor NOD2 variant in our study was 0.83 (95% CI, 0.58-1.2), which is inconsistent with the increased risk reported by Holler et al.3,5 The HR point estimate for the presence of any recipient NOD2 variant was 1.05 (95% CI, 0.73-1.51). The incidence of stage 2 to 4 gut GVHD in our cohort was ∼10%, whereas the incidence rate in the Regensberg cohorts reported by Holler et al was 20% to 25%. Power for testing these associations in our cohort was low. Our results do not exclude a true association, but the HR is unlikely to exceed 1.5. In addition, the risk associated with NOD variants may be influenced by factors such as donor type or use of in vivo T-cell depletion.8,9,20
Deficiency of ATG16L1 in the recipient exacerbates GVHD in mice due to increased dendritic cell numbers and costimulatory molecule expression,21 and expression of ATG16L1 by recipient intestinal epithelial cell protects against necroptosis mediated by donor T cells.11 Intestinal organoids from patients with the homozygous ATG16L1 T300A (rs2241880) variant allele have increased susceptibility to necroptosis induced by allogeneic T cells or tumor necrosis factor-α. Hubbard-Lucey et al21 described a trend suggesting an increased risk of nonrelapse mortality in recipients having one or two rs2241880 variant alleles, but the association with grade II to IV or grade III to IV GVHD or gut GVHD was not reported. Our results did not show a statistically significant association of the recipient rs2241880 variant with the risk of stage 2 to 4 gut GVHD (HR of 1.13 [95% CI, 0.9-1.5] in the dominant model, and HR of 1.19 [95% CI, 0.9-1.5] in the recessive model) (supplemental Table 6). Because power was suboptimal (62% for both), our results do not exclude a true association, but the HR is unlikely to exceed 1.5.
Our results suggest that rs1260326 and rs4665972 variants are associated with the risk of stage 2 to 4 gut GVHD. The T and C alleles of rs1260326, respectively, encode leucine and proline at codon 446 in glucokinase regulatory protein, an enzyme that inhibits phosphorylation of glucose by glucokinase. GCKR is expressed primarily in the liver, and this missense variant has no predicted consequences, making it unlikely that it accounts for the association with IBD or stage 2 to 4 gut GVHD. rs4665972 represents an intron variant in SNX17, a gene expressed in the small intestine and colon, among many other tissues.
GWAS studies have associated rs1260326 with Crohn’s disease and a variety of other phenotypes, including blood metabolite concentrations, cholesterol and triglyceride concentrations, renal function, blood cell counts, blood protein concentrations, and type 2 diabetes.22 In individuals of European ancestry, the rs1260326 T allele is associated with an increased risk of Crohn’s disease (odds ratio, 1.12; 95% CI, 1.10-1.15),14 corresponding with an increased risk of gut GVHD associated with the rs1260326 T allele or a protective effect associated with the C allele in the current study. The IBD locus containing rs1260326 extends from GRCh37 position 27 598 097 to position 27 752 871 on chromosome 2.12 rs4665972 is located at the left boundary of this locus and is included within the credible set of variants associated with Crohn’s disease, which also includes rs1260334, rs1313566,rs780095, rs780096, rs780093, rs780094, and rs6547692.
The highly correlated reference C alleles of rs1260326 and rs4664972 are associated with decreased expression of GPN1, NRBP1, PPM1G, SLC5A6, and IFT172 in the terminal ileum, transverse colon, or sigmoid colon and with increased expression of ATRAID, C2orf16, and FNCD4 in at least one of these tissues (Table 3). Murine intestinal phenotypes have been reported for NRBP1, PPM1G, SLC5A6, and FNDC4 but not for other genes whose expression is associated with rs1260326 or rs4664972 genotypes.18 Decreased or absent expression of NRBP1, PPM1G, and SLC5A6 causes adverse effects in the intestine in mice, which does not fit the protective effect associated with the C allele in the current study.
Association of rs1260326 and rs4665972 C alleles with intestinal gene expression
| Gene . | SNP ID . | Expression . | Intestinal phenotype induced by loss or decreased expression . | |||||
|---|---|---|---|---|---|---|---|---|
| Terminal ileum . | Transverse colon . | Sigmoid colon . | ||||||
| NES . | P . | NES . | P . | NES . | P . | |||
| GPN1 | rs1260326 | –0.163 | 4.0E-03 | –0.076 | 1.0E-02 | NS | NS | None |
| rs4665972 | –0.192 | 1.6E-03 | NS | NS | NS | NS | ||
| NPBP1 | rs1260326 | –0.214 | 2.2E-06 | –0.151 | 9.3E-09 | NS | NS | Detrimental |
| rs4665972 | –0.209 | 2.0E-05 | –0.144 | 6.7E-08 | NS | NS | ||
| PPM1G | rs1260326 | NS | NS | –0.065 | 9.1E-04 | –0.054 | 1.0E-02 | Detrimental |
| rs4665972 | NS | NS | –0.066 | 8.2E-04 | –0.059 | 7.0E-03 | ||
| SLC5A6 | rs1260326 | NR | NR | NR | NR | NR | NR | Detrimental |
| rs4665972 | NS | NS | NS | NS | –0.123 | 4.5E-04 | ||
| IFT172 | rs1260326* | NR | NR | –0.43 | 3.6E.09 | –0.47 | 2.1E-09 | None |
| rs4665972* | NR | NR | –0.41 | 1.7E-08 | –0.47 | 4.0E-09 | ||
| ATRIAD | rs1260326 | 0.143 | 3.7E-03 | 0.194 | 5.6E-08 | 0.114 | 4.1E-03 | None |
| rs4665972 | 0.166 | 1.8E-03 | 0.194 | 7.5E-08 | 0.119 | 2.9E-03 | ||
| C2ort16 | rs1260326 | 0.237 | 1.7E-03 | NS | NS | NS | NS | None |
| rs4665972 | 0.222 | 7.0E-03 | NS | NS | NS | NS | ||
| FNDC4 | rs1260326* | NS | NS | 0.27 | 1.9E-05 | NS | NS | Detrimental |
| rs4665972 | NS | NS | NS | NS | NS | NS | ||
| Gene . | SNP ID . | Expression . | Intestinal phenotype induced by loss or decreased expression . | |||||
|---|---|---|---|---|---|---|---|---|
| Terminal ileum . | Transverse colon . | Sigmoid colon . | ||||||
| NES . | P . | NES . | P . | NES . | P . | |||
| GPN1 | rs1260326 | –0.163 | 4.0E-03 | –0.076 | 1.0E-02 | NS | NS | None |
| rs4665972 | –0.192 | 1.6E-03 | NS | NS | NS | NS | ||
| NPBP1 | rs1260326 | –0.214 | 2.2E-06 | –0.151 | 9.3E-09 | NS | NS | Detrimental |
| rs4665972 | –0.209 | 2.0E-05 | –0.144 | 6.7E-08 | NS | NS | ||
| PPM1G | rs1260326 | NS | NS | –0.065 | 9.1E-04 | –0.054 | 1.0E-02 | Detrimental |
| rs4665972 | NS | NS | –0.066 | 8.2E-04 | –0.059 | 7.0E-03 | ||
| SLC5A6 | rs1260326 | NR | NR | NR | NR | NR | NR | Detrimental |
| rs4665972 | NS | NS | NS | NS | –0.123 | 4.5E-04 | ||
| IFT172 | rs1260326* | NR | NR | –0.43 | 3.6E.09 | –0.47 | 2.1E-09 | None |
| rs4665972* | NR | NR | –0.41 | 1.7E-08 | –0.47 | 4.0E-09 | ||
| ATRIAD | rs1260326 | 0.143 | 3.7E-03 | 0.194 | 5.6E-08 | 0.114 | 4.1E-03 | None |
| rs4665972 | 0.166 | 1.8E-03 | 0.194 | 7.5E-08 | 0.119 | 2.9E-03 | ||
| C2ort16 | rs1260326 | 0.237 | 1.7E-03 | NS | NS | NS | NS | None |
| rs4665972 | 0.222 | 7.0E-03 | NS | NS | NS | NS | ||
| FNDC4 | rs1260326* | NS | NS | 0.27 | 1.9E-05 | NS | NS | Detrimental |
| rs4665972 | NS | NS | NS | NS | NS | NS | ||
Expression results were obtained from the GTEx Portal, and intestinal phenotypes associated with loss or decreased expression were obtained from the Mouse Genome Database.18 Multi-tissue expression quantitative loci are reported except where indicated otherwise.
NES, normalized effect slope per C allele; NR, not reported; NS, not significant.
Single tissue splicing quantitative trait locus.
The rs1260326 C allele is associated with increased expression of FNDC4 (Fibronectin Type III Domain Containing 4) in the transverse colon (normalized effect size, 0.27; P = 2 × 10−5), although not in the terminal ileum or sigmoid colon (Table 3). Inflammatory mediators such as transforming growth factor-β upregulate expression and secretion of FNDC4 by murine and human intestinal epithelial cells.23 As a feedback regulatory mechanism to control inflammation, an FNDC4 cleavage product secreted by intestinal epithelial cells binds to macrophages, improves their survival in vitro, and reduces their expression of proinflammatory chemokines and their phagocytic activity.23 Based on the association of rs1260326 and rs4664972 genotypes with gut GVHD, the role of intestinal FNDC4 in regulating the inflammatory activity of macrophages, and studies implicating inflammatory macrophages in the pathogenesis of acute GVHD,24-28 we hypothesize that the C alleles of these SNPs could be associated with higher expression of FNDC4 by intestinal epithelial cells stimulated with transforming growth factor-β or other inflammatory mediators.
The genetic mechanism that accounts for the association of rs1260326 and rs4665972 with IBD and gut GVHD remains to be determined. Given that murine- and human-secreted FNDC4 cleavage products have 100% homology, further results showing that expression of FNDC4 by transforming growth factor-β–stimulated human intestinal epithelial cells varies according to rs1260326 and rs4664972 genotypes would suggest that targeting inflammatory macrophages with recombinant FNDC4 might offer an attractive avenue of clinical investigation for the management of IBD and gut GVHD. Finally, we acknowledge that genetic loci not associated with IBD could be associated with gut GVHD. A genome-wide association study could be used to identify additional genetic susceptibility loci that might have stronger associations than those observed in the current study.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI33484 and AI049213), the National Cancer Institute (CA015704 and CA18029), and the National Heart, Lung, and Blood Institute (HL087690, HL088201, HL094260, HL105914, and K23HL69860). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by the National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke. The results used for the analyses described in this article were obtained from the GTEx Portal on March 31, 2021.
Authorship
Contribution: All authors were responsible for the study concept and design, interpretation of results, and writing of the manuscript; and D.M.L. and B.E.S. were responsible for data acquisition, imputation, informatics analyses, quality control, and statistical analyses. All authors other than J.A.H. revised draft manuscripts and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
John A. Hansen died on 31 July 2019.
Correspondence: Paul J. Martin, Fred Hutchinson Cancer Research Center, PO Box 19024, Seattle, WA 98019; e-mail: pmartin@fredhutch.org.
References
Author notes
Deidentified individual donor and recipient genomic and disease data from our study are available indefinitely in the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov; accession number phs001918).
The full-text version of this article contains a data supplement.